Abstract
The fundamental role of the major histocompatibility complex (MHC) in immune recognition has led to a general consensus that the characteristically high levels of functional polymorphism at MHC genes is maintained by balancing selection operating through host–parasite coevolution. However, the actual mechanism by which selection operates is unclear. Two hypotheses have been proposed: overdominance (or heterozygote superiority) and negative frequency-dependent selection. Evidence for these hypotheses was evaluated by examining MHC–parasite relationships in an island population of water voles (Arvicola terrestris). Generalized linear mixed models were used to examine whether individual variation at an MHC class II DRB locus explained variation in the individual burdens of five different parasites. MHC genotype explained a significant amount of variation in the burden of gamasid mites, fleas (Megabothris walkeri) and nymphs of sheep ticks (Ixodes ricinus). Additionally, MHC heterozygotes were simultaneously co-infected by fewer parasite types than homozygotes. In each case where an MHC-dependent effect on parasite burden was resolved, the heterozygote genotype was associated with fewer parasites, and the heterozygote outperformed each homozygote in two of three cases, suggesting an overall superiority against parasitism for MHC heterozygote genotypes. This is the first demonstration of MHC heterozygote superiority against multiple parasites in a natural population, a mechanism that could help maintain high levels of functional MHC genetic diversity in natural populations.
Keywords: major histocompatibility complex, parasites, overdominance, natural selection, adaptive polymorphism, individual fitness
1. Introduction
The genes of the major histocompatibility complex (MHC) are central to the vertebrate immune system, encoding proteins geared towards the recognition and presentation of foreign antigens to initiate a host immune response (Klein 1986). In the majority of species where comparisons have been made, MHC loci display considerably greater levels of nucleotide diversity and heterozygosity than the genomic average (Gaudieri et al. 2000; Garrigan & Hedrick 2003; Robinson et al. 2003). This is perhaps intuitively expected, given that more functional diversity at the MHC means more antigenic epitopes can be recognized and thus more immune insults combated. It is generally accepted that such high levels of MHC diversity are maintained by balancing selection acting through antagonistic host–parasite coevolution. However, what is less clear is exactly how parasite-mediated balancing selection operates. Two predominant modes have been proposed: (i) overdominance (Doherty & Zinkernagel 1975), whereby MHC heterozygote individuals are able to recognize more parasite-derived peptides and therefore enjoy reduced parasitism, resulting in even MHC allele frequencies; and (ii) frequency-dependent selection (Takahata & Nei 1990; Slade & McCallum 1992), whereby cyclic relationships between susceptible/resistant MHC alleles and specific parasites maintain MHC alleles in flux. Testing which mechanism predominates to explain MHC variation in natural populations has proven problematic for several reasons. First, the two hypotheses are not mutually exclusive, in that there is a frequency-dependent component to overdominance, given that rare alleles occur disproportionately in heterozygous genotypes (Apanius et al. 1997), and some models of frequency dependence and overdominance predict the same allele frequency distributions, making the two empirically intractable (Denniston & Crow 1990; Takahata & Nei 1990). Second, the time scales that may be required to capture the dynamics between alleles and parasites under frequency-dependent coevolution may be considerable and well beyond the scope of most studies (Apanius et al. 1997). Third, the overdominance hypothesis can only be properly tested in the context of the effects of multiple parasites, because the advantages of being able to bind multiple parasite epitopes may only be realized when multiple parasite-mediated immune insults are prevalent (Hughes & Nei 1992; McClelland et al. 2003b). Additionally, the efficiency of overdominance in maintaining MHC diversity depends on the equivalence of fitness and overlap in peptide-binding specificity between alleles (Robertson 1962; Richman et al. 2001). Moreover, the fitness of a heterozygote is frequently greater than the average of two homozygotes, but not significantly greater than the most-fit homozygote, as fitness is a consequence of the presence of a dominant resistant allele that both the heterozygote and homozygote genotypes carry. In this case, there is a heterozygote advantage in the broadest sense, but this is not synonymous with overdominance, where the fitness of the heterozygote is superior to either homozygote. This is important because the two processes are predicted to be considerably different in their efficacy for retaining diversity in natural populations, with only heterozygote superiority able to maintain the balanced allele frequencies observed at MHC loci (Takahata & Nei 1990; Penn et al. 2002; McClelland et al. 2003b). For clarity, we will adopt the terminology of Penn et al. (2002) and use ‘heterozygote advantage’ in the broad sense to describe heterozygote fitness that is on average higher than homozygote fitness, which may be attributable to dominant or overdominant effects, and ‘heterozygote superiority’ to describe heterozygote fitness that is higher than either homozygote and strictly synonymous with overdominance.
Some disease studies in humans and experimental infections of captive-bred organisms provide support for heterozygote advantage (Thursz et al. 1997; Carrington et al. 1999; Arkush et al. 2002; Penn et al. 2002; McClelland et al. 2003a), whereas others find no evidence of heterozygote advantage, but rather show resistance that is attributable to single alleles and therefore more consistent with frequency-dependent selection (Hill et al. 1991, 1992; Thursz et al. 1995; Hohler et al. 1997; Grimholt et al. 2003; McClelland et al. 2003a; Wu et al. 2004). In some cases, there is evidence that heterozygotes actually have reduced fitness relative to homozygotes (Wedekind et al. 2005; Ilmonen et al. 2007). However, a potential criticism of laboratory tests of MHC and parasite associations is that some relationships may be context-dependent and therefore only emerge under particular environmental conditions, which are hard to reproduce artificially (Bernatchez & Landry 2003; Sommer 2005; Piertney & Oliver 2006).
The majority of studies of MHC–parasite associations in natural populations have reported associations between specific MHC alleles and parasites (Paterson et al. 1998; Froeschke & Sommer 2005; Harf & Sommer 2005; Meyer-Lucht & Sommer 2005; Schad et al. 2005; Loiseau et al. 2008; Tollenaere et al. 2008), with heterozygote advantage only rarely supported (Froeschke & Sommer 2005). The association between specific alleles and parasite species is often taken as a support for the frequency-dependence hypothesis, particularly when rare MHC alleles are associated with increased resistance and common alleles with greater susceptibility (Paterson et al. 1998; Froeschke & Sommer 2005; Schad et al. 2005). However, if a wide variety of parasite taxa were to be considered simultaneously, it may emerge that MHC heterozygotes have a lower cumulative parasite load than homozygotes in such systems. Indeed, it is intuitive to expect that this would be the case (McClelland et al. 2003b). To date, the studies of individual MHC and parasite loads in natural populations, particularly for mammals, have mostly focused on helminths (Paterson et al. 1998; Froeschke & Sommer 2005; Harf & Sommer 2005; Meyer-Lucht & Sommer 2005; Schad et al. 2005). Extending the scope of studies across a broader range of parasite taxa would enhance our understanding of MHC–parasite dynamics in natural populations.
While the studies that examine MHC–parasite associations in natural populations do so in a more ecologically meaningful context, they face a number of impediments that are not common to laboratory studies. The main impediment is that associations between the MHC and parasite burdens in wild animals are likely to be confounded by a number of additional environmental and ecological factors (such as seasonality and host–parasite population dynamics) that could obscure the detection of MHC effects. One way to overcome this is to employ linear models that allow the inclusion of multiple explanatory variables, thereby incorporating other individual (e.g. sex), population (e.g. host density) and environmental (e.g. season) effects (Paterson et al. 1998). A hindrance to this approach, however, is that studies tend to resolve a relatively high number of MHC alleles from a given sample of individuals, the result of which is severe over-parametrization of models that attempt to incorporate all MHC alleles/genotypes as terms in a single model (Apanius et al. 1997; de Eyto et al. 2007). Alternatively, if each allele or genotype is modelled separately, then the loss of statistical power from low sample sizes becomes a problem for rare alleles, and corrections must be made for multiple tests.
