Abstract
Background
Freshwater planktonic crustaceans of the genus Daphnia show a remarkable plasticity to cope with environmental changes in oxygen concentration and temperature. One of the key proteins of adaptive gene control in Daphnia pulex under hypoxia is hemoglobin (Hb), which increases in hemolymph concentration by an order of magnitude and shows an enhanced oxygen affinity due to changes in subunit composition. To explore the full spectrum of adaptive protein expression in response to low-oxygen conditions, two-dimensional gel electrophoresis and mass spectrometry were used to analyze the proteome composition of animals acclimated to normoxia (oxygen partial pressure [Po2]: 20 kPa) and hypoxia (Po2: 3 kPa), respectively.
Results
The comparative proteome analysis showed an up-regulation of more than 50 protein spots under hypoxia. Identification of a major share of these spots revealed acclimatory changes for Hb, glycolytic enzymes (enolase), and enzymes involved in the degradation of storage and structural carbohydrates (e.g. cellubiohydrolase). Proteolytic enzymes remained constitutively expressed on a high level.
Conclusion
Acclimatory adjustments of the D. pulex proteome to hypoxia included a strong induction of Hb and carbohydrate-degrading enzymes. The scenario of adaptive protein expression under environmental hypoxia can be interpreted as a process to improve oxygen transport and carbohydrate provision for the maintenance of ATP production, even during short episodes of tissue hypoxia requiring support from anaerobic metabolism.
Background
The planktonic crustacean Daphnia spp. is an important model organism for ecology, ecotoxicology and evolutionary genomics. This genus plays a central role in the planktonic food webs of standing freshwaters. These habitats exhibit pronounced variations in ambient variables such as oxygen content and temperature, both on a temporal and spatial scale. There are more or less distinctive diurnal and seasonal changes in these abiotic factors. In addition, vertical migrations expose daphnids to a wide range of different oxygen concentrations and temperatures as well. The physiology and metabolism of poikilothermic animals are strongly affected by both environmental factors [1]. Plastic adaptive responses to environmental changes include the differential regulation of gene expression, which provides specific sets of proteins for acclimation/acclimatization and, in consequence, for the maintenance of cellular function under the new ambient conditions.
A key protein of this adaptive gene control in Daphnia under varying oxygen and temperature conditions is hemoglobin (Hb) [2-6]. Under hypoxia or at warm temperatures, new Hb macromolecules of altered subunit composition and with an enhanced oxygen affinity [7-11] are synthesized in the fat cells and epithelial cells of the epipodites of D. magna (subgenus Ctenodaphnia) [12]. Depending on oxygen or temperature condition, seven Hb subunits are differentially expressed, which represents a remarkable example of phenotypic plasticity and functional isoform multiplicity [13]. The release of higher quantities of these newly synthesized Hb aggregates [14,15] into the hemolymph strongly improves oxygen transport from the ambient medium to the cells and restores cellular oxygen homeostasis after environmental change [16-18]
A hypoxic induction of Hb with the consequence of an improved hemolymph oxygen transport capacity under oxygen-poor conditions has also been shown for D. pulex (subgenus Daphnia sensu stricto) [19,20]. As both species, D. magna and D. pulex, inhabit similar habitats (smaller water bodies such as ponds and ditches) and show a high tolerance to hypoxic conditions, a plastic adaptive response of similar complexity as in D. magna may be supposed for D. pulex as well. So far, sequence information was only available for one globin gene in D. pulex[21], although biochemical studies indicate the presence of multiple subunit isoforms [22-24]. Moreover, the full spectrum of adaptive gene control under hypoxia beyond Hb expression has remained unexplored in both species, D. pulex and D. magna. The recent release of the Daphnia pulex genome sequence [25,26] offers the opportunity to identify these target genes. The present study aims to analyze the protein expression patterns of animals which are acclimated to normal and low ambient oxygen conditions, respectively. Two-dimensional gel electrophoresis and mass spectrometry are employed to identify a subset of the proteome induced by hypoxia with subsequent assignment of their functional role using bioinformatic tools.
Results
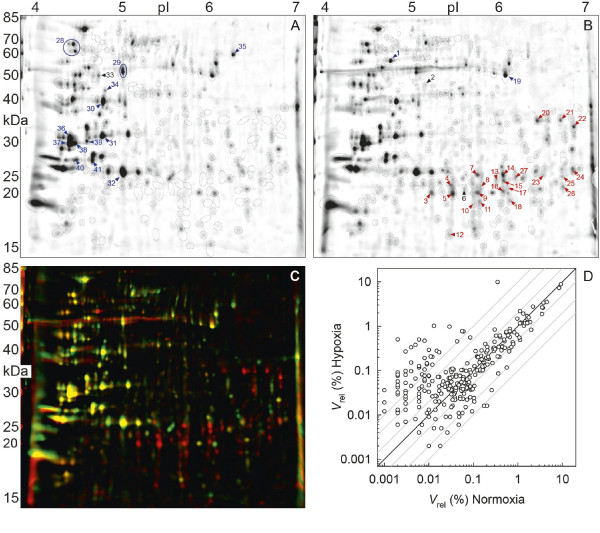
Two-dimensional gels were prepared from total soluble proteins extracted from normoxic or hypoxic cultures of Daphnia pulex (oxygen partial pressure, Po2: 20 kPa or 3 kPa, respectively). The high reproducibility of 2D gels from the same acclimation group allowed one to generate representative fusion images for each acclimation condition (Figure 1A, B). A total of 276 spots were detected on the two fusion gels (encircled spots). The dual-channel representation of both fusion gels (Figure 1C) revealed a distinct set of up-regulated protein spots in the hypoxia-acclimation group (red-colored spots; molecular-weight range: 15–40 kDa, pI range: 5–7). In contrast, down-regulated protein spots were less obvious in the hypoxia-acclimation group as indicated by the sparse occurrence of green-colored spots in the dual-channel representation (Figure 1C). Plotting the relative volumes of related spots from both acclimation groups against each other revealed a number of approximately 50 candidate proteins that were up-regulated in the hypoxia-acclimation group (Figure 1D).
Figure 1.
Two-dimensional protein gels from normoxia (A) and hypoxia (B) acclimated Daphnia pulex. Gel images represent fusion (average) images from a set of three (A) or two (B) biological replicates. Consensus spots used for comparison are encircled. Numbers indicate spots that were picked from the 2D gels for analysis by mass spectrometry. Spots identified as globin or non-globin material were labeled in red or blue. Black labels (spots 2, 6 and 33) indicate proteins that could not be identified. (C) Dual-channel representation of the gel images shown in (A) and (B). Protein spots of similar expression intensity appear in yellow. Red indicates that spots are much stronger or unique on the gel from hypoxia-acclimated animals, whereas green means that spots are much stronger or unique in the gel from normoxia-acclimated D. pulex. (D) Scatter plot showing the comparison of expression levels in the two fusion images (Vrel: relative spot volume). Protein spots that are strongly induced by hypoxia (approximately 50) are found in the upper left part of the graph.
A total number of 41 spots (labeled in Figure 1A, B) comprising differentially as well as constitutively expressed proteins were excised from representative 2D gels, subjected to in-gel tryptic digestion, and analyzed by tandem mass spectrometry (MS/MS). The MS/MS data were searched against the Daphnia pulex protein database ("Frozen Gene Catalog" as of 03/07/2007, [26]) using the MOWSE algorithm as implemented in the MS search engine Mascot (Matrix Science Ltd. London, UK)[27]. Only in three cases (spots 2, 6 and 33 in Figure 1A, B), the identification was ambiguous. Information on identified proteins is given in the Tables 1, 2, 3 together with the hypoxia-to-normoxia expression ratio, the number and sequence of matched peptides, the percentage sequence coverage, the Mascot score (a statistical measure of identification probability), and the theoretical and experimental molecular weight (Mr) and isolectric point (pI) (excluding the contribution of the signal peptide in case of extracellular proteins).
Table 1.
