Abstract
Cancer pathogenesis involves multiple genetic and epigenetic alterations, which result in oncogenic changes in gene expression. δ-Catenin (CTNND2) is overexpressed in cancer although the mechanisms of its upregulation are highly variable. Here we report that in prostate cancer the methylation of CpG islands in δ-catenin promoter was not a primary regulatory event. There was also no δ-catenin gene amplification. However, using Single-Strand Conformation Polymorphism analysis, we observed the increased nucleotide changes in the 5′-untranslated region of δ-catenin gene in human prostate cancer. At least one such change (-9 G>A) is a true somatic point mutation associated with a high Gleason score, poorly differentiated prostatic adenocarcinoma. Laser capture microdissection coupled with PCR analyses detected the mutation only in cancerous but not in the adjacent benign prostatic tissues. Using chimeric genes encoding the luciferase reporter, we found that this mutation, but not a random mutation or a mutation that disrupts an upstream open reading frame, resulted in a remarkably higher expression and enzyme activity. This mutation did not affect transcriptional efficiency, suggesting that it promotes δ-catenin translation. This is the first report of δ-catenin gene mutation in cancer and supports the notion that multiple mechanisms contribute to its increased expression in carcinogenesis.
Keywords: Single-Strand Conformation Polymorphism, Single nucleotide polymorphism, 5′UTR mutation, Methylation, Gene amplification
Introduction
Cancer pathogenesis is a long-term process that involves multiple genetic and epigenetic alterations. Extensive research has shown that many molecular abnormalities are involved in cancer development, including chromosomal amplification of oncogenes, gene methylation, gene silencing, and transcriptional and translational alterations.
δ-Catenin (gene designation as CTNND2) is an adhesive junction associated protein of the β-catenin superfamily (Paffenholz and Franke 1997; Zhou et al., 1997; Lu et al, 1999; Paffenholz et al., 1999). It was mapped to human chromosome 5p15, a region critical for the development of severe mental retardation phenotype in Cri-du-Chat syndrome (Medina et al., 2000). While 5p is variably deleted or duplicated in Cri-du-Chat syndrome and other developmental disorders (Marinescu et al, 1999; Medina et al, 2000; Cervera et al, 2005), it shows gene amplification in some cancers (Zheng et al, 2004; Huang et al, 2006; Fatima et al., 2006). δ-Catenin was initially identified as a neural specific protein in the brain (Paffenholz and Franke 1997; Zhou et al., 1997; Lu et al, 1999; Ho et al, 2000), interacting with Alzheimer’s disease protein presenilin (Zhou et al, 1997; Tanahashi and Tabira, 1999). Further studies show that δ-catenin promotes the disruption of E-cadherin based adherens junction to favor cell spreading upon stimulation by hepatocyte growth factor (Lu et al., 1999). Examination of human EST data bank revealed δ-catenin mRNA sequences in kidney, ovarian, brain, breast, and esophageal tumors. Other studies demonstrated that δ-catenin mRNA was overexpressed in prostate cancer compared to benign prostate hyperplasia (Burger et al., 2002). Recently, we showed that δ-catenin protein expression is upregulated in over 80% of prostatic adenocarcinomas, and its expression is correlated with increasing Gleason scores (Lu et al, 2005). An increased expression of δ-catenin is accompanied by the down regulation of tumor suppressor E-cadherin and p120ctn in primary prostatic adenocarcinomas. In addition, the forced overexpression of δ-catenin in cultured prostate cancer cells disrupted the distribution of E-cadherin and p120ctn, major cell-cell junction proteins whose inactivation is often linked to the aggressive phenotype of prostate cancer (Umbas et al, 1994; Lu et al, 2005).
The frequently increased expression of the neuronal protein δ-catenin in peripheral tissues of cancer raised important questions such as what mechanisms are in place in cancer to result in a high level of δ-catenin expression. Increased δ-catenin gene copy number was observed in cervical cancer (Huang et al, 2006) and bladder cancer (Zheng et al, 2004). In addition, transcription factor Pax6 was found to play an important role for regulation of δ-catenin expression in developing eye and central nervous system (Duparc et al, 2006). Recently, we showed that the ectopic overexpression of E2F1 and Pax6 positively up regulates δ-catenin expression in prostate cancer cells (Kim et al., 2008). In order to gain further insights into the mechanisms by which δ-catenin overexpression is controlled in cancer, we examined the regulation of δ-catenin expression in prostate cancer by methylation status and potential gene amplification. We also screened possible nucleotide sequence changes in the promoter and 5′-untranslated region (5′UTR) of δ-catenin gene by Single-Strand Conformation Polymorphism (SSCP) analyses. We discovered an increased incidence of single nucleotide polymorphisms (SNPs) in δ-catenin in human prostate cancer specimens. Importantly, a true somatic point mutation in 5′UTR region of δ-catenin was identified in a 70-year old patient with a high Gleason score, poorly differentiated prostatic adenocarcinoma. This point mutation was located close to the end of 5′UTR, at position -9 base pair with respect to the translation initiation ATG codon. Remarkably, the nucleotide fragment with this point mutation promotes the expression of a chimeric gene based luciferase reporter. Prostatic tissues bearing this mutation displayed an increased δ-catenin expression over normal tissues. Our results support the notion that somatic mutations in 5′UTR of δ-catenin can contribute to the alteration of δ-catenin expression and provide new insight into δ-catenin gene regulation in cancer pathogenesis.
