Abstract
The symbiosis between legumes and rhizobia results in the development of a new plant organ, the nodule. A role for polar auxin transport in nodule development in Medicago truncatula has been demonstrated using molecular genetic tools. The expression of a DR5::GUS auxin-responsive promoter in uninoculated M. truncatula roots mirrored that reported in Arabidopsis, and expression of the construct in nodulating roots confirmed results reported in white clover. The localization of a root-specific PIN protein (MtPIN2) in normal roots, developing lateral roots and nodules provided the first evidence that a PIN protein is expressed in nodules. Reduced levels of MtPIN2, MtPIN3, and MtPIN4 mRNAs via RNA interference demonstrated that plants with reduced expression of various MtPINs display a reduced number of nodules. The reported results show that in M. truncatula, PIN proteins play an important role in nodule development, and that nodules and lateral roots share some early auxin responses in common, but they rapidly differentiate with respect to auxin and MtPIN2 protein distribution.
Keywords: Medicago runcatula, PIN genes, Auxin transport, Nodulation, RNA interference, Auxin distribution
INTRODUCTION
Legumes are unique among crop plants in that they can grow in the absence of soil nitrogen. Although such growth is dependent on the presence of compatible species of Sinorhizobiaum in the soil, the symbiosis that is set up between the plant and the bacterium allows legumes to exploit niches where other plants cannot grow. The ability of these plants to utilize nitrogen fixed from the atmosphere by the bacteria living within specialized root structures (nodules) makes legumes ideally suited to soils where nitrogen is limiting.
The rhizobia-legume symbiosis is the subject of intense study directed at gaining insight into the molecular signals exchanged between the plant and the bacterium. Although there are slight differences in the process between species, most interactions follow a common pattern. In indeterminate nodulating plants such as alfalfa and M. truncatula, the presence of compatible rhizobia releasing a lipo-chitin oligosaccharide (Nod factor) begins a signal transduction cascade. Upon attachment of the rhizobia to the root hair tips, the tips curl tightly and entrap the bacteria in the curls. The plant cell forms a new structure, a tubular infection thread, through which the bacteria enter the plant. At the same time, a specific subset of cortical cells is mitotically activated and forms the nodule primordia. The infection thread grows toward these primordia through the outer cortex, the cells of which undergo structural modifications to allow passage of the thread. Once the thread reaches the nodule primordia, bacteria are released into the cytoplasm in membrane bound “symbiosomes” (Mylona and others 1995; Bladergroen and Spaink 1998; Cohn and others 1998; Schultze and Kondorosi 1998; Stougaard 2000; Limpens and Bisseling 2003).
Plant hormones have been suspected to be involved in nodule formation since 1936, when Thimann hypothesized that auxin was involved in pea root nodulation (Thimann 1936). Certain species of alfalfa can make spontaneous nodules in the absence of rhizobia, implying that a second signal independent of the Nod factor can cause spontaneous nodulation (Joshi and others 1991). Because inhibitors of polar auxin transport (PAT) can induce pseudonodules on alfalfa roots and non-nodulating mutants of white sweet clover (Hirsch and others 1989; Wu and others 1996), a change in auxin physiology could be involved in the early nodule primordia formation process (Hirsch and LaRue 1997). Changes in auxin concentration could be a second signal for nodule development after the recognition of a rhizobia-derived signal.
Auxin, cytokinin, gibberellin, and ABA all are present in nodules at higher concentrations than in uninfected roots (Torrey 1986). Of these hormones, only auxin is transported in a polar fashion. Because PAT plays an important role in the normal growth and development of plants, including lateral root development (Lomax and others 1995; Himanen and others 2004), and because the nodule is a new root organ with similarity to a lateral root, it is highly probable that PAT plays a role in nodule formation. Evidence that lateral roots and nodules share developmental programs is bolstered by the isolation of genes such as M. truncatula LATD, necessary for both nodule and lateral root development (Bright and others 2005).
Accumulating evidence also shows that endogenous auxin can be transported during nodulation in response to rhizobia (Boot and others 1999). In one study, auxin was observed to accumulate at the site of spot inoculation of L. japonicus with rhizobia, and a significant increase in auxin transport was reported 48 h after inoculation (Pacios Bras and others 2003). There is also evidence of auxin transport inhibition preceding nodulation in the early stages of root nodule formation in Vicia sativa (vetch) and white clover (Mathesius and others 1998; Boot and others 1999). Treatment with N-1-naphthylphthalamic acid (NPA) inhibited PAT in roots to a similar extent as during nodulation. Most importantly, this capacity of Nod factor to reduce PAT is restricted to the elongation zone, the part of the root that is susceptible to nodulation.