Identified hemoglobins and non-identified proteins from hypoxia-acclimated (Hyp) and normoxia-acclimated (Norm) Daphnia pulex
| Spot no. | Specificity Hyp: Norm | Matched peptide sequencesa) | Sequence coverageb) | Mascot scorec) | Mr predicted/Mr geld) | pI predicted/pI gele) | SP Length |
Functionf) Model name [Protein ID, Reference ID] |
| 20 | 19 | FVTAHPEYQK SGLAALVAGISK KSEDLADPQTK SEDLADPQTK LSPHMIGDVQR NAMVSDIFIK LFKETPR QVALVADR VDTIISALDDK LLVQSLAAK GVSSDDLDSWK |
30.7% | 635 | 35.3/34 | 5.80/6.38 | 17 | Hemoglobin (Hb4) SNAP_00002894 [234836, 42066] |
| 21 | 9.3 | FVTAHPEYQK SGLAALVAGISK NAMVSDIFIK LFKETPR QVALVADR VDTIISALDDK GAWDDFGR LLVQSLAAK GVSSDDLDSWK |
26.2% | 397 | 35.5/34 | 6.01/6.65 | 17 | Hemoglobin (Hb5) SNAP_00002895 [234837, 311665] |
| 22 | 11.3* | FVTAHPEYQK SGLAALVAGISK KSEDLVDPQTK SEDLVDPQTK LSGHMIGDVQR LFKETPR QVALVADR LDTMIAAMDDK LLLDVLNAK |
24.2% | 397 | 35.7/32 | 6.26/6.81 | 16 | Hemoglobin (Hb3) NCBI_GNO_0400436 [311662, 311662] |
| 3–5 7–18 23–27 |
proteolytic fragments of Hb | |||||||
| 2, 6, 33 | not identified | |||||||
Identification was based on 2D gel electrophoresis and nano-HPLC-ESI-MS/MS analysis of trypsin-digested proteins matched against the "Frozen Gene Catalog" of the D. pulex protein database [26], which contains all manual curations as of July 3, 2007 as well as automatically annotated models chosen from the "Filtered Models" v1.1 reference set. The compiled information includes the spot number (Figure 1A, B), the hypoxia-to-normoxia expression ratio, the number and sequences of matched peptides, the sequence coverage, the Mascot score as a statistical measure of identification probability, the theoretical and experimental molecular weight (Mr) and isolectric point (pI) of the mature protein (without signal peptide), the predicted length of the N-terminal signal peptide (SP) in extracellular proteins, the putative function of the protein, as well as the gene model name and protein identification number for the locus. The protein IDs may differ from those contained in the "Filtered Models v1.1" reference set. The Reference ID can be used to retrieve the corresponding models from this reference set. Underlined and bold-printed sequences indicate peptides that are specific for a globin gene.
a) Matched peptide sequences: tryptic peptide sequences identified via nano-HPLC-ESI-MS/MS.
b) Sequence coverage %: percentage of predicted protein sequence covered by matched peptides.
c) Probability based Mascot score: -10*Log(P), where P is the probability that the observed match is a random event. Scores > 38 indicate identity or extensive homology (p < 0.05). Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. The Mascot-score calculation was performed using whole-protein sequence (including the N-terminal signal peptide in case of extracellular proteins).
d) Mr predicted/Mr gel: molecular mass of predicted protein/of protein on gel.
e) pI predicted/pI gel: isoelectric point of predicted proteins/of proteins on gel.
f) Function of identified proteins was obtained either via automated blastp search provided by JGI or after manual curation of a gene model.
* p < 0.05 (t-Test)
Table 2.
Identified proteolytic enzymes from hypoxia-acclimated (Hyp) and normoxia-acclimated (Norm) Daphnia pulex
| Spot no. |
Specificity Hyp: Norm |
Matched peptide sequencesa) |
Sequence coverageb) |
Mascot scorec) |
Mr predicted/ Mr geld) |
pI predicted/ pI gele) |
SP Length |
Functionf) Model name [Protein ID, Reference ID] |
| 28 | 1.2 | DDLTETLK EPNNPDDAIPVNTAR ITTPAEDR ITTPAEDRR VIVTETDYLK VIVTETDYLKK KLVALLDATPTR LVALLDATPTR TIANYIHWR TETTLMIANLK TLVDDATWMDDGTK KDFLTLR LEEQILTDPHSPSR VIGPLSNNEDFAR |
18.0% | 876 | 75.4/72 | 4.36/4.4 | Peptidase M13 estExt_Genewise1Plus.C_750105 [200882, 200882] |
|
| IYGSYQACR YVELSNK ELHAYIR VLGVAPPVGR VWLEAENAK |
6.6% | 210 | 73.4/72 | 4.81/4.4 | 19 | Peptidase M2 PASA_GEN_6000071 [307230, 307230] |
||
| SGQAVEYLPGR TYTVAADDAR |
3.6% | 116 | 64.1/67 | 4. 66/4.4 | 19 (?) | Carboxylesterase, type B PASA_GEN_25200006 [304160, 304160] |
||
| NADEAVAEGCNNR IVTTDIADQSK |
4.6% | 135 | 58.4/67 | 4.34/4.4 | Sphingomyelin phosphodiesterase PASA_GEN_2900053 [304453, 304453] |
|||
| TYTVDGPR IVTTDIADQSK |
3.6% | 88 | 59.4/67 | 4.57/4.4 | Sphingomyelin phosphodiesterase PASA_GEN_13800028 [301526, 301526] |
|||
| 31 | 0.8* | TFENRDMPLVK KAIVVDGGIHAR AIVVDGGIHAR NRKPNAGIGGIPCIGTDMNR KPNAGIGGIPCIGTDMNR GGAGIPFSYTVEMR DEGTFGFQLPAR QILPNNEEVWEGVK VMAESLF |
22.4% | 475 | 44.8/30 | 4.82/4.75 | 16 | Carboxypeptidase A estExt_Genewise1Plus.C_150058 [195011, 195011] |
| KAIVVDGGIHAR AIVVDGGIHAR LTAVYGTR GGAGIPFSYTVEMR VMAESLF |
10.3% | 246 | 44.8/30 | 4.87/4.75 | 16 | Carboxypeptidase A NCBI_GNO_1500041 [315693, 315693] |
||
| GVTDLTIFR VVAGEHSLR |
6.5% | 135 | 29.1/30 | 4.88/4.75 | 15 | Trypsin SNAP_00016212 [231152, 248154] |
||
| 32 | 0.9 | IVGGTQASPNEFPYQISLR LGSHICGASIYK HEHVSYSSR GSYGTNAITDSMICAGFR |
22.7% | 177 | 26.7/23 | 5.43/4.98 | 17 | Trypsin estExt_fgenesh1_kg.C_230008 [230885, 230885] |
| 36 | 0.85 | VVAGEHSLR SVDVPVVDDDTCNR |
8.9% | 149 | 27.2/30 | 4.32/4.39 | Trypsin e_gw1.85.43.1 [59836, 59836] |
|
| LTAAEEPTRVEIR IRNDVALIK |
7.5% | 80 | 31.5/30 | 5.48/4.39 | 15 | Trypsin PASA_GEN_2900126 [304512, 304512] |
||
| 37 | 0.85 | GVTDLTIFR VVAGEHSLR VVAGEHSLRTDSGLEQNR |
9.8% | 159 | 29.1/29 | 4.88/4.39 | 15 | Trypsin SNAP_00016212 [231152, 248154] |
| VVAGEHSLR SVDVPVVDDDTCNR |
8.9% | 149 | 27.2/29 | 4.32/4.39 | Trypsin e_gw1.85.43.1 [59836, 59836] |
|||
| 38 | 1.18 | GLADADIAVFK LIWMGQYNR YYRDELAGK |
10.7% | 123 | 29.8/29 | 4.5/4.46 | 19 | Placental protein 11 PASA_GEN_12200001 [301221, 301221] |
| GLADADIAVFK LIWMGQYNR YYRDELAGK |
8.0% | 123 | 38.7/29 | 4.57/4.46 | 20 | Placental protein 11 PASA_GEN_6000032 [307196, 307196] |
||
| VVAGEHSLR SVDVPVVDDDTCNR |
8.9% | 149 | 27.2/29 | 4.32/4.46 | Trypsin e_gw1.85.43.1 [59836, 59836] |
|||
| GVTDLTIFR VVAGEHSLR |
6.5% | 80 | 29.1/29 | 4.88/4.46 | 15 | Trypsin SNAP_00016212 [231152, 248154] |
||
| 39 | 0.92 | VVAGEHSLR SVDVPVVDDDTCNR |
8.9% | 149 | 27.2/29 | 4.32/4.59 | Trypsin e_gw1.85.43.1 [59836, 59836] |
|
| GVTDLTIFR VVAGEHSLR |
6.5% | 120 | 29.1/29 | 4.88/4.59 | 15 | Trypsin SNAP_00016212 [231152, 248154] |
||
| 40 | 0.57 | TTEEYYVSVQK TGGGCYSYIGR |
6.5% | 112 | 26.9/25 | 5.32/4.47 | Astacin-like metalloprotease (ACN) FRA_fgenesh1_kg.C_scaffold_182000002 [347623, 93694] |
|
| GVTDLTIFR VVAGEHSLR |
6.5% | 109 | 30.7/25 | 4.82/4.47 | 15 | Trypsin SNAP_00016212 [231152, 248154] |
||
| 41 | 1.16 | LTAAEEPTR LTAAEEPTRVEVR IINDVALIK |
9.1% | 141 | 25.3/25 | 4.52/4.65 | Trypsin e_gw1.29.198.1 [52244, 52244] |
|
Identification was based on 2D gel electrophoresis and nano-HPLC-ESI-MS/MS analysis of trypsin-digested proteins matched against the "Frozen Gene Catalog" of the D. pulex protein database [26], which contains all manual curations as of July 3, 2007 as well as automatically annotated models chosen from the "Filtered Models" v1.1 reference set. The compiled information includes the spot number (Figure 1A, B), the hypoxia-to-normoxia expression ratio, the number and sequences of matched peptides, the sequence coverage, the Mascot score as a statistical measure of identification probability, the theoretical and experimental molecular weight (Mr) and isolectric point (pI) of the mature protein (without signal peptide), the predicted length of the N-terminal signal peptide (SP) in extracellular proteins, the putative function of the protein, as well as the gene model name and protein identification number for the locus. The protein IDs may differ from those contained in the "Filtered Models v1.1" reference set. The Reference ID can be used to retrieve the corresponding models from this reference set. Underlined and bold-printed sequences indicate peptides that are specific for a globin gene.