Results
δ-Catenin mRNA expression in prostate cancer cells
We examined δ-catenin mRNA levels in several widely used prostate cancer epithelial cell lines. CWR22Rv-1 was derived from a primary prostate tumor xenograft (Pretlow et al, 1993; Sramkoski et al, 1999). PC-3 was derived from prostate cancer bone metastasis (Kaighn et al., 1979). LNCaP was isolated from lymph node with confirmed diagnosis of metastatic prostate carcinoma (Horoszewicz et al., 1983). DU145 was isolated from brain metastasis of prostate cancer (Stone et al, 1978). PZ-HPV-7, a control cell line, was derived from epithelial cells of the peripheral zone of normal human prostate (Weijerman et al. 1994). RT-PCR detected δ-catenin mRNA in all of the five cell lines examined (Fig 1A); however, while δ-catenin mRNA level in PZ-HPV-7, LNCaP and DU145 cells was low, its expression in CWR22Rv-1 and PC-3 cells was considerably high (Fig 1B). Consistent with our previous report (Lu et al., 2005), we also detected δ-catenin mRNA overexpression in 8/8 cases of prostatic adenocarcinomas when compared to that of benign prostate tissue specimens (data not shown).
Figure 1.
Alteration of δ-catenin transcription and methylation states in prostate cancer. A. RT-PCR showing δ-catenin mRNA expression in prostate cancer and non-cancer prostate cells. B. Quantitative illustration of δ-catenin mRNA expression in prostate cancer and non-cancer prostate cells. Note that except for DU145 and LNCaP, prostate cancer cells showed increased mRNA expression when compared to non-cancer prostate epithelia cell PZ-HPV-7. *: p< 0.05. C. Schematic presentation of CpG island (red line) in promoter and 5′UTR regions of δ-catenin. D. δ-Catenin gene methylation status in prostate cnacer cells. Upper panel: δ-catenin gene methylation status in 4 prostate cancer cell lines CWR22Rv-1, PC-3, DU145, LNCaP) and 1 non-cancer prostate cell line PZ-HPV-7. PC-3 and DU145 showed heterozygous methylation in δ-catenin while it is unmethylated in all the other cell lines. Lower panel: Nested methylation-specific PCR analysis of prostate cancers (#1, #2, #3), normal prostate tissues (#4, #5), and normal bloods (#6, #7) showed no methylation in all of the samples except 1 case in which heterozygous methylation was detected (#3). U lane: unmethylation; M lane: methylation. D: Demethylation in CWR22Rv-1 and DU145 cells does not alter δ-catenin mRNA expression.
Methylation status of CpG islands around δ-catenin transcription initiation site in prostate cancer cells
Gene expression can be activated or inactivated by reversible epigenetic events such as DNA methylation alterations in the promoter region (Nakayama et al, 2001; Zhang et al, 2004). To determine if changes in DNA methylation could account for different expression levels of δ-catenin in normal and cancerous prostatic epithelial cell lines, we analyzed the CpG islands in δ-catenin promoter region. According to http://www.urogene.org/, regions of CpG islands were identified from −828 to −57 nucleotides in relation to the ATG start codon (Fig 1C). The software predicts the most prominent and PCR amplifiable CpG islands (− 281 to −95 nucleotides) around the transcription initiation site (−145 nucleotide). The methylation status of these CpGs islands in δ-catenin was examined by nest PCR with the methylation-specific primer (MSP) sets (Supplemental material S1 Table 1). δ-Catenin methylation was not observed in PZ-HPV-7, CWR22Rv-1, or LNCaP cells (Fig 1D, upper panel). A heterozygous status of δ-catenin methylation was found in DU145 cells, but was only weakly detectable in PC-3 cells (Fig 1D, upper panel). Since we did not detect δ-catenin methylation in the promoter region of non-cancer (PZ-HPV-7, low δ-catenin expression) or prostate cancer (CWR22Rv-1, high δ-catenin expression) cell lines, the methylation state in δ-catenin CpG islands did not seem to be involved in the regulation of δ-catenin expression in these cell lines. Furthermore, we examined a total of 25 cases of normal and prostate cancer tissue specimens but detected the heterozygous δ-catenin methylation in only one case (Fig. 1D, case#3, lower panel).
To investigate further if the methylation of δ-catenin CpG islands is functionally important for the regulation of δ-catenin expression, we compared the expression of δ-catenin gene in DU145 and CWR22Rv-1 cells by semi-quantitative RT-PCR. The expression of δ-catenin with heterozygous methylation status in DU145 was the lowest whereas the expression of δ-catenin with no methylation was highest in CWR22Rv-1 cells (Fig 1B). To test if demethylation affects the expression of δ-catenin, we treated DU145 and CWR22Rv-1 cells with methyltansferase inhibitor 5′Aza-DC. Our results showed that 5 μM 5′Aza-DC treatment had no effect on δ-catenin mRNA levels in either DU145 or CWR22Rv-1 cells (Fig 1E).
δ-Catenin gene copy number in prostate cancer cells
Gene amplification and deletion play an important role in the pathogenesis of solid tumors, including prostate cancer (Murthy et al, 2005; Kindich et al, 2005; Ren et al, 2006). To determine if gene amplification at chromosome 5p15 could be an underlying mechanism for δ-catenin upregulation in prostate cancer, we applied real-time PCR for quantitative DNA analysis using the primer sets illustrated in Supplemental materials S2 Table 2. We then applied the absolute quantification method to determine the input copy number by relating the PCR signal to a standard curve (Livak and Schmittgen, 2001; see also Supplemental materials S4).