Our working hypothesis is that, at the spot where cortical cells are dividing to form nodule primordium, auxin redistribution will occur as a result of Nod factor perception and in fact may be the cause of cell division. To investigate the possible involvement of PAT in nodule development, we examined auxin distribution in uninoculated and nodulating Medicago truncatula roots and examined the localization of a root-specific auxin transport protein in uninoculated roots, developing lateral roots, and nodules. Upon confirmation that changes in auxin distribution occurred during early nodule and lateral root development and that an auxin efflux transport protein was localized to developing nodules and lateral roots, we used RNAi to reduce expression of this and other specific MtPIN proteins. Plants with reduced expression of various MtPINs displayed a reduced number of nodules. Our results confirm that in M. truncatula PAT plays an important role in nodule development, and that nodules and lateral roots share some early auxin responses in common but rapidly differentiate with respect to auxin and MtPIN2 protein distribution.
MATERIALS AND METHODS
Bacterial Strains
Bacterial strain Escherichia coli DH5α was used for plasmid propagation and manipulation. Plants were inoculated with Sinorhizobium meliloti ABS7M (Leong and others 1985). Hairy root transformation was carried out with Agrobacterium rhizogenes strain ARqua l, a derivative of A. rhizogenes strain A4T (Quandt and others 1993). A. rhizogenes and S. meliloti strains were grown at 30°C in TY medium with kanamycin (50 μg/ml) and tetracycline (15 μg/ml) as for antibiotic selection, respectively.
Plant Material and Growth Conditions
M. truncatula cultivar Jemalong A17 seeds were scarified in sulfuric acid (A.C.S. 95–98%) for 8 min in 50-ml centrifuge tubes and washed in sterile water five times for 1 min each, followed by 1 min in straight bleach (12% sodium hypochlorite) and five washes in sterile water for 1 min each. Seeds were then imbibed in water with shaking for 3–4 h at room temperature and 50 rpm. Seeds were placed in a humid chamber and vernalized for 2 days at 4°C and then germinated in darkness at 25°C for 24 h. Plants used in the aeroponic apparatus experiments were then placed in the apparatus in a 25°C growth room with a 14-h photoperiod. Plants used for transformation were exposed to light for 24 h before being excised for transformation.
Plant Transformation
Transformation with A. rhizogenes followed a protocol published by Boisson-Dernier and others (2001) with the concentration of kanamycin in the selection media reduced to 12.5 μg/ml. Plants with transformed roots as determined by kanamycin resistance were transferred to a 25°C growth chamber (16 h photoperiod) for 3 weeks to allow roots to grow before being transferred onto Harrison Modified Farheus (HMF) media minus nitrate plates for inoculation. HMF media minus nitrate consists of 0.9 mM CaCl2.H2O, 0.5 mM MgSO4, 0.7 mM KH2PO4, 0.8 mM Na2HPO4.7H2O , 20 μM Fe-citrate, 33 μg/l MnCl2 · 4H2O, 33 μg/l CuSO4, 7 μg/l ZnCl2, 100 μg/l H3BO3, 33 μg/l Na2MoO4 · H2O, pH 7.4 prepared with 15 g/l washed agar (Sigma Chemical, St Louis, MO, USA).
Five milliliters of TY media containing 15 μg/ml tetracycline were inoculated with S. meliloti ABS7M and grown overnight at 30°C with shaking at 200 rpm to an OD600 of 0.9. The cells were then pelleted by centrifugation at 2600 rpm for 10 min at room temperature. Pelleted cells were resuspended in the same volume of sterilized water and used immediately. Plants that had been growing for 3 weeks on HMF medium were carefully washed in sterilized water to remove agar clinging to the roots and transferred to HMF minus nitrate plates containing 0.5 μM 2-aminoethoxyvinylglycine (AVG) (an inhibitor of ethylene biosynthesis) for 3–5 days to starve them of nitrate. Each plate contained at least one control plant transformed with empty vector and several plants carrying the construct being tested. S. meliloti suspension was applied to the elongation zone of the plants’ roots by pipet. Plants were then grown at 25°C with a 16-h photoperiod for 15 days. Nodules were counted and tissue was collected for GUS staining and reverse transcription-polymerase chain reaction (RT-PCR) at indicated times post-inoculation.