a) Matched peptide sequences: tryptic peptide sequences identified via nano-HPLC-ESI-MS/MS.
b) Sequence coverage %: percentage of predicted protein sequence covered by matched peptides.
c) Probability based Mascot score: -10*Log(P), where P is the probability that the observed match is a random event. Scores > 38 indicate identity or extensive homology (p < 0.05). Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. The Mascot-score calculation was performed using whole-protein sequence (including the N-terminal signal peptide in case of extracellular proteins).
d) Mr predicted/Mr gel: molecular mass of predicted protein/of protein on gel.
e) pI predicted/pI gel: isoelectric point of predicted proteins/of proteins on gel.
f) Function of identified proteins was obtained either via automated blastp search provided by JGI or after manual curation of a gene model.
* p < 0.05 (t-Test)
Table 3.
Identified carbohydrate-modifying enzymes from hypoxia-acclimated (Hyp) and normoxia-acclimated (Norm) Daphnia pulex
| Spot no. |
Specificity Hyp: Norm |
Matched peptide sequencesa) |
Sequence coverageb) |
Mascot scorec) |
Mr predicted/Mr geld) |
pI predicted/ pI gele) |
SP Length |
Functionf) Model name [Protein ID, Reference ID] |
| 1 | 7.2 | MFQLLNR WINGLANSK DGCDFASYR MNDHTFYGPGSTFK FYVQNGVR GLFGDLDDHK GLFGDLDDHKNK |
13.3% | 284 | 48.2/58 | 4.73/4.72 | 19 | Cellubiohydrolase (CEL7A) PIR_PASA_GEN_1000209 [347598, 300366] |
| 19 | 1.4 | GNPTVEVDLTTEK MGTETYHHLKK NGKYDLDFK NPASDPATYLESNK RIQMAVDCK ACNCLLLK VNQIGTVTESIAAHK LAKYNQILR IEEELGAAAK |
22.6% | 468 | 46.8/51 | 5.98/6.01 | Enolase (ENO) PIR_PASA_GEN_1500033 [347595, 301844] | |
| 29 | 1.2 | KSILFYEAQR SILFYEAQR NAYTAAGELDNGLAALR QLYDFAK MAGISVLLSR ILGDQKYK QQIDYALGSTGR SYVVGFGNNPPVK |
17.7% | 355 | 47.3/53 | 5.09/5.00 | 18 | Endo-β-1,4-Glucanase (CEL9A) PIR_estExt_fgenesh1_kg.C_70001 [347602, 230437] |
| VQLEEEAEAR LTHELDKTR KLGDENAELK LKTEIQR |
4.1% | 124 | 103.7/53 | 5.42/5.00 | Myosin estExt_Genewise1.C_2380001 [219409, 219409] |
|||
| 30 | 0.8 | DSILHIKPTLTEDR GGGNTINPAMAAR YGRVEVNAK SSTPGYNSAFHR YQLEWTPDYLK FSIDDVETGR |
19.8% | 327 | 38.5/39 | 4.76/4.77 | 19 | β-1,3-Glucan-binding protein (gram-negative bacteria-binding protein) PASA_GEN_0200102 [303036, 303036] |
| SFLDFAQSK FVNWQADGVK NYYTDSCLVAAGGK |
9.1% | 88 | 39.0 | 4.75 | 19 | Endo-β-1,4-Mannanase (MAN5A) PASA_GEN_8600009 [347627, 308762] |
||
| 34 | 0.3* | YLGHEVGDAR LKDYYLR DLINDCIMDPK |
7.3% | 160 | 43.1/44 | 4.75/4.76 | 19 | Exo-β-1,3-Glucanase (EXG5) PIR_PASA_GEN_1000289 [347606,300436] |
| 35 | 0.5 | WDDIAAECER YQPVSYK SGDENAFKSMVDR GKILEFLNK ILEFLNK LTSYGVAGFR HMWPGDLK KLSDVFHK LSDVFHK LSDVFHKK GHGGGGDLLTFR QIYNMAK |
17.2% | 536 | 54.9/62 | 6.03/6.30 | 19 | α-Amylase (AMY) PASA_GEN_2100059 [303445, 303445] |
Identification was based on 2D gel electrophoresis and nano-HPLC-ESI-MS/MS analysis of trypsin-digested proteins matched against the "Frozen Gene Catalog" of the D. pulex protein database [26], which contains all manual curations as of July 3, 2007 as well as automatically annotated models chosen from the "Filtered Models" v1.1 reference set. The compiled information includes the spot number (Figure 1A, B), the hypoxia-to-normoxia expression ratio, the number and sequences of matched peptides, the sequence coverage, the Mascot score as a statistical measure of identification probability, the theoretical and experimental molecular weight (Mr) and isolectric point (pI) of the mature protein (without signal peptide), the predicted length of the N-terminal signal peptide (SP) in extracellular proteins, the putative function of the protein, as well as the gene model name and protein identification number for the locus. The protein IDs may differ from those contained in the "Filtered Models v1.1" reference set. The Reference ID can be used to retrieve the corresponding models from this reference set. Underlined and bold-printed sequences indicate peptides that are specific for a globin gene.
a) Matched peptide sequences: tryptic peptide sequences identified via nano-HPLC-ESI-MS/MS.
b) Sequence coverage %: percentage of predicted protein sequence covered by matched peptides.
c) Probability based Mascot score: -10*Log(P), where P is the probability that the observed match is a random event. Scores > 38 indicate identity or extensive homology (p < 0.05). Protein scores are derived from ions scores as a non-probabilistic basis for ranking protein hits. The Mascot-score calculation was performed using whole-protein sequence (including the N-terminal signal peptide in case of extracellular proteins).
d) Mr predicted/Mr gel: molecular mass of predicted protein/of protein on gel.
e) pI predicted/pI gel: isoelectric point of predicted proteins/of proteins on gel.
f) Function of identified proteins was obtained either via automated blastp search provided by JGI or after manual curation of a gene model.
* p < 0.05 (t-Test)
In some cases, MS data suggest that more than one protein was present in the excised spot. For example, spot 29 corresponding to an apparent Mr of 53 kDa yielded two proteins, an endo-β-1,4 glucanase (predicted Mr: 47.3 kDa; identification based on 8 peptides) and myosin (predicted Mr: 103.7 kDa; 4 peptides). Accordingly, spot 29 contains the glucanase as the major protein with a minor amount of a myosin fragment. The deviation between predicted and experimental Mr/pI suggests that the fragment resulted from proteolytic cleavage during sample preparation.
The identified proteins can be classified into three groups with (i) a set of hemoglobin (Hb) subunits and fragments which were up-regulated at hypoxia acclimation, (ii) a set of proteases which were expressed in high amounts at both acclimation (oxygen) conditions, and (iii) a set of carbohydrate-modifying enzymes, for which a complex regulation pattern was observed including constant expressions as well as up- and down-regulations.