Based on this experimental design, we determined the gene copy number of δ-catenin in all aforementioned cell lines, normal bloods, normal prostate tissues and primary prostatic adenocarcinomas. We did not observe significant increases in δ-catenin gene copy number in normal or prostate cancer specimens (Fig 2A). On the other hand, c-myc gene, which was used as a positive control, showed an amplification of 3 fold in 1 prostatic adenocarcinoma in comparison to that of normal prostate tissues (Fig 2B, a 3 fold increase is indicated by an asterisk).
Figure 2.
Analysis of δ-catenin gene amplification in prostate cancer. A. Scatter plot of 25 prostate cancer and benign cases showing there is no significant gene copy number change in δ-catenin gene. B. Scatter plot of 25 prostate cancer and benign cases showing there is no significant gene copy number change except one case in myc gene. *: p<0.05.
Mutational analysis of δ-catenin coding and non-coding sequences
The presence of activating or inactivating mutations in the coding region of proteins can provide important functional implications. We prepared primer sets to cover the entire open reading frame (ORF) of δ-catenin (Supplemental materials S3. Table 3). These primer sets divided the full-length δ-catenin into 8 sequence segments. δ-Catenin cDNA was obtained by RT-PCR and the resulting PCR fragments were directly sequenced. From all the prostate cancer specimens described above and the 5 cases of esophageal cancer tissue samples, we were not able to detect any mutations or truncations. This result suggested that the primary sequence alterations in δ-catenin coding regions may not be common in prostate cancer.
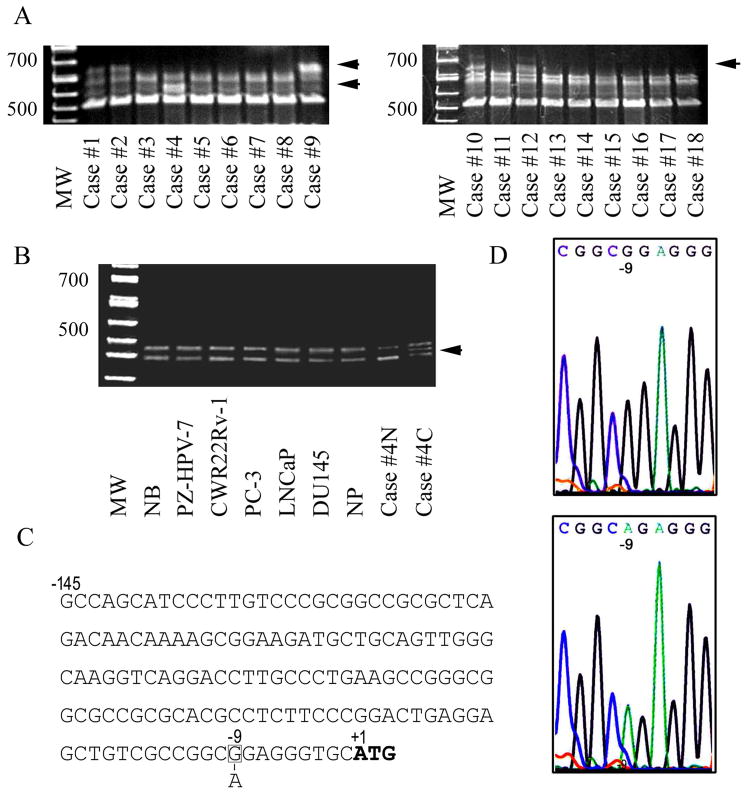
We further examined if any nucleotide sequence alterations are present in the regulatory elements of δ-catenin gene non-coding region. A 204bp DNA fragment of promoter and 5′UTR of δ-catenin genomic DNA upstream of initial translation site was amplified and analyzed using SSCP method (Fig 3A and B). Polymorphic nucleotide changes with the mobility shifts of DNA fragments were identified in 6 out of 18 prostate cancer tissue specimens (Fig 3A: Case # 1, 2, 4, 9, 10 and 12) with Gleason scores ranging from 6 to 9 (Supplemental materials S5. Table 4). No single nucleotide polymorphism was detected in 7 normal blood or benign prostatic tissues (data not shown). We then chose to focus on one prostatic adenocarcinoma (Case #4) to further verify the presence of polymorphism. A 133bp fragment of promoter and 5′UTR regions of δ-catenin genomic DNA upstream of initial translation site was amplified using a different primer set. Using different primer sets to amplify δ-catenin DNA of different sizes ruled out the possibility of PCR errors. The mobility shift was confirmed by SSCP experiment (Fig 3B, arrow). In this analysis, we included normal blood (NB) and normal prostatic tissue (NP) as well as the 5 prostate epithelial cell lines. Only the prostate cancer case #4 with confirmed tumor cell clusters (Case #4C) isolated by laser microdissection (LCM) technology consistently showed mobility shift in SSCP analysis (Fig 3B). Convincingly, this mobility shift in SSCP analysis was not detected in the adjacent benign tissue of case #4 (Case #4N) isolated by LCM. Direct DNA sequencing confirmed the presence of one single nucleotide polymorphism (SNP, -9 G>A) (Fig 3C and D, Case #4) at the position of -9 from the ATG start codon in a 70-year old patient with a high Gleason score of 9, poorly differentiated primary prostatic adenocarcinoma. Therefore, this SNP represents a true point mutation associated with prostate cancer. As an additional comparison, we verified that -58 C>T in one prostatic adenocarcinoma case is a SNP since δ-catenin gene sequences with the same change were present in both malignant and the matched adjacent benign prostatic glands isolated by LCM (date not shown).
Figure 3.