MtPIN2::GUS Plasmid Construction
MtPIN2 was identified in a survey of auxin transport genes in M. truncatula (Schnabel and Frugoli 2004). A 4.8 kb fragment (1980 bases upstream of the start of MtPIN2 to base 2789 of Genbank Accession Number AY115837) was amplified from a BAC clone by PCR using the Expand Long Template PCR System (Roche Applied Science, Mannheim, Germany), with 5′-CGGGATCCAATTGTTTTTACGTCTATTTAGCA CTTA-3′ and 5′- TTACCATGGCTACCCCCAGAAGCACATAGTATAGTATTG T-3′ as primers. The ends of the PCR product were blunted and ligated into the pGEM-T Easy Vector (Promega, WI, USA) and recombinant transformants selected by blue/white screening on selective media. Plasmids of the correct size were digested with BamH I and Nco I and ligated into a modified pCAMBIA 1381 vector termed pC2381ES. This modified vector was created by replacing the hygromycin resistance gene of pCAMBIA 1381 with the kanamycin resistance gene from pCAMBIA 2304 in which the upstream Bgl II and Nco I sites had been destroyed by PCR modification. The resulting plasmid contained MtPIN2 in frame with the GUS reporter gene driven by the MtPIN2 promoter and a kanamycin selection gene. The fidelity of the entire insert was confirmed by both sequencing and restriction analysis. The plasmid was then transferred to A. rhizogenes strain ARqua l by electroporation.
RNAi Plasmid Construction
Unique fragments of 141–230 bp from the first exons of various PIN genes were inserted into the vector pKANNIBAL (Smith and others 2000) in opposite orientations. The fragments used were MtPIN1 (GB# AY115836) bases 1239–1468; MtPIN2 (GB# AY115837) bases 1316–1467; MtPIN3 (GB# AY115838) bases 1497–1662, and MtPIN4 (GB# AY115839) bases 1080–1220. The silencing construct for each gene was then transferred to the binary vector pCAMBIA 2304 containing GUS and GFP reporter genes. The resulting plasmid contained the RNA interference construct driven by a 35S promoter from pKANNIBAL and GUS and GFP reporters driven by a 35S promoter from pCAMBIA 2304. The fidelity of the clones was verified by sequencing. Verified plasmids were then transferred to A. rhizogenes strain ARqua l by electroporation.
Phenotypic Analysis
Fifteen days after inoculation with S. meliloti, nodules were counted. Roots used in the analysis were verified as transgenic by GUS staining of an excised piece of tissue from roots with nodules. The remainder of the root tissue was frozen in liquid nitrogen after nodule counting and preserved at –80°C for later RNA extraction.
Histochemical Localization of GUS Activity
GUS activity was localized based on a protocol by Jefferson and others (1987). Samples were infiltrated with substrate under vacuum for 20 min and incubated at 37°C for 3 to 24 h until the blue color developed. The staining buffer was then removed and the samples were cleared with 75% ethanol and preserved at 4°C. The phenotypes of roots were recorded on a Nikon E600 microscope with a Retiga EXi FAST monochrome CCD 12-bit camera.
Isolation of RNA and RT-PCR
RNA from individual roots of transgenic plants was isolated using the Qiagen RNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA). RNA was eluted in 30 μl dH2O and quantified by measuring the absorbance of a 1:50 dilution at 260 nm. The RNA was then used as a template for single stranded cDNA synthesis with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Synthesis of cDNA was done in a 20-μl reaction using 0.2 to 0.5 μg RNA.
Gene-specific primers designed to amplify MtPIN genes and distinguish cDNA products from genomic products by size in previous work (Schnabel and Frugoli 2004) were used to detect gene expression. Detection occurred in a 20 μl PCR reaction consisting of 2.5× PCR Mastermix (Eppendorf AG, Hamburg, Germany), 0.625 μM each of gene specific primers, and 1–3 μl cDNA as template. Thermocycling conditions were 95°C for 4 min followed by 40 cycles of 95°C for 20 s, 61°C for 20 s, and 72°C for 30 s. Products were analyzed by electrophoresis on 1.5% (w/v) agarose gels in 0.5× Tris-borate-EDTA buffer and visualized with ethidium bromide staining.
RESULTS
Auxin Distribution during Nodule Development
Synthetic auxin responsive promoters such as DR5 (Ulmasov and others 1997; Hagen and Guilfoyle 2002) are useful tools for monitoring auxin response in plants, because the activity of these reporters correlates with auxin content in roots (Casimiro and others 2001). Using the DR5::GUS construct (gift of Tom Guilfoyle), we transformed M. truncatula roots and observed the pattern of GUS staining in uninoculated roots (Figure 1a, b). Staining was observed near the xylem and pericycle cells of the root, with a broader distribution around the root tip and in early lateral roots. In addition to staining in the columella cells and the primary root apex in agreement with the pattern of expression of this reporter in Arabidopsis (Sabatini and others 1999; Casimiro and others 2001), the staining observed in M. truncatula roots includes the vascular bundle, an effect seen in Arabidopsis roots when longer staining times were used (Sabatini and others 1999). Within 72 h of inoculation with S. meliloti, nodule primordia were visible in roots, arising from the division of inner cortical cells. Expression of the DR5::GUS construct in the vascular bundle was no longer detectable below the site of nodule development (Figure 1c, 1d). The observed patterns of expression match those of the GH3 auxin-responsive promoter observed by Mathesius and others (1998) in inoculated white clover and have been interpreted as a transient interruption in auxin transport at the site of nodule development. Nodule primordia can be distinguished from lateral root primordia because lateral roots arise from the pericycle, whereas indeterminate nodules arise from the inner cortex (Hirsch 1992), and DR5 staining patterns between emerging lateral roots and nodules differed (Figure 1c).