Hemoglobins
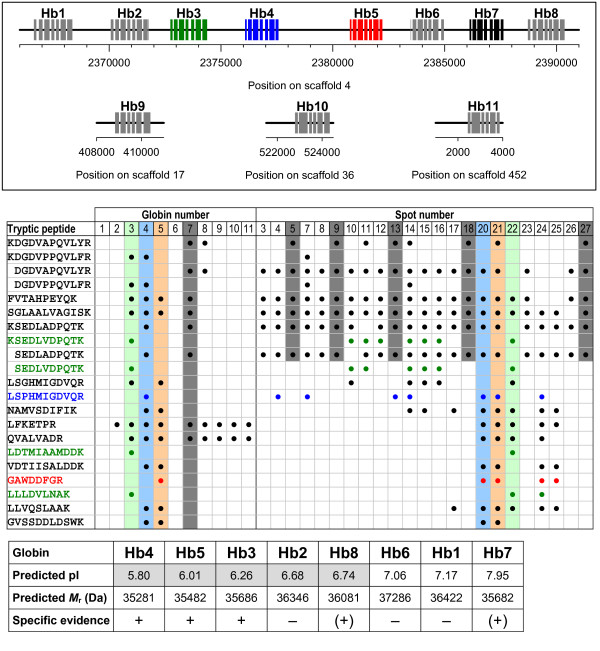
Among the proteins up-regulated in hypoxia-acclimated animals, 23 spots were identified to contain Hb (Figure 1B, spots 3–5, 7–18 and 20–27). The tryptic peptides (fragments) used for the identification of Hb are listed in Figure 2 in the order of their appearance in the globin genes. Peptide sequences that are specific for one globin gene, and which therefore allow for a discrimination between globin subunits, are printed in green, blue and red colors. Only the subunits Hb3, Hb4 and Hb5 received specific support by the MS analysis of fragments. An unambiguous discrimination was not possible for the subunits Hb7 and Hb8, which received the support by the same pair of tryptic peptides. However, the spots 5, 9, 13, 18 and 27 yielded the same set of six tryptic fragments (including the Hb7/Hb8-related pair) which could all be assigned to subunit Hb7. Subunit Hb8, in contrast, was only supported by the Hb7/Hb8-related pair. This suggests that subunit Hb7 rather than Hb8 is expressed under hypoxic conditions. The spots 20, 21 and 22 mainly contained tryptic peptides related to subunit Hb4 (11 of 13 fragments), Hb5 (9 of 14 fragments) or Hb3 (9 of 11 fragments), respectively, with a sequence coverage of up to 30% (Figure 2, Table 1). In addition, the experimental Mr of only these three spots (20, 21, 22) matched the expected size of an intact globin subunit (Table 1). Taking further into account the correlations between the observed and predicted pI patterns, then an assignment of subunits Hb4, Hb5, and Hb3 to the spots 20, 21 and 22 seems plausible (see Discussion). These subunits showed a 9-19-fold increase in expression under hypoxia.
Figure 2.
Assignment of protein spots to the globin genes (HbA-HbL) of D. pulex. Positional information on the globin genes (Hb1–Hb11) is given on top (boxes represent exons). Genes with specific proteomic support (Hb3, Hb4, Hb5) are shown in green, blue and red colors. The middle part lists the tryptic peptides in the order of their appearance in the globin sequences. Black circles indicate the occurrence of tryptic peptides in the globin sequences and in the analyzed spots. Colored sequences and circles indicate tryptic peptides that are specific for only one globin. Shaded in gray is a set of six tryptic fragments which were detected in several spots (5, 9, 13, 18, 27) and which could all be assigned to subunit Hb7. The lower part lists the characteristics of globins in terms of predicted isoelectric point (pI) and molecular mass (Mr). Shaded in gray are the predicted pI values which fall within the pH 4–7 gradient range used for isoelectric focussing.
Proteases and Peptidases
Proteolytic enzymes were identified in spots 28, 31, 32, and 36–41 (Table 2). All of these nine spots were expressed in comparable amounts under both oxygen conditions. The trypsin-containing spots (31, 32, 36–41) were expressed in large amounts (Figure 1A). Several other proteases were also present including the peptidases M13 and M2 (spot 28), a carboxypeptidase A (spot 31), and an astacin-like metalloprotease (spot 40).
Glycoside hydrolases
Several enzymes involved in carbohydrate metabolism were identified in the gels (Figure 1A, B and Table 3). A cellubiohydrolase (CEL7A, spot 1) showed the strongest differential expression with a seven-fold up-regulation under hypoxia. Acclimation to hypoxia was additionally associated with a slight up-regulation of the enolase (ENO, spot 19). The strongest reduction in protein expression was observed for an α-amylase (spot 35) and an exo-β-1,3-glucanase (EXG5, spot 34).
Discussion
Using a proteomic approach, the present study identified, aside from constitutively expressed proteins, a set of proteins, which is differentially expressed in normoxia or hypoxia acclimated Daphnia pulex. Identification and biochemical characterization of this proteome subset may contribute to the ongoing annotation process of the D. pulex genome as it provides complementary information on the protein level for predicted genes with EST support as well as information on key players of adaptive gene control involved in the adjustment of physiological functions under different oxygen conditions.
Methodical constraints
To improve resolution, proteins were separated on 2D gels using a relatively narrow pH gradient (pH 4–7). Although the pI of the bulk of soluble proteins falls into this pH region during isoelectric focusing, there is still a considerable number of polypeptides with pI values outside this range (unpublished data), which requires additional experiments in future with extended pH gradients for isoelectric focusing. In order to guarantee identical protein load per gel, a relatively low amount of protein was loaded (140 μg/gel). Therefore, protein identification by mass spectrometry was possible only for prominent spots representing high-copy proteins. The identification of differentially expressed proteins was impeded by a residual proteolytic activity, since several spots were identified as cleavage fragments of proteins (see below), despite of the use of protease inhibitors for protein extraction and the preparation at cold temperatures. The residual proteolytic activity in the crude extract is the consequence of the high abundance of proteases, which are equally expressed under both normoxic and hypoxic conditions. Hence, for future experiments a custom made inhibitor cocktail for specific and effective inhibition of D. pulex proteases has to be developed.
Oxygen transport and energy metabolism
The analysis of differential expression patterns revealed the hemoglobins as one major group that is induced under hypoxia. Eight genes coding for the globins AHb1–Hb8 are present in the genome of Daphnia pulex forming a cluster on scaffold 4, whereas three additional gene copies are spread across different scaffolds (Figure 2) [28]. Among the 23 Hb spots, sequences specific for the subunits Hb3, Hb4 and HB5 were detected by the MS analysis of fragments. The tryptic-peptide analysis further revealed that subunit Hb7 (but not Hb8) is very likely expressed under hypoxic conditions. The spots 20, 21 and 22 showed a dominance of tryptic peptides related to subunit Hb4 (11 of 13 fragments), Hb5 (9 of 14 fragments) and Hb3 (9 of 11 fragments), respectively, with a sequence coverage of up to 30% (Figure 2, Table 1). As the mature subunits have predicted pI values of 5.80–7.95 and Mr values of 35–37 kDa (Figure 2), they should distribute according to their pI values along a horizontal line in the order Hb4-Hb5-Hb3-Hb2-Hb8-Hb6-Hb1-Hb7. Due to the pH 4–7 gradient range used for isoelectric focusing, however, only the subunits Hb4, Hb5, Hb3, Hb2 and Hb8 would migrate into this pH range. In addition, the experimental pI values (Table 1) turned out to be shifted by 0.5–0.7 units towards higher values in comparison to the predicted pI values (Hb4: 5.80→6.38, Hb5: 6.01→6.65, Hb3: 6.26→6.81), which maybe due to posttranslational modifications of the Hb subunits [15]. Provided that such a pI shift applies to all other products of the globin gene cluster as well, then none of the remaining subunits (Hb2, Hb8) would have migrated into and would be visible in the pH 4–7 gradient range. Actually, only the spots 20–22 showed experimental Mr values (Table 1), which matched the expected size of intact globin subunits. Consequently, the spots 20, 21 and 22 most likely represent the subunits Hb4, Hb5 and Hb3. The contamination of these spots with small quantities of unrelated tryptic peptides could be the consequence of minor proteolytic cleavage of other subunits and the co-localization of cleavage products of similar pI at these spots.