Identification of δ-catenin gene mutation in prostate cancer. A. SSCP of δ-catenin 204bp PCR products amplified from prostate cancer and benign tissues. Although 25 cases were studied, single strand DNA patterns of 18 prostate cancer cases were displayed here. Arrows point to the shift of DNA bands due to single strand conformation change. B. SSCP of δ-catenin 133bp PCR fragments amplified from prostate cancer tissue case # 4, cell lines and normal control specimens to verify the mutation causing band shift initially observed in A. NB: normal blood. NP: normal prostate. Case #4N: adjacent benign tissue. Case #4C: cancerous tissue with mutation. C: Sequence illustration showing the position of mutation (-9 G>A) detected in 5′UTR of δ-catenin in prostate cancer case #4 by SSCP. D. δ-Catenin 5′UTR sequencing profile showing G (Upper panel, normal) to A (Lower panel, cancer) mutation at -9 nucleotide position.
Increased expression of luciferase reporter gene driven by δ-catenin promoter with mutation -9 G>A in 5′UTR
The analysis of δ-catenin promoter and 5′UTR sequence using Mat Inspector V2.2 predicted the minimal core promoter at −56 nt to −34 nt and a CCAAT/enhance site at −92 nt to −78 nt. The point mutation (−9 G>A) that we discovered was just downstream to these regions. Therefore, we took advantage of this structural feature and directly amplified the sequence of 5′UTR and exon1 from genomic DNA of δ-catenin and subcloned it into pGL3-basic luciferase reporter system (Fig 4A).
Figure 4.
Cancer specific -9 G>A mutation in δ-catenin 5′UTR promotes protein expression by increased translational but not transcriptional efficiency. A. Schematic illustration of the pGL3-basic reporter vector containing δ-catenin 5′UTR with minimal promoter sequence and luciferase gene. Grey box: CCAA/T enhancer. Black box: minimal promoter. X: position of point mutations. B. DNA construct with -9 G>A point mutation produced a 3–7 fold higher luciferase activity, in contrast to −32 or −97 point mutant as controls, as well as compared to the wild type. *: p < 0.005. C: Real time PCR showed that there was no significant differences in transcriptional efficiency among DNA constructs containing wild type, - 9 G>A, -32 G>A or -97 G>A mutation.
To examine if the -9 G>A point mutation specifically affects the expression of δ-catenin, we compared the ability of different sequence changes (wild type and mutated forms) in δ-catenin 5′UTR to drive luciferase reporter expression and activity. Luciferase expression vectors containing wild-type, M-9 (-9 G>A), M-32 (-32 G>A), and M-97 (-97 T>A) DNA sequence were constructed (Fig 4A). These vectors were then transfected into prostate cancer CWR22Rv-1 and non-cancer PZ-HPV-7 cells, respectively. The luciferase activities, as shown in Fig 4B, represent the average values of three independent experiments. Under this experimental condition, M-9 transfection produced a 6 to 14 fold higher luciferase activity than that of pGL3-basic vector control or a 3 to 7 fold higher luciferase activity than that of wild-type construct, depending on the different cell lines transfected (Fig 4B, left panel, CWR22Rv-1 cells; right panel, PZ-HPV-7 cells). In contrast, M-32 and M-97 transfected cells did not show significant increases in luciferase activity when compared to that of wild-type sequence.
In order to distinguish if the observed differences in luciferase activity between cells transfected with wild-type and mutant δ-catenin 5′UTR were caused by an alteration of transcription or translation efficiency of the mutant forms, we analyzed the levels of chimeric mRNAs in the transfected cells. Extracted mRNAs were treated with DNase to eliminate the possible contamination from plasmid DNA (date not shown). Real-time PCRs were performed using the primers covering the whole 5′UTR and the part of luciferase mRNA to ensure the transcription of inserted 5′UTR. No significant differences at the mRNA level were observed, indicating that the transcription efficiency and RNA stability were similar for all chimeric mRNAs (Fig. 4C).
δ-Catenin expression in prostatic adenocarcinomas with mutation -9 G>A in 5′UTR
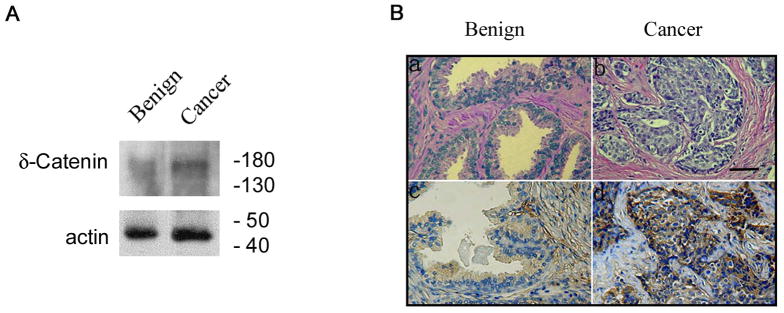
Our previous studies showed that the expression of δ-catenin mRNA and protein is increased in prostatic adenocarcinoma (Lu et al., 2005). Consistent with our earlier report, we found that δ-catenin expression in the prostatic adenocarcinoma harboring mutation -9 G>A in δ-catenin 5′UTR was also increased compared to that of benign prostatic tissues (Fig 5A). In addition, immunohistochemical (IHC) analysis showed that while anti-δ-catenin immunoreactivities were weak in normal prostatic glandular epithelial cells (Fig 5B, a-H&E and c-IHC), their activities were clearly increased in the prostatic adenocarcinoma harboring this mutation (Fig 5B, b-H&E and d-IHC).
Figure 5.
Increased δ-catenin immunoreactivity in prostate cancer specimen showing point mutation in 5′UTR of δ-catenin. A. Western blot showing increased δ-catenin expression in prostate cancer specimen with -9 G>A mutation in 5′UTR as compared to benign specimen without mutation. B. Immunohistochemistry showing increased δ-catenin expression in prostate cancer specimen with -9 G>A mutation in 5′UTR as compared to benign specimen without mutation. a and c, benign prostatic glands. b and d, prostatic adenocarcinoma. a and b, H & E staining. c and d. anti-δ-catenin immunohistochemistry. Bar. 100 μm.
Discussion
Understanding how gene expression is regulated plays important roles in the elucidation of gene functions in human cancer. While δ-catenin gene amplification was reported in cervical (Huang et al, 2006) and bladder cancer (Zheng et al, 2004), δ-catenin overexpression at the transcriptional level has been documented in prostate cancer (Burger et al., 2002; Lu et al., 2005). Our current studies show that multiple mechanisms probably account for the upregulation of δ-catenin expression in cancer, including the upregulation of transcription and increased translational efficiency caused by gene mutations.
Using RT-PCR, we have shown that δ-catenin expression was increased in prostate cancer cell lines such as CWR22Rv-1 and PC-3, when compared to that of non-cancer prostate epithelial cell PZ-HPV-7. However, two other prostate cancer cell lines LNCaP and DU145 did not show an increased δ-catenin expression. This raised questions as to whether δ-catenin overexpression is absolutely required in prostate cancer progression. Since not all prostatic adenocarcinomas showed δ-catenin overexpression, it seems more likely that δ-catenin is one of many oncoproteins that contribute to tumorigenesis. It is also possible that increased δ-catenin expression may occur at certain stages of prostate cancer progression.
Unlike all the other prostate cell lines we have tested, DU145 cells showed a heterozygous hypermethylation status in δ-catenin gene promoter. Treatment of cells with methyltransferase inhibitor 5-deoxyazacytidine did not increase δ-catenin expression, suggesting that hypermethylation is not the major mechanism for regulating δ-catenin gene expression in DU145 cells. We also did not find any significant increases in gene copy numbers in prostate cancer cell lines or from the prostate cancer tissue specimens we have tested. Therefore, while δ-catenin gene amplification occurs in cervical (Huang et al, 2006) and bladder cancer (Zheng et al, 2004), it cannot be a major mechanism for δ-catenin overexpression in prostate cancer. In contrast, transcriptional upregulation appears to play important roles in δ-catenin overexpression in at least some prostate cancer cases. Indeed, recently, we found that ectopic expression of transcription factors such as Pax6 and E2F-1 can upregulate δ-catenin expression in prostate cancer cells in culture (Kim et al., 2008). Interestingly, while Pax6 expression is correlated with pancreatic adenocarcinomas, and differentiation caused its downregulation (Lang et al., 2008), Pax6 expression suppresses glioblastoma cell invasiveness (Mayes et al., 2006). In the future, it will be important to determine how Pax6 may be involved in the regulation of δ-catenin overexpression in prostate cancer.
Besides the transcriptional regulation, translation efficiency also contributes to the expression of proteins, and this is especially true if the alteration of gene sequences occur at the promoter and 5′-UTR proximal to the translation initiation site. For example, high glucose condition increased the mRNA translation efficiency of CD36 in hyperglycemia (Griffin et al., 2001). On the other hand, it was found that a somatic mutation in the 5′UTR of BRCA1 gene in sporadic breast cancer caused down-modulation of translation efficiency (Signori et al., 2001). In our present study, we have discovered an increased incidence of gene mutations in δ-catenin promoter and 5′-UTR associated with prostate cancer. Remarkably, while we consistently detected the nucleotide changes by SSCP in prostate cancer specimens, we have not observed any such SNPs in normal control specimens, indicating that the changes were quite frequent in prostate cancer. However, while -9 G>A mutation occurred only in cancerous tissues, not all SNPs manifested cancer specific patterns. For example, -32 G>A sequence change occurred in both cancerous and adjacent benign tissues. Therefore, we speculate that δ-catenin 5′-UTR may be particularly susceptible to oxidative or other DNA damage attack to introduce mutations. Not all SNPs in δ-catenin 5′UTR show cancer related alteration of its gene expression. But the increased chances in nucleotide changes can increase the possibility of shifting the balance towards cancer specific expression. Future studies will be needed to explore what tumor microenvironment factors promote the occurrence of mutations in δ-catenin gene promoter and 5′UTR.
Materials and Methods
Cell lines and tissue specimens
All human prostate cancer or non-cancer cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and were cultured according to the manufacturer’s instruction at 37°C with 5% CO2 atmosphere.
A total of 25 cases of human prostate cancer and benign specimens were analyzed in this study. Total RNAs were obtained either from prostate cancer and matched adjacent non-cancer tissue commercially (Ambion, Austin, TX; Clontech, Mountain View, CA), or from fresh prostate cancer tissues in house. Fresh and paraffin embedded non-cancer prostate tissues and primary prostatic adenocarinoma samples for DNA analyses were obtained after radical prostatectomy from previously untreated patients at East Carolina University Brody School of Medicine. For some experiments, we also used normal blood as additional non-cancer specimen control. All samples were collected according to the East Carolina University Institutional Research Board (IRB) approved protocol and further verified by pathological analysis.