Figure 1.
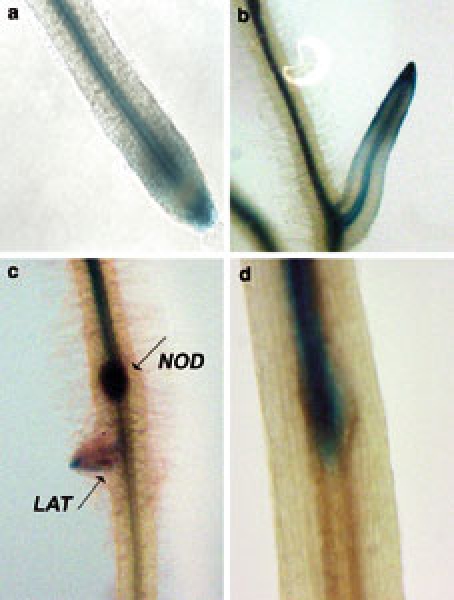
GUS staining patterns of M. truncatula roots carrying the DR5::GUS reporter. Root tip of plant growing in the absence of rhizobia (a); lateral root in the absence of rhizobia (b); emerging lateral root (LAT) and nodule (NOD) on M. truncatula roots approximately 120 h after inoculation with rhizobia (c); interruption of GUS staining pattern at point of nodule development 72 h post-inoculation (d).
Expression of MtPIN2
Previous characterization of PIN genes in M. truncatula identified one gene, the AtPIN2 ortholog MtPIN2, with expression detectable only in roots (Schnabel and Frugoli 2004). Of the first four PIN genes identified in M. truncatula, MtPIN2 is the only one for which a one-to-one correspondence with an Arabidopsis ortholog could be determined, so we chose to begin our study with this protein. In Arabidopsis, a protein:GUS fusion construct was effective in determining tissue-specific expression of EIR1 (AtPIN2) without disruption of function (Sieberer and others 2000). Use of a protein:GUS fusion is particularly important in the case of PIN proteins because the same work determined that AtPIN2 is subject to post-transcriptional regulation (Sieberer and others 2000). Therefore, M. truncatula roots were transformed with a protein:GUS fusion. The MtPIN2:GUS fusion was expressed under the control of the MtPIN2 promoter, and GUS staining was observed in uninoculated and inoculated roots.
Before inoculation with S. meliloti, the transgenic roots exhibited GUS activity at the root tip in a region spanning the root meristem and distal elongation zone, with strong expression at epidermal and cortical root cells (Figure 2a, 2b). Strong GUS activity was also noted in the early development of lateral root primordia (Figure 2c), and GUS activity spread into the central organ as it developed. Root tissues in which active cell division occurs (the base and at the tips of the lateral roots) showed the highest GUS activity (Figure 2d). However, as the lateral roots matured, GUS activity was no longer detected at the root base (Figure 2e).
Figure 2.
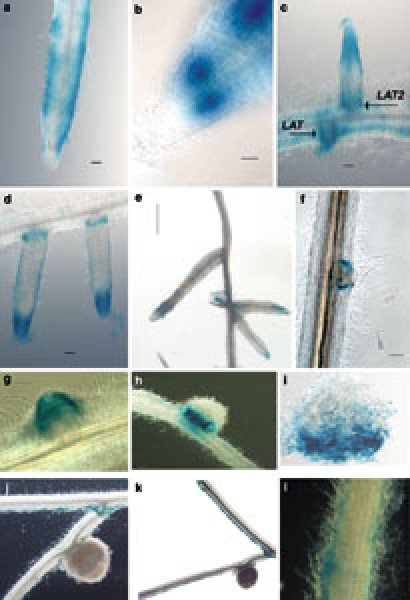
GUS staining patterns of M. truncatula roots carrying the MtPIN2 protein:GUS fusion construct. All scale bars are 100 μm unless indicated otherwise. Primary root in the absence of rhizobia (a); root tip in the absence of rhizobia (b); staining in emerging lateral root (LAT) and older lateral root (LAT2) (c); recently emerged lateral roots (d); maturing lateral roots revert to staining pattern of primary root (bar = 1 mm) (e); MtPIN2::GUS in developing nodules approximately 72 h post-inoculation with rhizobia (f); GUS staining in nodule 120 h post inoculation (g); staining in nodule 12 days post-inoculation (h); transverse section of nodule in h, root proximal side at bottom (i); staining in mature nodules (15–20 days post-inoculation) (j, k); staining in epidermis (15–20 days post-inoculation) (l).