It is clear that natural populations with relatively low numbers of alleles at MHC genes offer great potential for identifying the behavioural and ecological processes underlying MHC variation, as they suffer to a lesser extent from the analytical complications that arise from high allelic diversity (Richardson et al. 2005). Here, we analyse the associations between variation in individual parasite burden for multiple parasite taxa and MHC genotype in a population of water voles (Arvicola terrestris) on a small island on the west coast of Scotland. This population is an ideal natural system to undertake the analyses of host MHC–parasite relationships. The population displays limited MHC diversity (two alleles and thus three possible genotypes; Oliver & Piertney 2006), therefore making the association of MHC diversity amenable to statistical analyses. These alleles vary at 9 out of 17 (53%) of the amino acids involved in peptide binding (antigen recognition sites), and are therefore likely to present very different targets for selection (Oliver & Piertney 2006). Additionally, as an island, this population is effectively isolated and analyses can be undertaken without a need to incorporate the complexity of wide-scale spatial effects such as population structure and immigration. In this study, we construct separate models to investigate the relationship between MHC polymorphism and the prevalence or abundance of five types of water vole parasites, as well as co-infection status (i.e. the number of different parasite types simultaneously infecting an individual), while controlling for the effect of other potentially confounding variables. The parasites considered were microparasites of the bacterial genus Bartonella, and the following ectoparasites: (i) ticks (Ixodes ricinus), (ii) fleas (Megabothris walkeri and Ctenopthalmus nobilis) and (iii) mites of the genus Gamasidae. Characterization of the vertebrate immune response to extracellular parasites (including arthropods; Wikel 1996b), together with numerous examples of an acquired immune response to arthropod (and other) parasites in vertebrate hosts (Trager 1939; Allen & Kemp 1982; Wikel 1996a; Proctor & Owens 2000), implies a central role for MHC class II molecules in ectoparasite resistance. Individuals with functionally divergent MHC genotypes are expected to differ in their abilities to recognize particular parasite epitopes and could therefore be predicted to have different associated levels of parasitism. Here, the null hypothesis is that there is no difference in the level of parasitism associated with each MHC genotype. The null hypothesis will be evaluated against three alternative hypotheses: (i) heterozygote superiority (or overdominance), whereby the heterozygous MHC genotype has lower associated parasitism than both homozygous genotypes; (ii) non-additive single allele effects (or dominance), whereby MHC genotypes containing a specific allele have lower levels of parasitism than those without; and (iii) additive single allele effects (or co-dominance), whereby the single allele effect is quantitative, i.e. the level of resistance associated with the heterozygous genotype lies halfway between the two homozygous genotypes.
2. Material and methods
(a) Study site and sample collection
Arvicola terrestris is a large microtine rodent, weighing between 100 and 300 g, that is present across the Western Palaearctic. The focal population here is from Coiresa (56° 08′ N, 5°37′ W), a very small island (approx. 2 ha) in the Sound of Jura on the west coast of Scotland. The Coiresa population was surveyed intensively and sampled on six occasions (every May and October) between 2004 and 2006. A targeted trapping grid was established, consisting of four transects of 25–30 Elliot live traps baited with carrots and potatoes. These were positioned close to the signs of water vole activity and checked over three to four nights. Trap density was relatively constant throughout the suitable areas of habitat.
For each captured individual, its sex, reproductive status and weight were recorded and two ear punch samples taken and stored in 90 per cent ethanol. Males with abdominal testes were regarded as reproductively inactive, while those with descended or semi-descended testes were regarded as reproductively active. Females with medium or large nipples and/or vaginal perforation were considered as reproductively active, whereas those with small nipples and no vaginal perforation were regarded as reproductively inactive. Parasite loads were estimated for all individuals. Fleas were removed by combing the vole over water, which was then filtered and the filter paper sealed in a zip-lock bag to obtain subsequent accurate counts of numbers. Flea species were identified according to Smit (1976) by the examination of the genal comb. Total counts for each flea species were obtained per vole. The relationship between the number of fleas on individual hosts and the number of fleas in nests and burrows has been the subject of a meta-analysis by Krasnov et al. (2004). The two measurements were found to be highly correlated (r=0.83, p<0.0001) across a broad range of species (including A. terrestris and Megabothris). Gamasid mites (Gamasidae) were collected by combing the fur of individual voles as described above. The voles were thoroughly examined for ticks (I. ricinus), which were removed using forceps and placed directly into 2 ml screw cap Eppendorf tubes containing 100 per cent ethanol. The majority of ticks were either larvae or nymphs; these can be easily differentiated as larvae have six legs and nymphs have eight, and thus counts were made for each. The behaviour and dynamics of these two life stages may not be synchronized and they have also been shown to respond differently to acquired host immunity (Trager 1939). As such, larvae and nymphs were analysed separately.
Bartonella species are flea-transmitted haemoparasites that infect red blood cells. In May 2004 (n=69) and October 2004 (n=114), a 20–60 μl blood sample was taken from the tail tip of individual voles. Total DNA extractions were prepared by alkaline digestion (Bown et al. 2003). Bartonella DNA was detected using the genus-specific polymerase chain reaction (PCR) assay described in Telfer et al. (2005). PCR products differ in size between Bartonella species, and the examination of a subset of samples indicated that at least four species of Bartonella were present: Bartonella birtlesii; Bartonella doshiae; Bartonella taylorii; and Bartonella grahamii. However, as all species are thought to have similar life cycles within the mammalian hosts and are therefore predicted to interact with the immune system in a similar way, individual voles were described as either positive or negative for the presence of Bartonella.
The number of parasite types simultaneously co-infecting individual voles was also recorded. Since the presence of Bartonella was only assayed for a subset of voles captured, inclusion of Bartonella in the measurements of co-infection reduced the available sample size from 188 to 99 observations. Therefore, to help maximize statistical power, Bartonella was not included in the analysis of co-infection.
(b) Molecular analyses
DNA was extracted from ear punches using the Qiagen DNeasy Tissue Kit (Qiagen Ltd), according to the manufacturer's instructions. In all cases, the DNA was diluted to 10 ng μl−1. All individuals were genotyped at seven microsatellite loci (Stewart et al. 1998) using either radioactive genotyping according to Stewart et al. (1998) and Aars et al. (2006) or fluorescent detection on an ABI 3730 automated DNA sequencer with labelling of the 5′ end of forward primer with fluorescent dyes (loci AV7 and AV10 with NED; loci AV3, AV8, AV12 and AV14 with 6-FAM; loci AV9 with HEX; PerkinElmer Biosystems). All samples were also genotyped at the MHC class II locus Arte-DRB using single-strand conformation polymorphism analysis according to Oliver & Piertney (2006), with PCR conditions optimized to reduce the production of erroneous PCR products following Zylstra et al. (1998) and sequencing of multiple excisions of the same putative band to ensure consistency between gels. This locus was previously demonstrated to be expressed and to exhibit the patterns of nucleotide polymorphism that suggest a strong effect of positive selection (Oliver & Piertney 2006).