DNA extraction, RNA isolation and RT-PCR
Genomic DNA from fresh and paraffin-embedded tissues after deparaffinizing using xylene was isolated using DNeasy Tissue Kit (QIAGEN Science, Maryland). Genomic DNA of matched adjacent benign prostatic glandular epithelial cells captured from paraffin embedded sections using Laser Capture Microdissection (Zeiss, Germany) was extracted using PicoPure DNA Extraction Kit (Arcturus Bioscience, Mountain View, CA). Total RNAs were isolated using the RNeasy Mini kit (QIAGEN Science, MD). Reversed transcription was performed using SuperScript II (Invitrogen, TX). Semi-quantitative RT-PCR analysis was used to determine gene expression of d-catenin at mRNA level in cell lines, normal prostate tissues and prostate cancers and of luciferase in transfected cell lines. The PCR primers used for detection of δ-catenin mRNA expression level and sequencing for coding region were listed in Supplemental material (S3. Table 3). The expression of GAPDH was used as a control. For comparing the expression of luciferase after transfecting different DNA sequences in each cell line, the pairs of primers for luciferase included the forward sequence 5′-GCTCAGACAACAAAAGCGGAAGAT-3′ and reverse sequence 5′-TCCAGATCCACAACCTTCGCTTCA-3′. In these experiments the expression of β-galactosidase was used as a control.
Bisulfite modification and nest PCR using methylation-specific primers
Bisulfite treatment of the genomic DNA was performed using bisulfite modification kit (Active motif, CA). In the bisulfite reaction, all unmethylated cytosines were deaminated and converted to uracils, whereas 5-methyl cytosines remained unaltered. The modified DNA was subjected to PCR amplification using outer primers, and nest PCR was performed using the methylation–specific primers, which were designed using the MethPrimer website: http://www.urogene.org/methprimer/. The U primer sets were annealed to unmethylated DNA that had undergone chemical modification whereas M primer sets were annealed to modified, methylated DNA. W primer sets served as a control by annealing to wild-type DNA that had not undergone modification. All sequences of these primers were listed in Supplemental material (S1. Table 1). To determine if demethylation affects the expression of δ-catenin in prostate cancer cell lines, DU145 and CWR22Rv-1 cells were treated with 5 μM methyltansferease inhibitor 5-aza-2′ deoxycytidine (5′Aza-DC) (Sigma Corp, St. Louis, MO).
Quantitative real-time PCR and gene amplification
Real-time PCR was performed using SYBR Green PCR Master Mix in an iCycler iQ Multicolor Real-time PCR Detection System (Bio-Rad, Hercules, CA). For measurement of δ-catenin gene copy number in prostate cancer samples, we employed an absolute quantification method that determines the input copy number, by relating the PCR signal to a standard curve (Livak and Schmittgen, 2001). The primers used for the analysis were listed in Supplemental material (S2. Table 2). Primers were designed according to exon6 of GAPDH gene (12p13) as an internal reference gene, exon11 of δ-catenin gene (5p15) and exon3 of c-myc gene (8q24) by using a web-based program at www.idtdna.com and synthesized from IDT (Coralville, IA). Qiagen purified PCR products were subcloned into a TOPO-TA vector (Invitrogen, Carlsbad, CA). Positive clones were confirmed by digestion with EcoRI and by DNA sequencing. Qiagen purified DNA with the inserts was measured by UV absorbance, and copy number was calculated for each insert by formula: gene copy number= ng×10−9×6.02×1023/molecular weight. Serial dilutions ranging from 108 to 103 copies were prepared for real-time PCR of δ-catenin, GAPDH, and c-myc genes, respectively in duplication and individual standard curves were constructed (Supplemental material S4). These curves were used as standards for quantitative analysis of copy number of the unknown prostate cancer samples using threshold cycle (Ct) value by the iCycler. The PCRs for δ-catenin, GAPDH and c-myc gene were always run simultaneously using the same volume for each sample in duplication. The absolute gene copy number of δ-catenin and c-myc was determined by the ratio between δ-catenin and GAPDH, and between c-myc and GAPDH mean copy number in each duplicate.
PCR-SSCP analysis
Screening of potential mutations in δ-catenin promoter and 5′UTR was performed using two sets of primers for amplification. Forward primer 5′-TGC CGC CCG CCA GCA TCC CTT GT-3′ and reverse primer 5′-AGC CCC GCA ACT CAC CCA AAG GCG-3′ were used for PCR of DNA sequence with a size of 204bp located at 5′-UTR region and exon1. Forward primer 5′-GCT CAG ACA ACA AAA GCG GAA GAT- 3′ and reverse primer 5′-TTC CTC GCA AAC ATG CAC CCT- 3′ were used to obtain a PCR fragment with a size of 133bp located at 5′-UTR region and exon1. A mixture consisting of PCR product and SSCP stop dye (98% Deionized formamide, 0.5 M EDTA, 0.25% bromphenol blue and 0.25% xylene cyanol) were heated for 10 minutes, and then plunged into ice prior to loading onto the gel.
A 4–20% gradient polyacrylamide TBE gel electrophoresis (Invitrogen, Carlsbad, CA) was used to run samples at 300 volts for 5 minutes followed by running at 100 volts for 6 hours. The temperature of the buffer chamber was kept between 4~10°C and was monitored during the electrophoresis.
DNA sequence analysis
PCR fragments amplified from δ-catenin cDNAs were sequenced directly. PCR products of interest obtained from PCR-SSCP were ligated into TOPO-TA vector. Multiple clones were confirmed by digestion with EcoRI and sequenced. The DNA sequences of amplified PCR fragments obtained from prostate cancer tissues and the matched adjacent normal prostate cells were compared to δ-catenin DNA sequence as a reference published in the NCBI database using BLAST Program.
Plasmid construction of luciferase reporter system
The 133bp fragments containing 5′UTR and exon1 of wild type, M-9 (-9 point mutation G>A that resembles the gene mutation in Case #4 of prostatic adenocarcinoma), M-32 (-32 point mutation G>A that was randomly selected) and M-97 (-97 mutation T>A that disrupts the only upstream open reading frame) were PCR-amplified by using forward primer 5′-CCC TCG AGG CTC AGA CAA CAA AAG CGG AAG AT - 3′ and reverse primer 5′-CCA AGC TTT TCC TCG CAA ACA AGC ACC CT -3′. M-58, another fragment of 133bp of the same region (-58 point mutation C>T from the start codon ATG, which reflects a natural mutation in a prostatic adenocarcinoma) was also PCR-amplified but verified as polymorphism from a prostate cancer patient. These fragments were subcloned into a TOPO-TA vector and positive clones were confirmed by digestion with EcoRI and by DNA sequencing. The insert fragments were digested from TOPO-TA by HindIII and NcoI and ligated into pGL3-basic reporter vector (Promega, Madison, WI).
Transient cotransfection assay
CWR22Rv-1 and PZ-HVP-7 cells were transfected using Fugene 6 (Roche, Indianapolis, IN) with 1.0 μg of either pGL3-basic reporter vector, WT, M9, M-32, or M97 along with 0.5 ug of pCMV-β-gal plasmid (Promega, Madison, WI). The pGL3-basic reporter vector containing luciferase gene was used as a background control. pCMV-β-gal contains the β-galactosidase gene driven by CMV promoter and was used as a control for transfection efficiency. Luciferase and β-galactosidase activities were measured using the Luciferase and β-Galactosidase Assay Systems from Promega. Luciferase activity was read in a FLUROSKAN ASCENT FL (Thermo Electron Corporation, Vantaa, Finland). Luciferase values were normalized to β-galactosidase for each experiment, and adjusted for fold expression over that of the pGL3-basic reporter expression for each cell line tested.
Immunochemistry and Immunohistochemistry
Fresh prostate cancer and benign prostate tissues were homogenized in RadioImmuno Precipitation Assay (RIPA) buffer (150mM NaCl, 10mM HEPES pH 7.3, 2mM EDTA, 0.2% SDS, 0.5% Sodium deoxycholate, 1% Triton X-100) supplemented with protease inhibitors (Roche Complete Protease Inhibitor cocktail), followed by 30 minute incubation on ice. Cell debris was removed by centrifugation. Supernatant was collected and protein concentration was determined using Pierce BCA assay. Samples were heated and loaded on SDS-PAGE gels, transferred to nitrocellulose membrane and subject to Western blot analysis.
For tissue immunohistochemistry, 5 μm tissue sections of formalin-fixed, paraffin-embedded blocks were deparaffinized and rehydrated. Endogenous peroxidase was blocked by incubation with hydrogen peroxide. The sections were immunostained in Dako Autostainer (Carpinteria, CA) using rabbit anti-δ-catenin (1:100) (Abcam, Cambridge, MA) followed by streptavidin-biotin peroxidase method for detection.
Statistical analysis
For semi-quantitative RT-PCR and luciferase reporter experiments, all results were reported as mean ± SEM and statistical analysis were conducted using one-way ANOVA. For quantitative real-time PCR, all data were reported as mean ± SEM and the statistical analysis were conducted using Duncan’s test. The significance levels were set at either p <0.005 or p<0.05.
Supplementary Material
Acknowledgments
We thank Melissa Clark and GW Lanford for excellent technical assistance, and Lu laboratory members for many helpful discussions. This study was supported in part by NIH/NCI (CA111891) and the Department of Defense (PC040569) grants (Q.L.).
References
- Burger MJ, Tebay MA, Keith PA, Samaratunga HM, Clements J, Lavin MF, Gardiner RA. Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancer. Int J Cancer. 2002;100(2):228–237. doi: 10.1002/ijc.10468. [DOI] [PubMed] [Google Scholar]
- Cervera M, Sanchez S, Molina B, Alcantara MA, Del Castillo V, Carnevale A, Gonzalez-del Angel A. Trisomy of the short arm of chromosome 5 due to a de novo inversion and duplication (5) (p15.3 p13.3) Am J Med Genet A. 2005;136(4):381–385. doi: 10.1002/ajmg.a.30791. [DOI] [PubMed] [Google Scholar]
- Duparc RH, Boutemmine D, Champagne MP, Tétreault N, Bernier G. Pax6 is required for delta-catenin/neurojungin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300(2):647–55. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- Fatima S, Chui CH, Tang WK, Hui KS, Au HW, Li WY, Wong MM, Cheung F, Tsao SW, Lam KY, Beh PS, Wong J, Law S, Srivastava G, Ho KP, Chan AS, Tang JC. Transforming capacity of two novel genes JS-1 and JS-2 located in chromosome 5p and their overexpression in human esophageal squamous cell carcinoma. Int J Mol Med. 2006;17(1):159–170. [PubMed] [Google Scholar]
- Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS. A link between diabetes and atherosclerosis: Glucose regulates expression of CD36 at the level of translation. Nat Med. 2001;7(7):840–6. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- Ho C, Zhou J, Medina M, Goto T, Jacobson M, Bhide PG, Kosik KS. Delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J Comp Neuro. 2000;420(2):261–276. [PubMed] [Google Scholar]
- Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43(4):1809–1818. [PubMed] [Google Scholar]