Some protein spots (spots 5, 9, 13, 18, 27) within the molecular-weight range of 15–30 kDa (Figure 1B) yielded tryptic peptides that very likely represented cleavage products of subunit Hb7 (Figure 2). Further low-molecular weight spots in extracts from hypoxia-acclimated D. pulex contained fragments of conserved sequences that could originate from any of the subunits Hb3, Hb4, Hb5, and Hb7 (or Hb8) (Figure 2).
All three spots of intact hemoglobin subunits (spots 20–22) showed an increase in intensity in gels of hypoxia-acclimated animals. The induction intensity ranged from 9-fold (Hb5) to 19-fold (Hb4), which is comparable to the hypoxia-induced increase (5–21-fold) of Hb concentration in the hemolymph of Daphnia magna [9,10]. The support for the globins Hb3, Hb4, Hb5 and Hb7 suggests that these subunits are dominant in hypoxia-acclimated animals. A similar dominance of only a few subunits was found in hypoxia-acclimated Daphnia magna [10]. However, we cannot fully exclude the presence of other globins, because the migration position of intact chains of these subunits is outside the pH 4–7 gradient range, and the concentration of proteolytic cleavage products with pI values smaller than 7 could be too low to be detectable by protein gel staining or mass-spectrometric analysis.
The mechanism of hypoxic Hb induction involves the transcription factor HIF (hypoxia inducible factor) in Daphnia magna [29]. Similar as in mammalian cells [30], HIF is prevented from degradation under oxygen-poor conditions and binds to enhancing elements present in the intergenic regions of Daphnia's Hb gene cluster[9,28]. The target genes in vertebrates include proteins involved in oxygen homeostasis (EPO, VEGF) as well as key players of carbohydrate metabolism (for reviews, see [31,32]). The latter enzymes are involved in anaerobic metabolism which guarantees ongoing energy provision during oxygen deprivation. Since anaerobiosis is a less effective mode of ATP production, it requires a higher turnover rate of glycosides, which can be guaranteed by an increase in the concentration of glycolytic enzymes.
It is therefore reasonable to assume that glycolytic enzymes experience a comparable induction in animals exposed to environmental hypoxia. However, the present study identified only one element of the HIF-target genes involved in glycolysis, the enolase (ENO), which was only slightly induced (factor 1.4) in hypoxia-acclimated D. pulex. HIF-binding sites (hypoxia responsive elements: HRE) are present upstream of the enolase gene. The motif ACGTGT can be found in cis positions at -173 and -481. At least the first one is within the functional range where HIF-binding affects gene expression, as was documented for hypoxic D. magna hemoglobin induction [29]. The only moderate induction of enolase might be the consequence of the increase in oxygen-transport capacity arising from the strongly elevated Hb concentration. The successful restoration of oxygen homeostasis may reduce the need for adjustments in protein expression. A stronger induction than found here might be observed in animals from acute hypoxic exposure. The adjustment of the oxygen-transport system to environmental hypoxia via Hb induction, however, does not exclude the possibility of an occurrence of hypoxic states within certain cells and tissues. Episodes of higher energy demand, e.g. during enhanced activities, may drive the oxygen-transport system to the limit, thereby increasing the risk of oxygen lack in specific body regions. Moreover, the oxygen supply of cells depends on their size or location. Particularly in large cells (with small surface-to-volume ratio) or cells with a high metabolic rate, the Po2 threshold for the activation of anaerobic metabolism and the stabilization of HIF may be passed more or less frequently. The fat cells, for example, which constitute one major site of Hb synthesis in Daphnia [12], are likely to be the first candidates which suffer from hypoxia. The risk for undersupply with oxygen arises from their large size and their distribution in the body core region, where hemolymph Po2 values are low [18]. So, the difference in the up-regulation of Hb and other HIF target genes may be related to more frequent hypoxic episodes in Hb-synthesizing tissues.
Enolase is known to be one of the most abundantly expressed cytoplasmic proteins [33]. The dimeric magnesium-containing enzyme catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate. Besides its role in glycolysis, it has been characterized as a stress protein involved in hypoxia and thermal tolerance; even a heat-shock protein function has been reported [33]. In D. pulex, the enolase is present in high amounts (spot 19). The slight induction of this enzyme under hypoxia is well in line with its regulation by HIF, its role in anaerobiosis, and its possible function as a stress protein. Its high expression already in normoxia-acclimated animals might be interpreted as a pre-adaptive feature which renders a marked hypoxia response unnecessary.
Proteolytic enzymes
A group of proteolytic enzymes (particularly trypsin; spots 28, 31, 32, 36–41) was identified in large amounts in all 2D gels of D. pulex. Their expression was unaffected by hypoxia acclimation. In D. magna, the largest portion of proteases are trypsin- and chymotrypsin-like enzymes [34], which are endopeptidases characterized by the presence of a serine residue in the active site. More than 98% of the proteolytic activity of D. magna can be found in the gut. In the whole-animal extracts used in the present study, intestinal digestive enzymes are included in the preparation. Daphnia's serine proteases are targets of common inhibitors [34]. Specific inhibition of serine proteases is reported to reduce the total proteolytic activity of Daphnia to 15%, indicating that the residual proteolytic activity may originate from non-serine proteases [34]. Our identifications included indeed other classes of digestive enzymes such as the astacin-like zinc metalloendopeptidase (spot 40) [35,36], the zinc metallopeptidase M13 (spot 28), which is probably a membrane-bound enzyme because of the absence of a signal peptide in the predicted protein sequence, and the secretory zinc metallopeptidases M2 (spot 28), which carries signatures of a dipeptidyl carboxydipeptidase [37]. Strong expression was also observed for the zinc carboxypeptidase A (spot 31), which is secreted as an inactive proenzyme that becomes activated by the cleavage of an N-terminal propeptide [37]. This activating cleavage may explain the discrepancy between the predicted Mr (44.8 kDa for the mature protein with propeptide but without signal peptide) and the measured Mr of 30 kDa. While the protease-inhibitor cocktail used in the present study contained specific inhibitors to block serine proteases and metalloproteases, it seems that the inhibition was incomplete and that not all types of proteases were covered by the chosen inhibitors. Moreover, the extraction of proteins at cold temperatures might not have been as effective as expected. Since daphnids are confronted to large temperature fluctuations in the natural habitat, it is possible that their proteases are adapted to operate over a wide range of temperatures. Irrespective of these methodical aspects, the high representation of proteases in the D. pulex proteome documents an enormous digestive capacity, which probably guarantees an optimal exploitation of food resources to support the high growth and reproduction rates which are characteristic for these animals.
Polysaccharide-degrading enzymes
A set of polysaccharide-degrading enzymes was identified in the 2D gels. The putative enzymatic specificities, which could be assigned by sequence similarity with classified glycosyl hydrolases [38-40], comprise the hydrolytic cleavage of endoglycosidic bonds in α-1,4-glucans (α-amylase, spot 35), β-1,4-glucans (endo-glucanase, CEL9A; spot 29), and β-1,4-mannans (endo-mannanase, MAN5A; spot 30) as well as the exoglycosidic cleavage of β-1,4-glucans (cellubiohydrolase, CEL7A; spot 1) and β-1,3-glucans (exoglucanase, EXG5; spot 34). These different glycosidic bonds are characteristic of storage polysaccharides (starch: α-1,4-linked glucan) and structure polysaccharides (cellulose: β-1,4-glucans; hemicellulose: β-1,4-mannans and others) of plants including nanoplanktonic green algae, the typical food of daphnids [41]. β-1,3-glucans are structural components in the cell wall of fungi and algae. These functional assignments, the high degree of expression (Figure 1), and the presence of an N-terminal signal peptide (Table 3) strongly suggest that these candidate proteins are secretory digestive enzymes involved in the degradation of storage and structural polysaccharides.
The origin of cellulase activity in multicellular animals was formerly assigned to symbiotic microorganisms living in the host's gastrointestinal tract (see [42] for a review). The discrimination of cellulolytic enzymes from symbionts and their hosts by functional analyses is still a difficult task [43,44], but there are many indications for an endogenous (i.e. non-symbiontic) cellulolytic activity in metazoans including crustaceans [45-48]. Molecular biology techniques provided unequivocal support for the presence of cellulase genes in various metazoan lineages such as arthropods (crustaceans and insects), annelids, ascidian chordates, echinoderms and molluscs [42,49].