Approximately 72 h after inoculation with S. meliloti, the overall pattern of GUS staining was similar to that observed in uninoculated roots. However, expression of the MtPIN2::GUS fusion was also observed in the initiating nodule primordia (Figure 2f).
A later step of nodule development is the formation of meristem by dedifferentiated middle cortical cells located at the center of the nodule primordia. These cells divide many times in various directions, thus generating numerous islets of meristematic cells (Libbenga and others 1973). As mitosis continues, the nodule meristem grows away from the stele of the parent root. About 120 h after inoculation, staining was observed in the center of the young nodule (Figure 2g), but even stronger expression was detected at the nodule outer cortex. Twelve days after inoculation, GUS activity was restricted to the basal part of the nodule (Figure 2h), and cross sectioning of the nodule revealed GUS activity in the nodule base (Figure 2i).
Nodules became mature as indicated by the pink color of leghemoglobin necessary for nitrogen fixation conditions 15–20 days after inoculation. At these time points, GUS activity is no longer detected around the nodule base. However the staining pattern seen in uninoculated roots remains visible in the epidermal cells of the roots (Figure 2j, 2k, 2l).
Disruption of MtPIN Gene Expression by RNAi
The Arabidopsis genome encodes eight PIN genes, whereas 10 PIN genes have been identified in M. truncatula (Schnabel and Frugoli 2004). In Arabidopsis the PIN1 gene is expressed throughout the plant, whereas AtPIN2, AtPIN3, AtPIN4, AtPIN6, and AtPIN7 have demonstrated root functions (Galweiler and others 1998; Swarup and others 2001; Friml and others 2002a, 2002b; Benkova and others 2003). The M. truncatula orthologs of these genes are good candidates for involvement in nodulation; however, direct one-to-one orthology is not clear for every one of these genes (Schnabel and Frugoli 2004). Based on our previous work on gene orthology, we chose MtPIN1-MtPIN4 for initial RNAi experiments; although these are not the only PINs with the potential to be involved in nodulation, they are among the most closely related to AtPIN1, AtPIN2, AtPIN3, AtPIN4, and AtPIN7. Plasmids generating RNAi for MtPIN1-MtPIN4 were constructed (see Materials and Methods). Because other laboratories have confirmed that M. truncatula hairy roots grown on kanamycin are not all transgenic roots (Limpens and others 2004), and because multiple roots arise on a single plant, a constitutive GUS reporter gene was included in the RNAi construct so that transgenic roots could be confirmed by GUS staining. About 50%–70% of the roots stained positively for GUS activity, in line with transformation efficiencies reported by other labs using the protocol (Limpens and others 2004). Only nodules on roots that stained positively for GUS were used in the following analyses.
In different experiments conducted over several months, 37 GUS-positive plants carrying the empty vector control, nine carrying the PIN1i construct, 68 carrying the PIN2i construct, 17 carrying the PIN3i construct, and 23 carrying the PIN4i construct were generated. To ensure that plants included in the analysis were not only transgenic but also reduced in expression of the targeted MtPIN gene, qualitative RT-PCR was performed on cDNA isolated from each individual root. Statistical analysis of the effect of RNAi includes only the plants for which significant reduction could be verified by the inability to detect expression of the target gene. Thus, some of the plants excluded from the final analysis may have had reduced levels of expression that would not be detectable in our qualitative assay. Figure 3a shows representative RT-PCR results of what we classified as “silenced” single roots. Compared to control roots carrying only the GUS vector, these roots had undetectable levels of the targeted gene. In some cDNA preps, especially that for the root shown in PIN4i, evidence of genomic contamination (bands corresponding to the size of the product including introns) is indicated by an asterisk.
Figure 3.
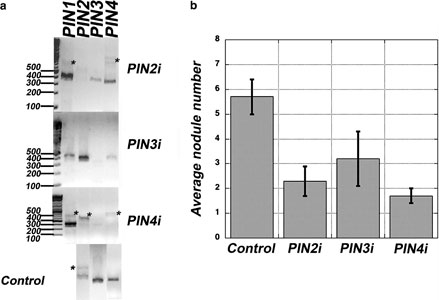
a. Reverse transcriptase polymerase chain reaction (RT-PCR) of MtPIN gene expression in roots expressing RNAi constructs indicated at the right of each gel. The migration of molecular weight markers (size indicated in bp) is marked at the left of each gel. Primers were chosen to generate gene-specific PCR products when amplifying cDNA from the root tested in each gel (see Materials and Methods). The expected size of product amplified from cDNA with the indicated primers should be: PIN2 384bp, PIN3 374bp, PIN4 365bp. Bands marked with an asterisk are consistent with the genomic DNA contamination (product size would include intron). b. Average nodule number on plants confirmed as silenced by RT-PCR (see text) compared to plants expressing the empty vector (control). Control, n = 37; PIN2i, n = 11; PIN3i, n = 6; PIN4i, n = 3.