- Huang FY, Chiu PM, Tam KF, Kwok YK, Lau ET, Tang MH, Ng TY, Liu VW, Cheung AN, Ngan HY, Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G. Semi-quantitative fluorescent PCR analysis identifies PRKAA1 on chromosome 5 as a potential candidate cancer gene of cervical cancer. Gynecol Oncol. 2006;103(1):219–225. doi: 10.1016/j.ygyno.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17(1):16–23. [PubMed] [Google Scholar]
- Kim K, Oh M, Ki H, Wang T, Bareiss S, Fini ME, Li D, Lu Q. Identification of E2F1 as a positive transcriptional regulator for delta-catenin. Biochem Biophys Res Commun. 2008;369(2):414–20. doi: 10.1016/j.bbrc.2008.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindich R, Florl AR, Jung V, Engers R, Müller M, Schulz WA, Wullich B. Application of a modified real-time PCR technique for relative gene copy number quantification to the determination of the relationship between NKX3.1 loss and MYC gain in prostate cancer. Clin Chem. 2005;51(3):649–52. doi: 10.1373/clinchem.2004.045013. [DOI] [PubMed] [Google Scholar]
- Lang D, Mascarenhas JB, Powell SK, Halegoua J, Nelson M, Ruggeri BA. PAX6 is expressed in pancreatic adenocarcinoma and is downregulated during induction of terminal differentiation. Mol Carcinog. 2008;47(2):148–56. doi: 10.1002/mc.20375. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu Q, Dobbs LJ, Gregory CW, Lanford GW, Revelo MP, Shappell S, Chen YH. Increased expression of d-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Human Pathology. 2005;36(10):1037–1048. doi: 10.1016/j.humpath.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Lu Q, Paredes M, Medina M, Zhou J, Cavallo R, Peifer M, Orecchio L, Kosik KS. Delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J Cell Biol. 1999;144(3):519–32. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu RC, Johnson EI, Dykens EM, Hodapp RM, Overhauser J. No relationship between the size of the deletion and the level of developmental delay in cri-du-chat syndrome. Am J Med Genet. 1999;86(1):66–70. doi: 10.1002/(sici)1096-8628(19990903)86:1<66::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK, Zhou YH. PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Cancer Res. 2006;66(20):9809–17. doi: 10.1158/0008-5472.CAN-05-3877. [DOI] [PubMed] [Google Scholar]
- Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63(2):157–164. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Magliocco AM, Demetrick DJ. Copy number analysis of c-erb-B2 (HER-2/neu) and topoisomerase IIalpha genes in breast carcinoma by quantitative real-time polymerase chain reaction using hybridization probes and fluorescence in situ hybridization. Arch Pathol Lab Med. 2005;129(1):39–46. doi: 10.5858/2005-129-39-CNAOCN. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Sasaki A, Mese H, Alcalde RE, Tsuji T, Matsumura T. The E-cadherin gene is silenced by CpG methylation in human oral squamous cell carcinomas. Int J Cancer. 2001;93(5):667–673. doi: 10.1002/ijc.1386. [DOI] [PubMed] [Google Scholar]
- Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61(5):293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250(2):452–64. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- Pretlow TG, Wolman SR, Micale MA, Pelley RJ, Kursh ED, Resnick MI, Bodner DR, Jacobberger JW, Delmoro CM, Giaconia JM. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85(5):394–398. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- Ren B, Yu G, Tseng GC, Cieply K, Gavel T, Nelson J, Michalopoulos G, Yu YP, Luo JH. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene. 2006;25(7):1090–8. doi: 10.1038/sj.onc.1209134. [DOI] [PubMed] [Google Scholar]
- Signori E, Bagni C, Papa S, Primerano B, Rinaldi M, Amaldi F, Fazio VM. A somatic mutation in the 5′UTR of BRCA1 gene in sporadic breast cancer causes down-modulation of translation efficiency. Oncogene. 2001;20(33):4596–600. doi: 10.1038/sj.onc.1204620. [DOI] [PubMed] [Google Scholar]
- Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35(7):403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145) Int J Cancer. 1978;21(3):274–281. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- Tanahashi H, Tabira T. Isolation of human delta-catenin and its binding specificity with presenilin 1. Neuroreport. 1999;10(3):563–568. doi: 10.1097/00001756-199902250-00022. [DOI] [PubMed] [Google Scholar]
- Umbas R, Isaacs WB, Bringuier PP, Schaafsma HE, Karthaus HF, Oosterhof GO, Debruyne FM, Schalken JA. Decreased E-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res. 1994;54(14):3929–3933. [PubMed] [Google Scholar]
- Weijerman PC, Konig JJ, Wong ST, Niesters HG, Peehl DM. Lipofection- mediated immortalization of human prostatic epithelial cells of normal and malignant origin using human papillomavirus type 18 DNA. Cancer Res. 1994;54(21):5579–5583. [PubMed] [Google Scholar]
- Zhang HT, Chen XF, Wang MH, Wang JC, Qi QY, Zhang RM, Xu WQ, Fei QY, Wang F, Cheng QQ, Chen F, Zhu CS, Tao SH, Luo Z. Defective expression of transforming primary non-small cell lung cancer. Clin Cancer Res. 2004;10(7):2359–2367. doi: 10.1158/1078-0432.ccr-0959-3. [DOI] [PubMed] [Google Scholar]
- Zheng M, Simon R, Mirlacher M, Maurer R, Gasser T, Forster T, Diener PA, Mihatsch MJ, Sauter G, Schraml P. TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am J Pathol. 2004;165(1):63–69. doi: 10.1016/S0002-9440(10)63275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liyanage U, Medina M, Ho C, Simmons AD, Lovett M, Kosik KS. Presenilin interaction in the brain with a novel member of the armadillo family. Neuroreport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.