Experimental support for a cellulolytic activity in daphnids was first provided by [43]. Toxicological studies in D. magna showed an inhibition of amylase/cellulase activities by cadmium and mercury as well as an activity increase upon chromium exposure [50]. Reduced activities of both enzymes were found under ultraviolet radiation [51]. Microarray studies [52] revealed an up-regulated expression of cellulase and amylase genes under cadmium stress. In the present study, the acclimation of D. pulex to hypoxic conditions was associated with a strong increase in cellubiohydrolase expression (spot1) and a moderate decrease in α-amylase (spot 35) and exo-β-1,3-glucanase (spot 34) expression. The presence of six glycosyl hydrolases among the spots of major intensity shows a large capacity for carbohydrate digestion, which obviously adapts D. pulex to hypoxic conditions. As suggested for protein digestion, the high potential for carbohydrate degradation may reflect a high turnover of nutrients for the animals' fast growth and reproduction rates. This suggestion might be in conflict with the 20% reduction in the oxygen-consumption rate of hypoxia-acclimated D. magna compared to normoxia-acclimated animals [53]. However, reduction in oxygen uptake does not necessarily imply a reduced need for digestive processes. In case of anaerobic energy production, the metabolic flux rate through the glycolytic pathway has to be increased due to the lower ATP yield of anaerobic glycolysis, which leads to an enhanced demand for carbohydrates. Concerning the regulation of the whole set of carbohydrate-degrading enzymes, the complex pattern of adaptive gene control certainly needs further investigation.
Conclusion
Adjustments of protein expression due to hypoxia acclimation in Daphnia pulex include a strong induction of Hb to adapt the oxygen-transport system to an oxygen-depleted environment. Other HIF target genes such as that for enolase, which is involved in anaerobic metabolism, are induced to a lower extent. This may reflect, on the one hand, the general restoration of oxygen-transport capacity by Hb induction and, on the other hand, tissue-specific variations in cellular oxygen supply with more frequent episodes of tissue hypoxia, especially in the body core region. In contrast to carbohydrate-degrading enzymes, the set of proteolytic enzymes does not respond to hypoxia. Independent of ambient oxygen conditions, the animals maintain a high level of proteolytic power, which is probably related to the high energy demands for activity, growth and reproduction. Among the complex pattern of adaptive gene control for carbohydrate hydrolysis, the enhanced need for carbohydrates during periods of anaerobiosis is probably related to the strong hypoxic induction of cellubiohydrolase, which may serve for a degradation of structural polysaccharides.
Methods
Acclimation conditions
Water fleas, Daphnia pulex, were originally obtained from a flooded eutrophic quarry at Gräfenhain (near Dresden, Germany) [54] and have been kept in the laboratory since 2002. The animals were cultured in 1.5L M4 medium [55] in 2–3L preserving jars under a 16 h:8 h L:D photoperiod as previously described [53]. The animals were acclimated at least for three weeks (mostly months) to normoxia (100% air saturation; oxygen partial pressure, Po2: 20 kPa) or hypoxia (15% air saturation; Po2: 3 kPa) at 20°C. Normoxic medium was obtained by mild aeration using an aquarium pump, whereas hypoxic conditions were established by reducing the atmospheric pressure in the residual air space of the closed preserving jar to 15% of standard atmospheric pressure using a vacuum pump (PC 511, Vacuubrand, Wertheim, Germany). Animals were fed with green algae (Desmodesmus subspicatus) ad libitum (> 1 mg C L-1) every second day. Three-quarter of the medium was renewed once weekly. Any males and ephippial females were sorted out to maintain parthenogenetic reproduction.
Protein extraction
Total (soluble) proteins were extracted from shock-frozen D. pulex (150–200 mg fresh weight per biological replicate). Before freezing, the animals had not been fed with algae for 12 h. The biological material was mixed 1:3 (w/v) with a freshly prepared rehydration solution containing 8 M urea, 2 M thiourea, 4% (w/v) CHAPS, 65 mM DTT, 0.5% (v/v) ampholyte-containing IPG buffer pH 4–7 (GE Healthcare, Munich, Germany), and a protease-inhibitor cocktail (Complete Mini, Roche, Mannheim, Germany) (one tablet per 10 mL solution). The biological material was disrupted using a tissue grinder (Pellet Pestle; Kimble/Kontes, Vineland, NJ, USA) for 1 min on ice, and the insoluble fraction was then removed by centrifugation at 17900 × g for 15 min at 4°C. The supernatant containing the soluble protein fraction was then subjected to ultrafiltration (17900 × g for 45 min at 4°C) using centrifugal filter devices with a molecular mass cut-off of 300 kDa (Microcon YM-300, Millipore, Schwalbach, Germany). The proteins in the lower-molecular-weight filtrate were precipitated with 13% TCA, incubated on ice for 70 min, and then centrifuged at 17900 × g for 15 min at 4°C. The protein pellet was repeatedly washed with ice-cold 80% acetone and centrifuged (17900 × g for 5 min at 4°C) ten times, and then resuspended in 200 μl rehydration solution. Protein quantification was performed using the Bradford assay [56].
Two-dimensional gel electrophoresis
Isoelectric focussing (IEF) was performed with 142 μg of protein extract diluted in 350 μl rehydration solution using 18-cm linear pH 4–7 IPG gradients (GE Healthcare) and the Ettan IPGphor II isoelectric focusing unit (Amersham Biosciences, Uppsala, Sweden). Rehydration of the IPG strips was performed at 50 V for 11 h at 20°C. The voltage settings of the IEF comprised a 50–100 V gradient for 1 min, 100 V for 2 h, 100–1000 V gradient for 10 min, 1000 V for 30 min, 1000–4000 V gradient for 1 h, 4000 V for 30 min, 4000–8000 V gradient for 45 min, 8000 V for 4.5 h, to a final setting of approximately 46000 Vh. After IEF, the strips were equilibrated for 15 min in equilibration solution (0.05 M Tris, 6 M urea, 30% glycerol, 2% SDS, pH 8.8) containing 65 mM DTT followed by 15 min in equilibration solution containing 135 mM iodoacetamide to block free thiol groups. For the second dimension, protein separation on the basis of the molecular mass was performed using 12% polyacrylamide gels (0.56 M Tris, 0.1% SDS, pH 8.8; 20 × 18 × 0.1 cm3) and the Protean II xi Cell apparatus (Bio-Rad Laboratories, Munich, Germany). The PageRulerTM Protein Ladder (Fermentas, Burlington, Canada) covering a molecular mass range from 10 kDa to 200 kDa was used for molecular mass calibration. Electrophoresis was performed at 15 mA per gel for 18–21 h. After electrophoresis, gels were stained with SYPRO Ruby protein gel stain (Bio-Rad) according to the manufacturer's instructions. Stained gels were scanned with a Typhoon 9400 fluorescence imager (GE Healthcare) and analyzed with Delta2D software, version 3.5 (DECODON, Greifswald, Germany) [57]. Gels were warped manually using the exact warp mode prior to spot detection and editing.
Statistical analysis of protein expression
Protein expression was quantified by translating the normalized intensity of candidate spots in 2D gels into relative spot volumes. Statistical differences in protein expression between the two acclimation groups were assessed by t-tests after differences in variance had been checked by F-tests.
nano-HPLC-ESI-MS/MS
Spots of sufficient size and staining intensity (relative spot volume, Vrel > 0.1%) were chosen for subsequent mass-spectrometric analyses if they were identified as differentially expressed between normoxia-acclimated and hypoxia-acclimated animals. Some spots of high but constitutive expression were also excised from representative gels. They were subjected to in-gel digestion using trypsin (sequencing grade, Promega, Mannheim, Germany) overnight at 37°C. Reversed-phase nano-LC-MS/MS was performed using an Ultimate nanoflow LC system (Dionex LC Packings, Idstein, Germany) containing the components Famos (autosampler), Switchos (loading pump and switching valves), and Ultimate (separation pump and UV-detector). The LC system was coupled to a QSTAR Pulsar i hybrid QqTOF mass spectrometer (Applied Biosystems/MDSSciex, Darmstadt, Germany), equipped with a nanoelectro-spray ion source (Column Adapter [ADPC-PRO] and distal coated SilicaTips [FS360-20-10-D-20], both from New Objective, Woburn, USA). Briefly, the tryptic peptide mixtures were autosampled at a flow rate of 30 μl/min in 0.1% aqueous trifluoroacetic acid, and desalted on a PepMap C18 trapping cartridge (LC Packings). The trapped peptides were eluted and separated on the analytical column (PepMap C18, 75 μm i.d. × 15 cm; LC Packings) using a linear gradient of 7–50% solvent B (acetonitrile 84% [v/v] in 0.1% [v/v] formic acid) for 27 min at a flow rate of 220 nl/min, and ionized by an applied voltage of 2200 kV to the emitter. The mass spectrometer was operated in the data-dependent acquisition mode to automatically switch between MS and MS/MS. Survey MS spectra were acquired for 1.5 s, and the three most intense ions (doubly or triply charged) were isolated and sequentially fragmented for 1.5 s by low-energy collision-induced dissociation. All MS and MS/MS spectra were acquired with the Q2-pulsing function switched on, and optimized for enhanced transmission of ions in the MS (m/z 400–1000) and MS/MS (m/z 75–1300) mass ranges. All results from 2-dimensional electrophoresis and mass spectrometry as well as all search results where stored in a LIMS-database (Proteinscape 1.3, Bruker Daltonics, Bremen, Germany).