The effectiveness of the RNAi construct varied, and RT-PCR suggested some cross silencing may have occurred. Even in plants for which reduction of expression appears specific, no attempt was made to quantify actual expression levels of targeted or non-targeted genes. Additionally, no plant in which MtPIN1 was undetectable was identified by RT-PCR, whereas in some plants carrying the MtPIN1 silencing construct, MtPIN2, MtPIN3, or MtPIN4 were also undetectable. Thus, although MtPIN1i roots had significantly fewer nodules than control roots, because of the observed effect on levels of other PIN genes, they are not reported in Figure 3. There was a significant difference in nodule number between plants carrying RNAi for MtPIN2 and the control plants carrying only the empty vector (Student’s t-test, p < 0.001; Figure 3b).
There was also a significant difference in nodule number between control plants and MtPIN3 and MtPIN4 RNAi plants (Student’s t-test, 0.01 > p > 0.001; Figure 3b). Observations of the number of hairy roots arising from the wound on each plant, the number of root hairs per mm, and the overall shape of the nodules revealed no differences between plants carrying the MtPIN1-MtPIN4 silencing constructs and the control plants; only the nodule number was different.
DISCUSSION
We have demonstrated that changes in auxin distribution in M. truncatula roots during nodulation mirror those reported for white clover (Mathesius and others 1998). The changes in auxin distribution observed during early nodule and lateral root development imply the involvement of auxin-transport proteins. We have also shown that disruption of expression of MtPIN2, MtPIN3, and MtPIN4 through RNAi leads to a reduction in the number of nodules, suggesting that these components of auxin-transport machinery play a role in nodule development. We have demonstrated that MtPIN2, the ortholog of AtPIN2, is expressed in the same pattern as AtPIN2 during the development of lateral roots. In addition, we have shown that MtPIN2 is expressed in developing but not mature nodules in a pattern suggesting that lateral roots and nodules share some common developmental pathways. Previously, three MtLAX genes (auxin permeases) had been localized to developing nodules (de Billy and others 2001), and multiple MtPINs are expressed in roots (Schnabel and Frugoli 2004). However, this is the first report of a PIN protein localized to nodules in M. truncatula. The changing localization of the MtPIN2 protein within the nodule during nodule development suggests that polar auxin transport, and particularly MtPIN2-mediated auxin transport, is part of the developmental program leading to nodules.
The organogenesis of lateral roots and that of nodules share a number of similarities. For instance, the majority of nodules and lateral roots are initiated in front of protoxylem poles (Libbenga and others 1973; Sussex and others 1995). In lateral root formation the pericycle cells in front of protoxylem poles are the most sensitive to the action of auxin (Sussex and others 1995; Himanen and others 2004). Nodules and lateral roots also show a similar pattern of GH3::GUS expression (another auxin reporter construct) during comparable developmental stages, suggesting that the requirement for auxin is similar in both organs (Mathesius and others 1998).
In uninoculated M. truncatula roots, the localization of MtPIN2 is consistent with that of AtPIN2 in Arabidopsis (Muller and others 1998), further confirming the orthology described in Schnabel and Frugoli (2004). However, MtPIN2 has a distinct pattern of expression in the nodule during nodule development different from that in lateral roots. We were able to detect MtPIN2 expression in areas of active cell division in lateral root tips throughout the time course of our experiments (for example, Figure 2c, d, e), yet despite the indeterminate nature of the M. truncatula nodule meristem, we were unable to detect MtPIN2 in mature nodules (compare Figure 2f, g, h with Figure 2j, k). This suggests that although nodule and lateral root development in M. truncatula shares many developmental steps, the two structures have distinct ontogenies.
The expression pattern of MtPIN2 changed as nodule development proceeded. The initial expression in the nodule primordia bore a strong resemblance to the pattern of expression in lateral root primordia—strong expression in the outer layers of the primordia that will become the peripheral tissues (Figure 2c, 2f). Expression of an auxin transporter in this tissue suggests that both lateral roots and nodules need PAT to establish an axis of growth. A large number of experiments indicate that auxin transport is required for the formation of vascular strands and patterning in the roots (reviewed in Doerner 2000; Berleth and Sachs 2001), and both nodules and lateral roots form a new meristem and require new vasculature.