(c) Statistical analyses
Statistical modelling was carried out using R (R Development Core Team 2006). An initial graphical data exploration was conducted to ascertain the spread and distribution of the data, to identify outliers and to examine the relationships between variables. Actions taken in the cases where outliers were identified are described as appropriate in the results.
The principle focus of these analyses was to examine the relationships between MHC genotype and variation in the burden of separate parasite species, while controlling for the confounding influence of other factors. Twelve explanatory variables were used to predict parasite burden, of which eight were continuous (numbers of C. nobilis, M. walkeri, I. ricinus larvae, I. ricinus nymphs, gamasid mites, weight, individual average observed microsatellite heterozygosity and population size) and four were nominal (MHC genotype, month, sex and reproductive status). Additionally, interactions between MHC and sex, and between MHC and reproductive status, were included in the models. The MHC variable consisted principally of three levels: the genotypes Arte-DRB*0101; Arte-DRB*0105; and Arte-DRB*0505. However, to fully address the hypotheses being tested, and for parsimony, in the cases where an MHC effect was identified, the model was refitted with MHC described as either the presence or absence of either allele, or as heterozygous or homozygous. The genotype Arte-DRB*0101 was absent from the sample taken in May 2005; therefore, the models that included MHC were refitted with these observations (n=8) removed to ensure that any extreme effects from this sample did not overtly bias the model. Any changes in the directionality, strength or error of parameter estimates that were observed following refitting with these data removed are reported in the results.
Collinearity among explanatory variables can confound analyses by making parameter estimates unstable and inflating standard errors (Quinn & Keough 2002). Collinearity was identified between vole weight and reproductive status, and between reproductive status and month, as well as vole weight and month (on average, voles were heavier in May than October). Furthermore, on average, population size tended to be lower in May (mean=78) than in October (mean=108). The potential confounding effect of these explanatory variables was considered during model selection and interpretation. The models that included collinear variables were refitted with each variable removed in turn to ascertain misleading changes in coefficients. The only notable case of collinearity affecting a parameter estimate was in the model explaining variation in mite burden, where the directionality of the coefficient for reproductive success changed according to the inclusion of month as a covariate. Both the variables were retained as the removal of either caused a decline in residual-based model validation, and the removal of additional background variance was seen as beneficial for clarifying the relationship between MHC variation and parasite burden.
In all cases, the error structure of the parasite response variable was not normally distributed. As such, and since the data were either counts or presence/absence, a generalized linear modelling (GLM) approach was used. Additionally, all datasets included repeat measurements from some individuals. In order to incorporate this pseudoreplication without introducing bias, generalized linear mixed models (GLMM) were employed, where ‘individual’ was included as a random factor. All models implemented the function glmmPQL (Breslow & Clayton 1993), using the packages MASS (Venables & Ripley 2002) and nlme (Pinheiro & Bates 2000). The models were fitted to either negative binomial, quasi-Poisson or binomial error structures with either a log-link function (negative binomial and quasi-Poisson) or a logit-link function (binomial).
A general principle of parsimony was applied during the modelling process, and model selection was performed by beginning with a model containing all main effects and the interactions. Variables were then removed in reverse order of significance and only significant parameters were retained. Models were validated by the examination of the plots of standardized residuals against fitted values and each retained explanatory variable on the criteria that residuals should be free of trends and evenly distributed. When encountered, overdispersion in the residuals from the analyses performed on count data was accounted for by fitting the model using a quasi-Poisson error structure. When outliers were identified in residual plots, the models were refitted with these observations removed to ascertain their effects on parameter estimation. Any actions taken following residual-based model validation are reported in the results.
One potential issue with the analyses is that a spurious relationship might be identified between MHC variation and individual parasite burdens as a result of spatial autocorrelation between related individuals and parasites within samples, or temporal autocorrelation between MHC gene frequencies and parasite abundance between sampling events. To address this, we examined the spatial distribution of MHC variation within the samples and found that MHC genotypes were well mixed within and between transects. We also calculated that, overall, individuals caught in the same trap had a less than 50 per cent chance of sharing the same MHC genotype, which corresponded closely to the random expected probability within the samples (40–50%). To account for potential temporal autocorrelation, we refitted the models with ‘sampling event’ rather than individual as a random effect, so that the parameters were estimated while accounting for variance attributable only to sampling event. This had a negligible impact on MHC parameter values and did not alter the conclusions of our study.
3. Results
Population size, and hence the number of voles trapped, varied greatly between sampling events, from 135 in October 2004 to 8 in May 2005. There was also a considerable variation in the abundance and prevalence of parasites.
(a) Mites (Gamasidae)
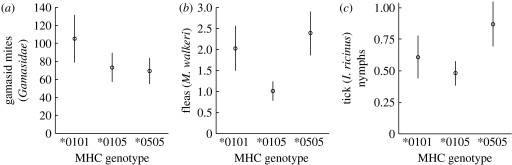
Individual mite burdens varied from 0 to 208 individuals. The mite data were modelled using a negative binomial error structure. Simplification carried out by removing variables in the order of non-significance derived the model: mites∼MHC+month+reproductive status+sex. Observations=152; groups (i.e. the number of individuals)=135. The mite model could be further simplified by replacing the term MHC (including all Arte-DRB genotypes) with Arte-DRB*05 (present or absent). The coefficients for the mite model, including both alternative MHC terms, are summarized in table 1. Individuals with the MHC genotypes Arte-DRB*0105 and *0505 had significantly fewer mites than individuals with the *0101 genotype, but did not vary significantly from each other. Individuals with the *05 allele had significantly fewer mites than individuals without the *05 allele. This term represented the most parsimonious biological description for how variation in mite burden could be explained by Arte-DRB variation. The predicted values for each MHC type are shown in figure 1a. Among the other explanatory variables, there were significantly fewer mites in October than in May, and females had significantly fewer mites than males.
Table 1.
Summary of retained terms and coefficients (with standard errors and p values in parentheses) of the final models for individual burdens of mites (Gamasidae), the flea M. walkeri, the nymphs of the tick I. ricinus and the number of co-infections.
| terms | comparisons | coefficients |
|---|---|---|
| mite(Gamasidae)burden | ||
| Arte-DRB | *0105 versus *0101 | −0.374 (s.e. 0.176, p=0.036) |
| *0505 versus *0101 | −0.428 (s.e. 0.194, p=0.029) | |
| *0505 versus *0105 | −0.054 (s.e. 0.160, p=0.734) | |
| Arte-DRB*05 | present versus absent | −0.395 (s.e. 0.165, p=0.018) |
| month | October versus May | −1.833 (s.e. 0.218, p<0.001) |
| sex | female versus male | −0.331 (s.e. 0.141, p=0.034) |
| reproductive status | not active versus active | 0.459 (s.e. 0.162, p=0.013) |
| flea(M. walkeri)burden | ||
| weight | 0.011 (s.e. 0.005, p=0.049) | |
| density | −0.006 (s.e. 0.002, p=0.022) | |
| C. nobilis | 0.027 (s.e. 0.007, p=0.004) | |
| Arte-DRB | *0105 versus *0101 | −0.818 (s.e. 0.297, p=0.006) |
| *0505 versus *0101 | −0.010 (s.e. 0.287, p=0.722) | |
| *0505 versus *0105 | 0.611 (s.e. 0.234, p=0.003) | |
| heterozygote versus homozygote | −0.748 (s.e. 0.219, p<0.001) | |
| month | October versus May | −1.223 (s.e. 0.362, p=0.005) |
| sex | female versus Male | −0.697 (s.e. 0.231, p=0.002) |
| reproductive status | not active versus active | 0.957 (s.e. 0.396, p=0.034) |
| tick(I. ricinus)nymph burden | ||
| larvae | 0.062 (s.e. 0.027, p=0.030) | |
| Arte-DRB | *0105 versus *0101 | −0.290 (s.e. 0.319, p=0.372) |
| *0505 versus *0101 | 0.438 (s.e. 0.328, p=0.182) | |
| *0505 versus *0105 | 0.525 (s.e. 0.264, p=0.048) | |
| month | October versus May | −1.122 (s.e. 0.246, p<0.001) |
| number of co-infections | ||
| density | −0.002 (s.e. 0.001, p=0.006) | |
| Arte-DRB | *0105 versus *0101 | −0.160 (s.e. 0.062, p=0.011) |
| *0505 versus *0101 | −0.050 (s.e. 0.066, p=0.454) | |
| *0505 versus *0105 | 0.110 (s.e. 0.053, p=0.039) | |
| heterozygote versus homozygote | −0.128 (s.e. 0.046, p=0.006) | |
| sex | female versus male | −0.125 (s.e. 0.047, p=0.008) |
| month | October versus May | −0.374 (s.e. 0.048, p<0.001) |
Figure 1.
Modelled (i.e. accounting for and holding constant all other significant covariates) individual burdens of three types of parasite: (a) gamasid mites (Gamasidae), (b) the flea M. walkeri and (c) the nymphs of the tick I. ricinus, according to the genotype at the MHC class II locus Arte-DRB. Bars represent standard errors.
(b) Fleas
(i) Megabothris walkeri
Individual burdens of M. walkeri varied between 0 and 10 and showed a strong right skew, where 58 per cent of the data were of zero value. The final model was fitted to a quasi-Poisson error structure and defined as follows: M. walkeri∼density+MHC+month+reproductive status+sex+weight+C. nobilis. Observations=201; groups=185 (table 1). A clear MHC effect was observed, with Arte-DRB*0105 individuals having significantly fewer M. walkeri than either *0101 or *0505 individuals, while no significant difference was observed between the two homozygous genotypes. This relationship could be simplified biologically to an MHC heterozygosity term, where Arte-DRB heterozygotes had significantly lower burdens of M. walkeri than Arte-DRB homozygotes. Refitting the model without the May 2005 sample did not affect the directionality, but increased the strength of the coefficient between Arte-DRB heterozygotes and homozygotes. Predicted M. walkeri burdens for Arte-DRB homozygotes and heterozygotes are shown in figure 1b. Megabothris walkeri burden was also positively correlated with C. nobilis burden and vole weight, and negatively correlated with density. Male voles had higher M. walkeri burdens than female voles, and reproductively inactive voles had higher burdens than those that were reproductively active. A seasonal effect was also detected where M. walkeri burdens were higher in May than in October.
(ii) Ctenopthalmus nobilis
Burdens of the flea, C. nobilis, ranged from 0 to 82 and had a right-skewed distribution with 17 per cent of observations being zero. An extreme value was identified, the value of which (82) was so far away from the next highest value (42) that it was removed on the basis that it was highly non-representative and could strongly bias the modelling process. The final model was fitted to a quasi-Poisson error structure and was defined as follows: C. nobilis∼month+M. walkeri. Observations=207; groups=190. There were fewer C. nobilis in October than May (intercept=1.84, β=−1.08, s.e.=0.12, p<0.001), and the number of C. nobilis was positively correlated with M. walkeri burden (β=0.10, s.e.=0.04, p=0.015).
(c) Ticks (I. ricinus)
An additional term was included in I. ricinus models, which described the presence or absence of sheep in the previous 12 months. This is likely to be biologically important as I. ricinus are thought to require a large mammal as a host for the adult life stage (Hillyard 1996) and, consequently, the abundance of early life stages in the environment is predicted to depend on the previous availability of suitable hosts.
(i) Larvae
The numbers of I. ricinus larvae varied from 0 to 19 and had an extremely skewed distribution with 73 per cent of the data being zero and a small number of relatively high values (n=10 (3%), range=6–19). The final model for I. ricinus larvae was fitted with quasi-Poisson errors and defined as follows: larvae∼weight+month+I. ricinus nymphs+sheep presence in the previous 12 months. Observations=295; groups=271. No MHC effect was observed. A positive association was found between the numbers of larvae and the number of I. ricinus nymphs (p=0.003). There was a highly significant positive association between the number of larvae found on voles and the presence of sheep on Coiresa in the previous year (p<0.001). The month effect was also highly significant with fewer larvae present in October relative to May (p<0.001). Refitting this model as presence/absence with a binomial error structure retained all three covariates with the same directionality in all relationships.
(ii) Nymphs
Individual burdens for the nymphs of the tick I. ricinus ranged from 0 to 4 and were also heavily skewed, with 77 per cent of the observations represented by zero. The final model for I. ricinus nymphs was fitted with quasi-Poisson errors and defined as follows: nymphs∼MHC+month+larvae. Observations=305; groups=280 (table 1). There was an Arte-DRB genotype effect, where *0105 individuals had significantly fewer nymphs than *0505 individuals, but there was no significant difference between *0101 and either of the other two genotypes. The predicted nymph burden for each Arte-DRB type is shown in figure 1c. Nymph burdens were positively correlated with larvae burdens, and were lower in October than in May. The diagnostic plot of the standardized residuals against fitted values revealed one potentially influential point. Removing this point had an impact on the standard error but not the directionality of the coefficients, altering the significance level associated with the coefficient for Arte-DRB*0105 versus *0505 to p=0.036.
(d) Bartonella
Bartonella was a binary response variable recorded as the presence or absence. In total, 93 (51%) of the voles from which blood was tested were positive. However, there was a marked difference between the two samples with 7 per cent prevalence in May 2004 and 77 per cent prevalence in October 2004. The final model was defined as follows: Bartonella∼month+weight. The probability of being infected with Bartonella was negatively correlated with weight (β=−0.033, s.e.=0.009, p=0.038) and was significantly higher in October than in May (β=3.26, s.e.=0.548, p=0.009).
(e) Co-infection
Individual voles were simultaneously co-infected with between zero and four different ectoparasite types. The final model of co-infection was fitted with quasi-Poisson errors and defined as follows: number of co-infections∼density+MHC+sex+month. Observations=188; groups=171 (table 1). Individuals with the heterozygous MHC genotype Arte-DRB*0105 had significantly fewer co-infections than those with either of the homozygous genotypes, while there was no significant difference between the two homozygotes. Females had fewer co-infections than males, individuals had fewer co-infections in October than in May and the number of co-infections was negatively correlated with vole population density.
4. Discussion
Individual MHC genotype was significantly associated with the number of parasite types simultaneously co-infecting individuals, as well as variation in the burdens of three of the six parasites (namely gamasid mites, the flea M. walkeri and nymphs of the tick I. ricinus). No significant association was identified between Arte-DRB polymorphism and I. ricinus larvae, Bartonella or the flea C. nobilis. For gamasid mites, variation in burden was best explained by the presence or absence of the Arte-DRB*05 allele, where the individuals that carried at least one copy of the allele had lower associated burdens than those that did not. Variation in the burdens of both M. walkeri and I. ricinus nymphs were associated with MHC heterozygosity. However, while individuals with the Arte-DRB*0105 genotype had lower predicted burdens of M. walkeri than either of the homozygote genotypes, this was not the case for I. ricinus nymphs, where predicted burdens for Arte-DRB*0105 individuals were significantly lower than those predicted for Arte-DRB*0505 individuals, but not Arte-DRB*0101 individuals. MHC heterozygotes were co-infected by fewer parasite types than homozygotes.
The results of the mite analysis are consistent with the effects of allelic dominance, with individuals carrying the Arte-DRB*05 allele in either homozygote or heterozygote form having significantly lower parasite burdens. By contrast, the M. walkeri data provide support for an overdominance/heterozygote superiority mode of selection where the heterozygote is fitter than either homozygote (McClelland et al. 2003b). However, the I. ricinus model is most compatible with MHC heterozygote advantage, where the heterozygote is fitter than one homozygote genotype, but not both (Penn et al. 2002). Although these models may independently provide evidence that different modes of selection act concomitantly on Arte-DRB diversity, the fitness of an individual is dictated by its ability to deal with all of the challenges it faces in its environment, and if all models are considered together, it is apparent that in this study there is overall MHC heterozygote superiority. In each case where an MHC-dependent effect on parasite burden was resolved, the heterozygote genotype was in the group that was associated with fewer parasites. Moreover, when compared with each homozygote, the heterozygote had statistically significant lower burdens in two out of three cases and was comparable in the third. Therefore, the associated cumulative fitness inferred from these models is greater for the Arte-DRB heterozygous genotype than for either homozygote (Hughes & Nei 1992). This is significant as heterozygote superiority is a mechanism that can maintain genetic diversity. No evidence for either an additive single MHC allele effect (co-dominance; de Eyto et al. 2007) or an effect of general heterosis (i.e. variation that could be explained by average microsatellite heterozygosity; Acevedo-Whitehouse et al. 2003) on parasite burden was apparent in any parasite model.
Interestingly, as well as enjoying reduced cumulative parasite burdens, MHC heterozygotes were also simultaneously co-infected by fewer parasite types than homozygotes. Interactions between parasites may influence the pathology of an infection with detrimental consequences for host fitness (Telfer et al. 2008). Indeed, available evidence suggests that the immune response to simultaneous multi-parasite insults may impose an immunopathological cost that is greater than the additive pathogenic effects of individual parasite species (Graham 2002; Pullan & Brooker 2008). Therefore, MHC heterozygotes may gain an additional fitness benefit in the form of reduced immunopathology through lower co-infection rates.
While it is clear from these results that, in this system, MHC heterozygotes have reduced cumulative parasite burdens and are simultaneously infected by fewer parasite species, it is difficult to predict whether similar results would be obtained from other systems. Indeed, the study population is relatively unique in that there are only two, albeit highly divergent, MHC alleles at the Arte-DRB locus. Although these circumstances present an excellent opportunity to test MHC evolutionary theory, much of which is based upon simple two-allele systems, higher levels of MHC variation are typically assayed in many other natural populations. Whereas our findings support an overdominant mode of MHC evolution, some theoretical studies predict that overdominance is inadequate (Borghans et al. 2004) or only capable of maintaining high levels of MHC variation under assumptions that are arguably unrealistic, such as all heterozygotes being of equivalent fitness (De Boer et al. 2004). In a given system, the evolutionary dynamics between MHC variation and parasites is likely to depend on a number of parameters, including the number of MHC alleles, allelic divergence and parasite variability (Richman et al. 2001; Hedrick 2002; Stoffels & Spencer 2008). Therefore, there exists a considerable empirical challenge to make the findings between studies comparable, both in the interpretation of results and using appropriate analytical techniques. For example, the presence of one more MHC allele in the focal population would increase the number of potential MHC genotypes from three to six, which would effectively require twice the current sample size to maintain statistical power. Thus, as MHC allelic diversity increases, sample sizes required for the robust statistical analytical techniques used here quickly become practically unfeasible.
Other factors correlated with the burden of the different parasite species were largely consistent with previous studies. Seasonality in infestation rates has been demonstrated earlier for ticks (Randolph et al. 2002), mites (Howell et al. 1957), fleas (Krasnov et al. 2002; Vashchenok & Tret'iakov 2003) and Bartonella species (Telfer et al. 2007). Male-biased parasitism rates, as observed here for mites and M. walkeri, have been recorded in a number of blood-feeding arthropods and may be explained by the effect of testosterone, which is higher in males and is known to depress the immune response to parasites (Harder et al. 1994; Hughes & Randolph 2001; Josabel Belliure 2004; Cox & John-Alder 2007). Previous studies have also found correlations in parasite burden between different flea species (Krasnov et al. 2005) and between different life stages of ticks (Randolph et al. 1999). Such associations may be due to facilitation by the suppression of the immune system (Krasnov et al. 2005) or correlated small-scale spatial variation in risk due to microclimatic conditions. Interestingly, from the point of view of the MHC, an alternative explanation is that the resistance or susceptibility profiles of individual hosts are similar for closely related parasite life stages or species (Ditchkoff et al. 2005).
The cost of parasitism by blood-feeding arthropods can have a demonstrable effect on host fitness parameters (Fitze et al. 2004). For example, the levels of haematophagous mites in nests were associated with reduced nestling mass in house finches (Carpodacus mexicanus; Stoehr et al. 2000) and reduced reproductive success in barn swallows (Hirundo rustica; Moller 2002). Hatching success, clutch survival and adult survival were improved for female pheasants (Phasianus colchicus) that had their tick (I. ricinus) burdens removed relative to those that did not (Hoodless et al. 2003). Juvenile gerbils (Gerbillus andersoni) with natural infestation levels of fleas exhibit slower weight gain and more rapid weight loss than uninfected individuals (Hawlena et al. 2006b). Additionally, parasitized juveniles were shown to have 48 per cent lower survival relative to individuals that had their flea burdens removed (Hawlena et al. 2006a).
When considering the benefit of enhanced resistance, the cost to the host of mounting an effective immune response also has to be taken into account. This has been investigated in great tits (Parus major), where field experiments showed that, in parasite-free broods, chicks that had been fed an immune stimulant had reduced growth relative to non-immune-stimulated chicks (Tschirren & Richner 2006). However, this trend was reversed in the broods where an ectoparasitic flea was present, indicating an overall energetic advantage to an enhanced immune response in the presence of fleas. In male sand lizards (Lacerta agilis), individuals with MHC-dependent resistance to ticks had marginally higher reproductive success at the end of the breeding season relative to non-resistant individuals, despite suffering higher perceived immune-energetic costs (measured as leucocyte/erythrocyte ratio) and higher levels of haemoprotid parasitaemia (Olsson et al. 2005). These examples illustrate how variation in the natural levels of parasitism can impact on important host fitness traits. It is easy to conceive how this could have evolutionary consequences as the success of differentially effective genotypes will directly influence the frequency at which they are represented in the population.
This is one of a small number of studies to investigate the importance of MHC polymorphism for ectoparasite resistance. A few studies have examined the importance of MHC in determining tick burden. Of these, associations have been found between DRB loci and resistance/susceptibility to ticks in cattle (although these associations were lost after Bonferroni correction for multiple tests; Acosta-Rodriguez et al. 2005) and white-tailed deer (Ditchkoff et al. 2005). MHC-dependent resistance to ticks has also been reported for sand lizards (L. agilis; Olsson et al. 2005).
In natural populations, MHC diversity is defined by a diverse array of neutral and selective forces, which is likely to include multiple parasites. Similarly, the prevalence and abundance of parasites in host populations is influenced by a complex variety of factors. To understand the coevolutionary relationship between MHC diversity and parasitism, it is therefore necessary to identify an appropriate system, apply suitable analytical techniques and obtain sufficient sample sizes across multiple parasite taxa. Here, we illustrated associations between individual parasite burdens and Arte-DRB genotype, which are consistent with a parasite-mediated mechanism of MHC evolution. From the specific relationships observed between MHC genes and parasites, it is clear that, in a two-allele, multi-parasite context, heterozygote superiority seems to prevail overall through the reduced cumulative parasite load and the number of co-infections associated with MHC heterozygosity. Such a mechanism could contribute to maintaining even allele frequencies, high nucleotide diversity and the levels of heterozygosity in excess of Hardy–Weinberg equilibrium as observed for MHC loci in some natural populations.
Acknowledgements
We thank Sarah Burthe, Alex Douglas, Elaine Fraser, Suzanne Hogg, Xavier Lambin, Juan Luque Larena, Vicki Saint, Laura Taylor, Edoardo Tedesco and Jamie Urquhart for their assistance with sample collection; Freda Marshall for microsatellite genotyping; Dave Jones and Nat Jones for Bartonella PCR assays; and Thomas Cornulier, Alex Douglas and Janine Illian for their comments on statistical analyses. This work was carried out under the tenure of a Natural Environment Research Council (NERC) postgraduate studentship to M.K.O. The blood sampling conforms to the UK regulations and was carried out under the home office project licence PPL 40/1813.
References
- Aars J., Dallas J.F., Piertney S.B., Marshall F., Gow J.L., Telfer S., Lambin X. Widespread gene flow and high genetic variability in populations of water voles Arvicola terrestris in patchy habitats. Mol. Ecol. 2006;15:1455–1466. doi: 10.1111/j.1365-294X.2006.02889.x. doi:10.1111/j.1365-294X.2006.02889.x [DOI] [PubMed] [Google Scholar]
- Acevedo-Whitehouse K., Gulland F., Greig D., Amos W. Disease susceptibility in California sea lions. Nature. 2003;422:35. doi: 10.1038/422035a. doi:10.1038/422035a [DOI] [PubMed] [Google Scholar]
- Acosta-Rodriguez R., Alonso-Morales R., Balladares S., Flores-Aguilar H., Garcia-Vazquez Z., Gorodezky C. Analysis of BoLA class II microsatellites in cattle infested with Boophilus microplus ticks: class II is probably associated with susceptibility. Vet. Parasitol. 2005;127:313–321. doi: 10.1016/j.vetpar.2004.10.007. doi:10.1016/j.vetpar.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Allen J.R., Kemp D.H. Observations of the behaviour of Dermacentor andersoni larvae infesting normal and tick resistant guinea-pigs. Parasitology. 1982;84:195–204. doi: 10.1017/s0031182000044760. [DOI] [PubMed] [Google Scholar]
- Apanius V., Penn D., Slev P.R., Ruff L.R., Potts W.K. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- Arkush K.D., Giese A.R., Mendonca H.L., McBride A.M., Marty G.D., Hedrick P.W. Resistance to three pathogens in the endangered winter-run chinook salmon (Oncorhynchus tshawytscha): effects of inbreeding and major histocompatibility complex genotypes. Can. J. Fish. Aquat. Sci. 2002;59:966–975. doi:10.1139/f02-066 [Google Scholar]
- Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. doi:10.1046/j.1420-9101.2003.00531.x [DOI] [PubMed] [Google Scholar]
- Borghans J.A.M., Beltman J.B., De Boer R.J. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. doi:10.1007/s00251-003-0630-5 [DOI] [PubMed] [Google Scholar]
- Bown K.J., Begon M., Bennett M.W.Z. Seasonal dynamics of Anaplasma phagocytophila in a rodent-tick (Ixodes trianguliceps) system, United Kingdom. Emerg. Infect. Dis. 2003;9:63–70. doi: 10.3201/eid0901.020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N.E., Clayton D.G. Approximate inference in generalized linear models. J. Am. Stat. Assoc. 1993;88:9–25. doi:10.2307/2290687 [Google Scholar]
- Carrington M., et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. doi:10.1126/science.283.5408.1748 [DOI] [PubMed] [Google Scholar]
- Cox R.M., John-Alder H.B. Increased mite parasitism as a cost of testosterone in male striped plateau lizards Sceloporus virgatus. Funct. Ecol. 2007;21:327–334. doi:10.1111/j.1365-2435.2007.01251.x [Google Scholar]
- De Boer R.J., Borghans J.A.M., van Boven M., Kesmir C., Weissing F.J. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics. 2004;55:725–731. doi: 10.1007/s00251-003-0629-y. doi:10.1007/s00251-003-0629-y [DOI] [PubMed] [Google Scholar]
- de Eyto E., et al. Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc. R. Soc. B. 2007;274:861–869. doi: 10.1098/rspb.2006.0053. doi:10.1098/rspb.2006.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denniston C., Crow J.F. Alternative fitness models with the same allele frequency dynamics. Genetics. 1990;125:201–205. doi: 10.1093/genetics/125.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchkoff S.S., Hoofer S.R., Lochmiller R.L., Masters R.E., Van Den Bussche R.A. MHC-DRB evolution provides insight into parasite resistance in white-tailed deer. Southwest. Nat. 2005;50:57–64. doi:10.1894/0038-4909(2005)050<0057:MEPIIP>2.0.CO;2 [Google Scholar]
- Doherty P.C., Zinkernagel R.M. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. doi:10.1038/256050a0 [DOI] [PubMed] [Google Scholar]
- Fitze P.S., Clobert J., Richner H. Long-term life-history consequences of ectoparasite-modulated growth and development. Ecology. 2004;85:2018–2026. doi:10.1890/03-0138 [Google Scholar]
- Froeschke G., Sommer S. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the Southern Kalahari. Mol. Biol. Evol. 2005;22:1254–1259. doi: 10.1093/molbev/msi112. doi:10.1093/molbev/msi112 [DOI] [PubMed] [Google Scholar]
- Garrigan D., Hedrick P.W. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. doi:10.1554/02-732 [DOI] [PubMed] [Google Scholar]
- Gaudieri S., Dawkins R.L., Habara K., Kulski J.K., Gojobori T. SNP profile within the human major histocompatibility complex reveals an extreme and interrupted level of nucleotide diversity. Genome Res. 2000;10:1579–1586. doi: 10.1101/gr.127200. doi:10.1101/gr.127200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A.L. When T-helper cells don't help: immunopathology during concomitant infection. Q. Rev. Biol. 2002;77:409–434. doi: 10.1086/344414. doi:10.1086/344414 [DOI] [PubMed] [Google Scholar]
- Grimholt U., Larsen S., Nordmo R., Midtlyng P., Kjoeglum S., Storset A., Saebo S., Stet R.J.M. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. doi:10.1007/s00251-003-0567-8 [DOI] [PubMed] [Google Scholar]
- Harder A., Danneschewski A., Wunderlich F. Genes of the mouse H-2 complex control the efficacy of testosterone to suppress immunity against the intestinal nematode Heterakis spumosa. Parasitol. Res. 1994;V80:446–448. doi: 10.1007/BF00932386. doi:10.1007/BF00932386 [DOI] [PubMed] [Google Scholar]
- Harf R., Sommer S. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the Southern Kalahari. Mol. Ecol. 2005;14:85–91. doi: 10.1111/j.1365-294X.2004.02402.x. doi:10.1111/j.1365-294X.2004.02402.x [DOI] [PubMed] [Google Scholar]
- Hawlena H., Abramsky Z., Krasnov B.R. Ectoparasites and age-dependent survival in a desert rodent. Oecologia. 2006;V148:30–39. doi: 10.1007/s00442-005-0345-4. doi:10.1007/s00442-005-0345-4 [DOI] [PubMed] [Google Scholar]
- Hawlena H., Khokhlova I.S., Abramsky Z., Krasnov B.R. Age, intensity of infestation by flea parasites and body mass loss in a rodent host. Parasitology. 2006;133:187–193. doi: 10.1017/S0031182006000308. doi:10.1017/S0031182006000308 [DOI] [PubMed] [Google Scholar]
- Hedrick P.W. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. doi:10.1554/0014-3820(2002)056[1902:PRAGVA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hill A.V.S., et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. doi:10.1038/352595a0 [DOI] [PubMed] [Google Scholar]
- Hill A.V.S., et al. Molecular analysis of the association of Hla-B53 and resistance to severe malaria. Nature. 1992;360:434–439. doi: 10.1038/360434a0. doi:10.1038/360434a0 [DOI] [PubMed] [Google Scholar]
- Hillyard P.D. Synopses of the British Fauna (New Series) Field Studies Council Publications; London, UK: 1996. Ticks of north-west Europe. [Google Scholar]
- Hohler T., et al. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J. Hepatol. 1997;26:503–507. doi: 10.1016/s0168-8278(97)80414-x. doi:10.1016/S0168-8278(97)80414-X [DOI] [PubMed] [Google Scholar]
- Hoodless A.N., Kurtenbach K., Nuttall P.A., Randolph S.E. Effects of tick Ixodes ricinus infestation on pheasant Phasianus colchicus breeding success and survival. Wildl. Biol. 2003;9:171–178. [Google Scholar]
- Howell J.F., Allred D.M., Beck D.E. Seasonal population fluctuations of mites in desert wood rat nests in central Utah. Ecology. 1957;38:82–88. doi:10.2307/1932129 [Google Scholar]
- Hughes A.L., Nei M. Models of host–parasite interaction and MHC polymorphism. Genetics. 1992;132:863–864. doi: 10.1093/genetics/132.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes V.L., Randolph S.E. Testosterone depresses innate and acquired resistance to ticks in natural rodent hosts: a force for aggregated distributions of parasites. J. Parasitol. 2001;87:49–54. doi: 10.1645/0022-3395(2001)087[0049:TDIAAR]2.0.CO;2. doi:10.1645/0022-3395(2001)087[0049:TDIAAR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ilmonen P., Penn D.J., Damjanovich K., Morrison L., Ghotbi L., Potts W.K. Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice. Genetics. 2007;176:2501–2508. doi: 10.1534/genetics.107.074815. doi:10.1534/genetics.107.074815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josabel Belliure L.S.G.S. Effect of testosterone on T cell-mediated immunity in two species of mediterranean lacertid lizards. J. Exp. Zoolog. A Comp. Exp. Biol. 2004;301A:411–418. doi: 10.1002/jez.a.20068. doi:10.1002/jez.a.20068 [DOI] [PubMed] [Google Scholar]
- Klein J. Wiley; New York, NY: 1986. Natural history of the major histocompatibility complex. [Google Scholar]
- Krasnov B.R., Burdelova N.V., Shenbrot G.I., Khokhlova I.S. Annual cycles of four flea species in the central Negev desert. Med. Vet. Entomol. 2002;16:266–276. doi: 10.1046/j.1365-2915.2002.00374.x. doi:10.1046/j.1365-2915.2002.00374.x [DOI] [PubMed] [Google Scholar]
- Krasnov B.R., Khokhlova I.S., Shenbrot G.I. Sampling fleas: the reliability of host infestation data. Med. Vet. Entomol. 2004;18:232–240. doi: 10.1111/j.0269-283X.2004.00500.x. doi:10.1111/j.0269-283X.2004.00500.x [DOI] [PubMed] [Google Scholar]
- Krasnov B.R., Mouillot D., Shenbrot G.I., Khokholva I.S., Poulin R. Abundance patterns and coexistence processes in communities of fleas parasitic on small mammals. Ecography. 2005;28:453–464. doi:10.1111/j.0906-7590.2005.04182.x [Google Scholar]
- Loiseau C., Zoorob R., Garnier S., Birard J., Federici P., Julliard R., Sorci G. Antagonistic effects of a MHC class I allele on malaria-infected house sparrows. Ecol. Lett. 2008;11:258–265. doi: 10.1111/j.1461-0248.2007.01141.x. doi:10.1111/j.1461-0248.2007.01141.x [DOI] [PubMed] [Google Scholar]
- McClelland E.E., Granger D.L., Potts W.K. Major histocompatibility complex-dependent susceptibility to Cryptococcus neoformans in mice. Infect. Immun. 2003;71:4815–4817. doi: 10.1128/IAI.71.8.4815-4817.2003. doi:10.1128/IAI.71.8.4815-4817.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland E.E., Penn D.J., Potts W.K. Major histocompatibility complex heterozygote superiority during coinfection. Infect. Immun. 2003;71:2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. doi:10.1128/IAI.71.4.2079-2086.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lucht Y., Sommer S. MHC diversity and the association to nematode parasitism in the yellow-necked mouse (Apodemus flavicollis) Mol. Ecol. 2005;14:2233–2243. doi: 10.1111/j.1365-294X.2005.02557.x. doi:10.1111/j.1365-294X.2005.02557.x [DOI] [PubMed] [Google Scholar]
- Moller A.P. Temporal change in mite abundance and its effect on barn swallow reproduction and sexual selection. J. Evol. Biol. 2002;15:495–504. doi:10.1046/j.1420-9101.2002.00386.x [Google Scholar]
- Oliver M.K., Piertney S.B. Isolation and characterization of a MHC class II DRB locus in the European water vole (Arvicola terrestris) Immunogenetics. 2006;58:390–395. doi: 10.1007/s00251-006-0121-6. doi:10.1007/s00251-006-0121-6 [DOI] [PubMed] [Google Scholar]
- Olsson M., Wapstra E., Madsen T., Ujvari B., Rugfelt C. Costly parasite resistance: a genotype-dependent handicap in sand lizards? Biol. Lett. 2005;1:375–377. doi: 10.1098/rsbl.2005.0339. doi:10.1098/rsbl.2005.0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson S., Wilson K., Pemberton J.M. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.) Proc. Natl Acad. Sci. USA. 1998;95:3714–3719. doi: 10.1073/pnas.95.7.3714. doi:10.1073/pnas.95.7.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn D.J., Damjanovich K., Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA. 2002;99:11 260–11 264. doi: 10.1073/pnas.162006499. doi:10.1073/pnas.162006499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. doi:10.1038/sj.hdy.6800724 [DOI] [PubMed] [Google Scholar]
- Pinheiro J.C., Bates D.M. Statistics and computing. Springer; New York, NY: 2000. Mixed-effects models in S and S-plus. [Google Scholar]
- Proctor H., Owens I. Mites and birds: diversity, parasitism and coevolution. Trends Ecol. Evol. 2000;15:358–364. doi: 10.1016/s0169-5347(00)01924-8. doi:10.1016/S0169-5347(00)01924-8 [DOI] [PubMed] [Google Scholar]
- Pullan R., Brooker S. The health impact of polyparasitism in humans: are we under-estimating the burden of parasitic diseases? Parasitology. 2008;135:783–794. doi: 10.1017/S0031182008000346. doi:10.1017/S0031182008000346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn G.P., Keough M.J. Cambridge University Press; Cambridge, MA: 2002. Experimental design and data analysis for biologists. [Google Scholar]
- Randolph S.E., Miklisova D., Lysy J., Rogers D.J., Labuda M. Incidence from coincidence: patterns of tick infestations on rodents facilitate transmission of tick-borne encephalitis virus. Parasitology. 1999;118:177–186. doi: 10.1017/s0031182098003643. doi:10.1017/S0031182098003643 [DOI] [PubMed] [Google Scholar]
- Randolph S.E., Green R.M., Hoodless A.N., Peacey M.F. An empirical quantitative framework for the seasonal population dynamics of the tick Ixodes ricinus. Int. J. Parasitol. 2002;32:979–989. doi: 10.1016/s0020-7519(02)00030-9. doi:10.1016/S0020-7519(02)00030-9 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2006 R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing.
- Richardson D., Komdeur J., Burke T., Von Schantz T. MHC-based patterns of social and extra-pair mate choice in Seychelles warbler. Proc. R. Soc. B. 2005;272:759–767. doi: 10.1098/rspb.2004.3028. doi:10.1098/rspb.2004.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman A.D., Herrera L.G., Nash D. MHC class II beta sequence diversity in the deer mouse (Peromyscus maniculatus): implications for models of balancing selection. Mol. Ecol. 2001;10:2765–2773. doi: 10.1046/j.0962-1083.2001.01402.x. doi:10.1046/j.0962-1083.2001.01402.x [DOI] [PubMed] [Google Scholar]
- Robertson A. Selection for heterozygotes in small populations. Genetics. 1962;47:1291–1300. doi: 10.1093/genetics/47.9.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., Marsh S.G. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucl. Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. doi:10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad J., Ganzhorn J.U., Sommer S. Parasite burden and constitution of major histocompatibility complex in the Malagasy mouse lemur, Microcebus murinus. Evolution. 2005;59:439–450. doi:10.1554/04-312 [PubMed] [Google Scholar]
- Slade R.W., McCallum H.I. Overdominant vs. frequency-dependent selection at MHC loci. Genetics. 1992;132:861–862. doi: 10.1093/genetics/132.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit F.G.A.M. Handbooks for the identification of British insects. Royal Entomological Society of London; London, UK: 1976. Siphonaptera. [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. doi:10.1186/1742-9994-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W.A., Piertney S.B., Dallas J.F. Isolation and characterization of highly polymorphic microsatellites in the water vole, Arvicola terrestris. Mol. Ecol. 1998;7:1258–1259. [PubMed] [Google Scholar]
- Stoehr A.M., Nolan P.M., Hill G.E., McGraw K.J. Nest mites (Pellonyssus reedi) and the reproductive biology of the house finch (Carpodacus mexicanus) Can. J. Zool. 2000;78:2126–2133. doi:10.1139/cjz-78-12-2126 [Google Scholar]
- Stoffels R.J., Spencer H.G. An asymmetric model of heterozygote advantage at major histocompatibility complex genes: degenerate pathogen recognition and intersection advantage. Genetics. 2008;178:1473–1489. doi: 10.1534/genetics.107.082131. doi:10.1534/genetics.107.082131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer S., Bown K.J., Sekules R., Begon M., Hayden T., Birtles R. Disruption of a host–parasite system following the introduction of an exotic host species. Parasitology. 2005;130:661–668. doi: 10.1017/s0031182005007250. doi:10.1017/S0031182005007250 [DOI] [PubMed] [Google Scholar]
- Telfer S., Clough H., Birtles R., Bennett M., Carslake D., Helyar S., Begon M. Ecological differences and coexistence in a guild of microparasites: Bartonella in wild rodents. Ecology. 2007;88:1841–1849. doi: 10.1890/06-1004.1. doi:10.1890/06-1004.1 [DOI] [PubMed] [Google Scholar]
- Telfer S., Birtles R., Bennett M., Lambin X., Paterson S., Begon M. Parasite interactions in natural populations: insights from longitudinal data. Parasitology. 2008;135:767–781. doi: 10.1017/S0031182008000395. doi:10.1017/S0031182008000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz M.R., Kwiatkowski D., Allsopp C.E.M., Greenwood B.M., Thomas H.C., Hill A.V.S. Association between an MHC class-II allele and clearance of hepatitis-B virus in the Gambia. New Engl. J. Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. doi:10.1056/NEJM199504203321604 [DOI] [PubMed] [Google Scholar]
- Thursz M.R., Thomas H.C., Greenwood B.M., Hill A.V.S. Heterozygote advantage for HLA class-II type in hepatitis B virus infection. Nat. Genet. 1997;17:11–12. doi: 10.1038/ng0997-11. doi:10.1038/ng0997-11 [DOI] [PubMed] [Google Scholar]
- Tollenaere C., et al. Multiple parasites mediate balancing selection at two MHC class II genes in the fossorial water vole: insights from multivariate analyses and population genetics. J. Evol. Biol. 2008;21:1307–1320. doi: 10.1111/j.1420-9101.2008.01563.x. doi:10.1111/j.1420-9101.2008.01563.x [DOI] [PubMed] [Google Scholar]
- Trager W. Acquired immunity to ticks. J. Parasitol. 1939;25:57–81. doi:10.2307/3272160 [Google Scholar]
- Tschirren B., Richner H. Parasites shape the optimal investment in immunity. Proc. R. Soc. B. 2006;273:1773–1777. doi: 10.1098/rspb.2006.3524. doi:10.1098/rspb.2006.3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashchenok V.S., Tret'iakov K.A. The seasonal dynamics of fleas (Siphonaptera) on bank voles (Clethrionomys glareolus) in the north part of Novgorod region. Parazitologiia. 2003;37:177–190. [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. Statistics and computing. Springer; New York, NY: 2002. Modern applied statistics with S. [Google Scholar]
- Wedekind C., Walker M., Little T.J. The course of malaria in mice: major histocompatibility complex (MHC) effects, but no general MHC heterozygote advantage in single-strain infections. Genetics. 2005;170:1427–1430. doi: 10.1534/genetics.105.040683. doi:10.1534/genetics.105.040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel S.K. Host immunity to ticks. Annu. Rev. Entomol. 1996;41:1–22. doi: 10.1146/annurev.en.41.010196.000245. doi:10.1146/annurev.en.41.010196.000245 [DOI] [PubMed] [Google Scholar]
- Wikel S.K. Immunology of the tick-host interface. In: Wikel S.K., editor. The immunology of host-ectoparasite arthropod relationships. CAB International; Wallingford, UK: 1996. pp. 204–231. [Google Scholar]
- Wu Y.F., Wang L.Y., Lee T.D., Lin H.H., Hu C.T., Cheng M.L., Lo S.Y. HLA phenotypes and outcomes of hepatitis B virus infection in Taiwan. J. Med. Virol. 2004;72:17–25. doi: 10.1002/jmv.10557. doi:10.1002/jmv.10557 [DOI] [PubMed] [Google Scholar]
- Zylstra P., Rothenfluh H., Weiller G.F., Blanden R.V., Steele E.J. PCR amplification of murine immunoglobulin germline V genes: strategies for minimization of recombination artefacts. Immunol. Cell Biol. 1998;76:395–405. doi: 10.1046/j.1440-1711.1998.00772.x. doi:10.1046/j.1440-1711.1998.00772.x [DOI] [PubMed] [Google Scholar]