Identification and characterization of proteins
Proteins were identified by correlating the ESI-MS/MS spectra with the "Frozen Gene Catalog" of the D. pulex protein database [26] using the MOWSE-algorithm as implemented in the MS search engine (Matrix Science Ltd., London, UK) [27]. The "Frozen Gene Catalog" contains all manual curations as of July 3, 2007 as well as automatically annotated models chosen from the "Filtered Models" v1.1 set. "Filtered Models" is the filtered set of models representing the best gene model for each locus. The putative function of identified proteins was inferred by sequence homology either from the automated blastp search provided by Joint Genome Institute [26] or after manual curation of gene models. Derived protein sequences were checked for the presence of N-terminal signal sequences [58,59]. The theoretical molecular weight (Mr) and isolectric point (pI) of mature proteins (without N-terminal signal peptide) was calculated using the ExPASy proteomics tool "Compute pI/MW" [60-62].
Abbreviations
EPO: erythropoetin; Mr: molecular weight; pI: isolectric point; Po2: oxygen partial pressure; VGEF: vascular endothelial growth factor.
Authors' contributions
SS and MK were involved in the culturing of animals and performed the protein extraction as well as the 2D-PAGE. 2D-gel image analysis was carried out by TL, WS, BZ and RP. JM and CF were responsible for mass spectrometry and protein identification. RP and SS retrieved the information contained in the Tables. Figures were designed by RP. The annotation of identified genes was performed by FN and RP. BZ, RJP, and RP conceived and coordinated the study, and prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors would like to thank Simone König from the Integrated Functional Genomics (University of Münster), and Marco Matthes (at that time at the Technical University Dresden) for providing the animals used for the clonal culture. We are grateful for support from DECODON GmbH (BioTechnikum Greifswald). The Proteom Centrum Tübingen is supported by the Ministerium für Wissenschaft und Kunst, Landesregierung Baden-Württemberg.
The sequencing and portions of the analyses were performed at the DOE Joint Genome Institute under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231, Los Alamos National Laboratory under Contract No. W-7405-ENG-36 and in collaboration with the Daphnia Genomics Consortium, DGC [63]. Additional analyses were performed by wFleaBase, developed at the Genome Informatics Lab of Indiana University with support to Don Gilbert from the National Science Foundation and the National Institutes of Health. Coordination infrastructure for the DGC is provided by The Center for Genomics and Bioinformatics at Indiana University, which is supported in part by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. Our work benefits from, and contributes to the Daphnia Genomics Consortium.
Contributor Information
Bettina Zeis, Email: zeis@uni-muenster.de.
Tobias Lamkemeyer, Email: tobias.lamkemeyer@uni-tuebingen.de.
Rüdiger J Paul, Email: paulr@uni-muenster.de.
Frank Nunes, Email: nunes@uni-muenster.de.
Susanne Schwerin, Email: susanne.schwerin@freenet.de.
Marita Koch, Email: marita@uni-muenster.de.
Wolfgang Schütz, Email: wolfgang.schuetz@uni-tuebingen.de.
Johannes Madlung, Email: johannes.madlung@uni-tuebingen.de.
Claudia Fladerer, Email: claudia.fladerer@uni-tuebingen.de.
Ralph Pirow, Email: pirow@uni-muenster.de.
References
- Hochachka PW, Somero GN. Biochemical adaptation: Mechanism and process in physiological evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Fox HM, Hardcastle SM, Dresel EIB. Fluctuations in the haemoglobin content of Daphnia. Proc R Soc Lond B Biol Sci. 1949;136:388–399. doi: 10.1098/rspb.1949.0032. [DOI] [PubMed] [Google Scholar]
- Fox HM, Phear EA. Factors influencing the haemoglobin synthesis by Daphnia. Proc R Soc Lond B Biol Sci. 1953;141:179–189. doi: 10.1098/rspb.1953.0034. [DOI] [PubMed] [Google Scholar]
- Chandler A. Causes of variation in the haemoglobin content of Daphnia (Crustacea: Cladocera) Proc R Soc Lond B Biol Sci. 1954;124:625–630. [Google Scholar]
- Kobayashi M, Hoshi T. Relationship between the haemoglobin concentration of Daphnia magna and the ambient oxygen concentration. Comp Biochem Physiol A Physiol. 1982;72:247–249. doi: 10.1016/0300-9629(82)90040-8. [DOI] [Google Scholar]
- Paul RJ, Zeis B, Lamkemeyer T, Seidl M, Pirow R. Control of oxygen transport in the microcrustacean Daphnia: regulation of haemoglobin expression as central mechanism of adaptation to different oxygen and temperature conditions. Acta Physiol Scand. 2004;182:259–275. doi: 10.1111/j.1365-201X.2004.01362.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Fujiki M, Suzuki T. Variation in and oxygen-binding properties of Daphnia magna hemoglobin. Physiol Zool. 1988;61:415–419. [Google Scholar]
- Kobayashi M, Ishigaki K-I, Kobayashi M, Igarashi Y, Imai K. Oxygen transport efficiency of multiple-component hemoglobin in Daphnia magna. Can J Zool. 1994;72:2169–2171. doi: 10.1139/z94-289. [DOI] [Google Scholar]
- Kimura S, Tokishita S, Ohta T, Kobayashi M, Yamagata H. Heterogeneity and differential expression under hypoxia of two-domain hemoglobin chains in the water flea, Daphnia magna. J Biol Chem. 1999;274:10649–10653. doi: 10.1074/jbc.274.15.10649. [DOI] [PubMed] [Google Scholar]
- Zeis B, Becher B, Goldmann T, Clark R, Vollmer E, Bölke B, Bredebusch I, Lamkemeyer T, Pinkhaus O, Pirow R, Paul RJ. Differential haemoglobin gene expression in the crustacean Daphnia magna exposed to different oxygen partial pressures. Biol Chem. 2003;384:1133–1145. doi: 10.1515/BC.2003.126. [DOI] [PubMed] [Google Scholar]
- Lamkemeyer T, Zeis B, Paul RJ. Temperature acclimation influences temperature-related behaviour as well as oxygen-transport physiology and biochemistry in the water flea Daphnia magna. Can J Zool. 2003;81:237–249. doi: 10.1139/z03-001. [DOI] [Google Scholar]
- Goldmann T, Becher B, Wiedorn KH, Pirow R, Deutschbein ME, Vollmer E, Paul RJ. Epipodite and fat cells as sites of hemoglobin synthesis in the branchiopod crustacean Daphnia magna. Histochem Cell Biol. 1999;112:335–339. doi: 10.1007/pl00007905. [DOI] [PubMed] [Google Scholar]
- Weber RE, Vinogradov SN. Nonvertebrate hemoglobins: Functions and molecular adaptations. Physiol Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- Lamkemeyer T, Paul RJ, Stöcker W, Yiallouros I, Zeis B. Macromolecular isoforms of Daphnia magna haemoglobin. Biol Chem. 2005;386:1087–1096. doi: 10.1515/BC.2005.125. [DOI] [PubMed] [Google Scholar]
- Lamkemeyer T, Zeis B, Decker H, Jaenicke E, Waschbüsch D, Gebauer WMJ, Meissner U, Rousselot M, Zal F, Nicholson GJ, Paul RJ. Molecular mass of macromolecules and subunits and the quarternary structure of hemoglobins from the microcrustacean Daphnia magna. FEBS J. 2006;273:3393–3410. doi: 10.1111/j.1742-4658.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamagata H. Structure and function of hemoglobin in Daphnia magna. Trends Comp Biochem Physiol. 2000;6:163–174. [Google Scholar]