As the nodule began to emerge from the root around 5 days after inoculation, and the expression of the MtPIN2::GUS fusion expanded throughout the nodule, but the highest expression was still in the peripheral tissue, just as in lateral roots. Later, as the nodule meristem continued to divide and the nodule elongated, MtPIN2 protein was located only at the base of the nodule, a pattern that bears striking similarity to the expression at the base of lateral roots (Figure 2c, 2d, 2f, 2h). We interpret this need for PAT at the base of the new organ as a need to divert auxin from the basal-acropetal flow in the main root into a proximal-distal gradient along the new organ in the initial stages of growth. Support for a diversion of auxin from the main gradient in the root during nodule development also comes from work by Mathesius and others (1998) and Boot and others (1999) indicating a disruption of normal auxin distribution at the point of nodule initiation. Interestingly, in Arabidopsis, monitoring the expression of the DR5::GUS auxin-responsive reporter suggests the establishment of a new concentration gradient of auxin in the early stages of lateral root formation as well (Benkova and others 2003). As both nodules and lateral roots mature, this “ring” disappears, suggesting that as the new meristem becomes established, it produces its own auxin, or that the transport of auxin shifts to a transporter other than MtPIN2. Using different methods, both Mathesius and others (1998) and Boot and others (1999) also observed a reestablishment of normal auxin distribution after nodule development, and our results with RNAi suggest that more than one PIN protein may be involved in nodule development.
The use of RNAi to reduce expression of MtPIN2 confirmed that the protein is important for normal nodule development because reducing expression levels significantly reduced nodule numbers when compared to plants expressing the empty vector. A similar result was observed for plants in which MtPIN3 or MtPIN4 were silenced. The involvement of PIN proteins in organ development is not surprising, given their central role in establishing and maintaining specific patterns of auxin distribution (reviewed in Paponov and others 2005). Although a reduction in nodule number may appear to be a mild phenotype, it should be noted that in Arabidopsis single PIN mutants often have very subtle phenotypes or no phenotype at all, due to the ability of the various PIN proteins to influence each other’s transcription (Blilou and others 2005), suggesting the reduced nodule number phenotype is significant. The changing pattern of auxin distribution bears some resemblance to the auxin redistribution that occurs in Arabidopsis roots during the development of lateral roots, and given the similar patterns of PIN protein expression, we postulate that PIN proteins control this auxin redistribution in nodule development just as they have been shown to do in lateral root development (Benkova and others 2003).
Therefore, although the involvement of auxin and its polar transport in developing nodules is not unexpected given its complex role in plant development, little molecular genetic evidence in legumes has been presented. Our work demonstrates that polar auxin transport is involved in regulating nodule development. The MtPIN2 protein in particular is localized to nodules and required for normal nodule development, and RNAi evidence suggests MtPIN3 and MtPIN4 may be important as well.
Acknowledgments
The authors thank Harry D. Kurtz, Jr., for the use of his microscope, Tom Guilfoyle for the DR5:GUS construct, Bill Marcotte for helpful comments on the manuscript, and Clemson University and the SC Life Howard Hughes Medical Institute grant 52003722 for financial support. This manuscript is Technical Contribution No. 5123 of the Clemson University Experiment Station.
References
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, and others. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602 [DOI] [PubMed]
- Berleth T, and Sachs T. 2001. Plant morphogenesis: long-distance coordination and local patterning. Curr Opin Plant Biol 4:57–62 [DOI] [PubMed]
- Bladergroen MR, and Spaink HP. 1998. Genes and signal molecules involved in the rhizobia-Leguminoseae symbiosis. Curr Opin Plant Biol 1:353–359 [DOI] [PubMed]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml, and others. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44 [DOI] [PubMed]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, and others. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant-Microbe Interact 14:695–700 [DOI] [PubMed]
- Boot KJM, van Brussel AAN, Tak T, Spaink HP, and Kijne JW. 1999. Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Mol Plant Microbe Interact 12:839–844
- Bright LJ, Liang Y, Mitchell D, and Harris J. 2005. The LATD gene of Medicago truncatula is required for both nodule and root development. Mol Plant Microbe Interact 18:521–532 [DOI] [PubMed]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, and others. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13:843–852 [DOI] [PMC free article] [PubMed]
- Cohn J, Day RB, Stacey G. 1998. Legume nodule organogenesis. Trends Plant Sci 3:105–110 [DOI]
- de Billy F, Grosjean C, May S, Bennett MJ, and Cullimore JV. 2001. Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14:267–277 [DOI] [PubMed]
- Doerner P. 2000. Root patterning: does auxin provide positional cues? Curr Biol 10:R201–R203 [DOI] [PubMed]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, and others. 2002a. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108:661–673 [DOI] [PubMed]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. 2002b. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415:806–809 [DOI] [PubMed]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, and others. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282:2226–2230 [DOI] [PubMed]
- Hagen G, Guilfoyle T. 2002. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385 [DOI] [PubMed]
- Himanen K, Vuylusteke M, Vanneste S, Vercruysse S, Boucheron E, and others. 2004. Transcript profiling of early lateral root initiation. Proc Natl Acad Sci USA 101:5146–5151 [DOI] [PMC free article] [PubMed]
- Hirsch AM. 1992. Develpmental biology of legume nodulation. New Phytol 122:211–237 [DOI] [PubMed]
- Hirsch AM, Bhuvaneswari TV, Torrey JG, Bisseling T. 1989. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 86:1244–1248 [DOI] [PMC free article] [PubMed]
- Hirsch AM, LaRue TA. 1997. Is the legume nodule a modified root or stem or an organ sui generis? Crit Rev Plant Sci 16:361–392
- Joshi PA, Caetano-Anolles G, Graham ET, and Gresshoff PM. 1991. Ontogeny and ultrastructure of spontaneous nodules in alfalfa (Medicago sativa). Protoplasma 162:1–11 [DOI]
- Leong SA, Williams PH, Ditta GS. 1985. Analysis of the 5′ regulatory region of the gene for delta- aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res 13:5965–5976 [DOI] [PMC free article] [PubMed]
- Libbenga KR, Van Iren F, Bogers RJ, Schraag-Lammers MF. 1973. The role of hormones and gradients in the initiation of cortex proliferation and nodule formation in Pisum sativa L. Planta 114:29–39 [DOI] [PubMed]
- Limpens E, and Bisseling T. 2003. Signaling in symbiosis. Curr Opin Plant Biol 6:343–350 [DOI] [PubMed]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, and others. 2004. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992 [DOI] [PubMed]
- Lomax TL, Muday GK, and Rubery PH. 1995. Auxin transport. In Davies PJ. Ed., Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Dordrecht, the Netherlands, Kluwer, pp. 508–530
- Mathesius U, Schlaman HR, Spaink H, Sautter C, Rolfe B, and others. 1998. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavanoids and derivatives of chitin oligosaccharides. Plant J 14: 23–34 [DOI] [PubMed]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijer P, and others. 1998. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17:6903–6911 [DOI] [PMC free article] [PubMed]
- Mylona P, Pawlowski K, and Bisseling T. 1995. Symbiotic nitrogen fixation. Plant Cell 7:869–885 [DOI] [PMC free article] [PubMed]
- Pacios Bras C, Schlaman HRM, Boot KJM, Admiraal P, and others. 2003. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol Biol 52:1169–1180 [DOI] [PubMed]
- Paponov I, Teale WD, Trebar M, Blilou I, and Palme K. 2005. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci 10:170–177 [DOI] [PubMed]
- Quandt HJ, Puhler A, and Broer I. 1993. Transgenic root-nodules of vicia-hirsuta – a fast and efficient system for the study of gene-expression in indeterminate-type nodules. Mol. Plant-Microbe Interact 6:699–706
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle TJ, and others. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99:463–472 [DOI] [PubMed]
- Schnabel E, and Frugoli J. 2004. The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol Gen Genom 272:420–432 [DOI] [PubMed]
- Schultze M, and Kondorosi A. 1998. Regulation of symbiotic root nodule development. Annu Rev Genet 32:33–57 [DOI] [PubMed]
- Sieberer T, Seifert GJ, Hauser MT, Grisafi P, Fink GR, and others. 2000. Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr Biol 10:1595–1598 [DOI] [PubMed]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, and others. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407:319–320 [DOI] [PubMed]
- Stougaard J. 2000. Regulators and regulation of legume root nodule development. Plant Physiol 124:531–540 [DOI] [PMC free article] [PubMed]
- Sussex IM, Godoy JA, Kerk NM, Laskowski MJ, Nusbaum HC, and others. 1995. Cellular and molecular events in a newly organising lateral root meristem. Phil Trans R Soc Biol Sci 359:39–43 [DOI] [PubMed]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, and others. 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15:2648–2653 [DOI] [PMC free article] [PubMed]
- Thimann KV. 1936. On the physiology of the formation of nodules on legume roots. Proc Natl Acad Sci USA 22:511–514 [DOI] [PMC free article] [PubMed]
- Torrey JG. 1986. Endogenous and exogenous influences on the regulation of lateral root formation. In: Jackson MB, Editor, New Root Formation in Plants and Cuttings. Hingham, MA, USA, Martinus Nijhoff pp. 31–66
- Ulmasov T, Murfett J, Hagen G, and Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971 [DOI] [PMC free article] [PubMed]
- Wu CF, Dickstein R, Cary AJ, and Norris JH. 1996. The auxin transport inhibitor N-(1-naphthyl)phthalamic acid elicits pseudonodules on nonnodulating mutants of white sweetclover. Plant Physiol 110:501–510 [DOI] [PMC free article] [PubMed]