- Pirow R, Bäumer C, Paul RJ. Benefits of haemoglobin in the cladoceran crustacean Daphnia magna. J Exp Biol. 2001;204:3425–3441. doi: 10.1242/jeb.204.20.3425. [DOI] [PubMed] [Google Scholar]
- Pirow R, Bäumer C, Paul RJ. Crater landscape: two-dimensional oxygen gradients in the circulatory system of the microcrustacean Daphnia magna. J Exp Biol. 2004;207:4393–4405. doi: 10.1242/jeb.01318. [DOI] [PubMed] [Google Scholar]
- Kring RL, O'Brien WJ. Effect of varying oxygen concentrations on the filtering rate of Daphnia pulex. Ecology. 1976;57:808–814. doi: 10.2307/1936194. [DOI] [Google Scholar]
- Sell AF. Adaptation to oxygen deficiency: Contrasting patterns of haemoglobin synthesis in two coexisting Daphnia species. Comp Biochem Physiol A Mol Integr Physiol. 1998;120:119–125. doi: 10.1016/S1095-6433(98)10019-3. [DOI] [Google Scholar]
- Dewilde S, Van Hauwaert ML, Peeters K, Vanfleteren J, Moens L. Daphnia pulex didomain hemoglobin: Structure and evolution of polymeric hemoglobins and their coding genes. Mol Biol Evol. 1999;16:1208–1218. doi: 10.1093/oxfordjournals.molbev.a026211. [DOI] [PubMed] [Google Scholar]
- Dangott LJ, Terwilliger RC. The subunit structure of Daphnia pulex hemoglobin. Comp Biochem Physiol B Biochem Mol Biol. 1980;67:301–306. doi: 10.1016/0305-0491(80)90148-0. [DOI] [Google Scholar]
- Wolf GH, Smet J, Decleir W. Oxygen binding properties of hemoglobins from Daphnia pulex (De Geer) Comp Biochem Physiol A Physiol. 1983;75:261–265. doi: 10.1016/0300-9629(83)90080-4. [DOI] [Google Scholar]
- Peeters K, Mertens J, Hebert P, Moens L. The globin composition of Daphnia pulex hemoglobin and the comparison of the amino acid composition of invertebrate hemoglobins. Comp Biochem Physiol B Biochem Mol Biol. 1990;97:369–381. doi: 10.1016/0305-0491(90)90295-5. [DOI] [Google Scholar]
- wFleaBase http://wFleaBase.org
- Joint Genome Institute http://www.jgi.doe.gov/Daphnia/
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence data bases using mass spectrometric data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Tokishita S, Ohta T, Yamagishi H. Genomic organization and expression of Daphnia hemoglobin genes. BMC Biology. 2008.
- Gorr TA, Cahn JD, Yamagata H, Bunn HF. Hypoxia-induced synthesis of hemoglobin in the crustacean Daphnia magna is hypoxia-inducible factor-dependent. J Biol Chem. 2004;279:36038–36047. doi: 10.1074/jbc.M403981200. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Ann Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB Journal. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Elert E, Agrawal MK, Gebauer C, Jaensch H, Bauer U, Zitt A. Protease activity in gut of Daphnia magna: evidence for trypsin and chymotrypsin enzymes. Comp Biochem Physiol B Biochem Mol Biol. 2004;137:287–296. doi: 10.1016/j.cbpc.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Stöcker W, Gomis-Rüth F-X, Bode W, Zwilling R. Implications of the three-dimensional structure of astacin for the structure and function of the astacin family of zinc-endopeptidases. Eur J Biochem. 1993;214:215–231. doi: 10.1111/j.1432-1033.1993.tb17915.x. [DOI] [PubMed] [Google Scholar]
- Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Protein Sci. 1995;4:1247–1261. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolutionary families of metalloproteases. Methods Enzymol. 1995;248:138–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1999;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- CAZy database http://www.cazy.org/
- Lampert W. Feeding and nutrition in Daphnia. In: Peters RH, DeBernardi R, editor. Memorie dell'Istituto Italiano di Idrobiologia, Daphnia. Vol. 45. Pallanza: Istituto Italiano di Idrobiologia; 1987. pp. 143–192. [Google Scholar]
- Watanabe H, Tokuda G. Animal cellulases. Cell Mol Life Sci. 2001;58:1167–1178. doi: 10.1007/PL00000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg SA, Maccubbin AE, Hodson RE. Cellulose digestion by freshwater microcrustacea. Limnol Oceanogr. 1984;29:1132–1136. [Google Scholar]
- Zimmer M, Bartholmé S. Bacterial endosymbionts in Asellus aquaticus (Isopoda) and Gammarus pulex (Amphipoda) and their contribution to digestion. Limnol Oceanogr. 2003;48:2208–2213. [Google Scholar]
- Monk DC. Digestion of cellulose and other dietary components, and pH of gut in amphipod Gammarus pulex L. Freshwater Biol. 1977;7:431–440. doi: 10.1111/j.1365-2427.1977.tb01692.x. [DOI] [Google Scholar]
- McGrath CC, Matthews RA. Cellulase activity in the freshwater amphipod Gammarus lacustris. J North Amer Benthol Soc. 2000;19:298–307. doi: 10.2307/1468072. [DOI] [Google Scholar]
- Crawford AC, Richardson NR, Mather PB. A comparative study of cellulase and xylanase activity in freshwater crayfish and marine prawns. Aquacult Res. 2005;36:586–592. doi: 10.1111/j.1365-2109.2005.01259.x. [DOI] [Google Scholar]
- Linton SM, Greenaway P. A review of feeding and nutrition of herbivorous land crabs: adaptations to low quality plant diets. J Comp Physiol B. 2007;177:269–286. doi: 10.1007/s00360-006-0138-z. [DOI] [PubMed] [Google Scholar]
- Davison A, Blaxter M. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol Biol Evol. 2005;22:1273–1284. doi: 10.1093/molbev/msi107. [DOI] [PubMed] [Google Scholar]
- De Coen WM, Janssen CR. The use of biomarkers in Daphnia magna toxicity testing. II. Digestive enzyme activity in Daphnia magna exposed to sublethal concentrations of cadmium, chromium and mercury. Chemosphere. 1997;35:1053–1067. doi: 10.1016/S0045-6535(97)00172-0. [DOI] [Google Scholar]
- Zellmer ID, Arts MT, Šustr V. Food chain effects of sublethal ultraviolett radiation on subarctic Daphnia pulex – a field and laboratory study. Arch Hydrobiol. 2006;167:515–531. doi: 10.1127/0003-9136/2006/0167-0515. [DOI] [Google Scholar]
- Soetaert A, Vandenbrouck T, Ven K van der, Maras M, van Remortel P, Blust R, De Coen WM. Molecular responses during cadmium-induced stress in Daphnia magna: Integration of differential gene expression with higher-level effects. Aquat Toxicol. 2007;83:212–222. doi: 10.1016/j.aquatox.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Seidl MD, Paul RJ, Pirow R. Effects of hypoxia acclimation on morpho-physiological traits over three generations of Daphnia magna. J Exp Biol. 2005;208:2165–2175. doi: 10.1242/jeb.01614. [DOI] [PubMed] [Google Scholar]
- Matthes M. Low genotypic diversity in a Daphnia pulex population in a biomanipulated lake: the lack of vertical and seasonal variability. Hydrobiologia. 2004;526:33–42. doi: 10.1023/B:HYDR.0000041616.86446.01. [DOI] [Google Scholar]
- Elendt BP, Bias WR. Trace nutrient deficiency in Daphnia magna cultured in standard medium for toxicity testing. Effects of the optimization of culture conditions on life history parameters of D. magna. Water Res. 1990;24:1157–1167. doi: 10.1016/0043-1354(90)90180-E. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Berth M, Moser FM, Kolbe M, Bernhardt J. The state of the art in the analysis of two-dimensional gel electrophoretic images. Appl Microbiol Biotechnol. 2007;76:1223–1243. doi: 10.1007/s00253-007-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- SignalP 3.0 http://www.cbs.dtu.dk/services/SignalP/
- Compute pI/Mw tool http://www.expasy.ch/tools/pi_tool.html
- Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu