Abstract
Exaggerated sexual displays are often supposed to indicate the indirect benefits females may receive from sexual reproduction with displaying males, but empirical evidence for positive relationships between the genetic quality and sexual trait quality is scant. The explanation for this might lie in the fact that mixing of reproductive individuals whose development has been influenced by genotype-by-environment interactions (GEIs) can blur the relationship between the individual male genetic quality and phenotype as perceived by females. Strong GEIs can generate an ecological crossover, where different genotypes are superior in environments that are separated either in space or time. Here, we use a stochastic simulation model to show that even a weak GEI, which does not generate an obvious ecological crossover, can neutralize or even reverse the relationship between genetic quality and sexual trait size in the presence of environmental heterogeneity during development. Our model highlights the importance of developmental selection in evolution of traits and allows us to predict the situations in which sexual displays might not be reliable indicators of genetic quality.
Keywords: developmental selection, genotype-by-environment interactions, good genes hypothesis, lek paradox, sexual selection
1. Introduction
Selection of mates has long been known to have profound effects on the evolution of organisms, and it is generally females that choose among competing males (Darwin 1871). Mate choice by females has led to the evolution of exaggerated sexual displays in males, which are often thought to indicate the indirect (i.e. genetic) benefits females may receive from sexual reproduction with the displayer. Indirect benefits may accrue if male offspring are particularly attractive to potential mates because they share the characteristics of their father's display (the ‘runaway’ process; Fisher 1930), or if offspring are highly viable because the father's display is an honest signal of his genetic quality (the ‘good genes’ model; Zahavi 1975). The good genes model is supported by the widespread findings that developmental stress impacts on the development of traits and so might affect the attractiveness of the male signaller (Emlen 1994; Knell et al. 1999; Spencer et al. 2004). Since models of the so-called Fisherian runaway process often assume, at least initially, the importance of selection for good genes, it has been argued that the two theories are not truly distinct (Kokko et al. 2006).
Despite the widespread assumption that indirect selection for sexual traits occurs, correlations between reproductive success and sexual trait quality are usually weak (Jennions et al. 2001), and clear examples are rare (e.g. Qvarnström et al. 2006). The shortage of empirical evidence of this kind may result from the fact that heavy investment in traits leads to reduced survival or parental investment (Kokko et al. 2006). A more fundamental problem with the good genes model of sexual selection becomes apparent when one considers that trait quality (which for the purposes of this paper we shall consider as synonymous with trait size) must ultimately be limited by natural selection (Darwin 1871). If genotypes that correspond to high quality and high viability are consistently chosen, such genotypes should come to dominate in the population, and genetic variation on which selection can act should be reduced to the point where the benefits of choosing are insignificant (the ‘lek paradox’; Borgia 1979; Kirkpatrick & Ryan 1991). Such a reduction in the heritability of trait characteristics and viability is expected to be particularly acute in poor environments. By contrast, in benign environments, in which most individuals survive regardless of genotype, the opposite problem might occur: selective pressure on genetic quality, and hence mate choice, might be very low (Wilson et al. 2006).
Recent efforts to understand the lek paradox have concentrated on the influence the environment may have on genetic expression (Greenfield & Rodriguez 2004; Danielson-François et al. 2006). Models have shown that strong genotype-by-environment interactions (GEIs) can generate an ‘ecological crossover’, where different genotypes are superior in environments that are separated either in space or time (Gillespie & Turelli 1989; Ellner & Hairston 1994; Qvarnström 1999; Danielson-François et al. 2006; Wilson et al. 2006, 2007). Kokko & Heubel (2008) have recently shown that, if there is limited mixing of reproductive individuals who have developed in different environments, ecological crossover, and weaker forms of GEI, can help maintain additive genetic variation in male viability and trait characteristics, and hence support the persistence of female preference. However, the same authors also show that extensive mixing of individuals from different developmental environments can lead to the opposite result: GEIs can make female choice less likely to evolve. The explanation for this lies in the fact that mixing of reproductive individuals whose development has been influenced by GEIs can blur the relationship between the individual male genetic quality and the phenotype as perceived by females. It is this ‘blurring’ that we explore in detail in this paper.
Using a stochastic simulation model, we examine the effect of weak GEIs, brought about by non-random mortality of some individuals during development (‘developmental selection’; Moller 1997), on the extent to which phenotype (trait size) is informative about male quality. Specifically, we test the hypothesis that when mortality during development is common, and is biased towards individuals of lower genetic quality, and when some individuals experience more severe developmental conditions than others, the positive relationship between genetic quality and trait size that would be expected in a homogeneous environment will be weakened and/or reversed.
Attempts to explore the impacts of genetic and environmental factors on sexual selection and mate choice often involve the simultaneous modelling of the evolution of male traits and female choice. Such models by necessity make assumptions about processes occurring in both ecological and evolutionary time, and it can be difficult to disentangle cause and effect. Here, we present a very simple model with which we seek to clarify the impact of GEIs on a single generation of trait-bearing males. We do not explicitly consider the consequences of GEIs on the coevolutionary dynamics of female mate choice and male trait size, and instead focus on the information content of the male ‘signal’, asking how environmental heterogeneity can erode the intuitive value of the ability of females to assess male phenotype. The results reveal patterns that appear to underlie the conclusions of recent coevolutionary modelling efforts.
2. The model
(a) Model construction
We constructed a stochastic model in which n individuals are born, and each individual i has a genetic quality ai at birth and is subjected to a local environment of harshness hi during development. Genetic ‘quality’ is here defined as both positively related to the ability to survive in the face of developmental stress and the ability to produce a large sexual trait, and therefore represents the base additive genetic effect on phenotype. We assume that each individual experiences a unique developmental environment, with both ai and hi being randomly sampled from a normal distribution with a mean of 10, and standard deviations σa and σh. We justify this assumption on the grounds that young animals are faced with stochasticity in a suite of interacting factors, such as microclimate as an egg or nutritional conditions in utero, damaging attacks by predators, infectious disease and food availability. No two individuals are likely to experience identical conditions as they develop.
It is possible to imagine the types of environmental variability that affect some individual genotypes in a population positively and some negatively, or in which the relationship between environment and survival or the development of sexual traits is strongly nonlinear. In our model, however, we consider the simple case of an axis in environmental parameter space that is monotonically related to survival and trait size for all genotypes. Hence, the probability of survival during development and the ability to produce a large sexual trait are both assumed to be negatively related to environmental harshness. The qualities of ‘quality’ and ‘harshness’ are necessarily abstract, and it is difficult to find empirical evidence to support the choice of specific functions for survival and trait size. Our approach is therefore to construct simple and generic but plausible relationships.
First, we assume that a simple sigmoidal function determines the probability of survival to maturity for each individual:
| (2.1) |
where α and β control the impact on the probability of survival of genetic quality and environmental harshness, respectively. This function describes a logistic equation where higher values of hi and ai cause a reduction and an increase in the probability of survival, respectively. In the simulations, individual i survives if Pi is greater than a uniformly distributed pseudorandom number between 0 and 1. Default values for all parameters are shown in table 1.
Table 1.
Default values of parameters used in simulations.
| description | symbol | default value |
|---|---|---|
| number of individuals | n | 5000 |
| mean genetic quality | 10 | |
| standard deviation of genetic quality | σa | 4 |
| mean environmental harshness | 10 | |
| standard deviation of environmental harshness | σh | 4 |
| standard deviation of trait size | σS | 0.2 |
| effect of quality on survival | α | 0.5 |
| effect of environment on survival | β | 0.5 |
| effect of quality on trait size | δ | 0.25 |
| effect of environment on trait size | λ | 0.1 |
Next, we calculate the size Si of the sexual trait of each individual, based on the genetic quality ai and the environmental harshness hi. Initially, we assume that the trait size is described by a exponential function that tends towards one and zero at high values of ai and hi, respectively, and we assume a multiplicative interaction between ai and hi:
| (2.2) |
where δ and λ control the impact on sexual trait size of genetic quality and environmental harshness, respectively. Both δ and λ were set not only to control the relative impact of quality and environment on trait size (as modifiers) but also to scale the various functions to equivalent ranges for trait size to enable comparisons among functional forms (i.e. zero to one over the ranges of a and h). Preliminary simulations suggested that our results were to some extent sensitive to the shape of this function (equation (2.2)). In the absence of clear empirical support for one particular functional form, we also chose to explore a number of alternatives (see below), but the first results we describe are from simulations employing equation (2.2), because this function was computationally convenient. In all the functions we used, trait size approaches an asymptote as genetic quality increases and the height of the asymptote is reduced by environmental harshness. The following two properties reflect our assumption: (i) the costs of a trait accelerate with increasing size, which is likely to be true of traits for which elaboration is opposed by natural selection, and (ii) the maximum trait size is determined in part by the environment. For example, the ornate, long tails found in males of a variety of bird species are commonly used in sexual displays and can impair flight (Andersson 1994). In this case, we expect that the impacts of increases in tail length on flight are greater for birds with longer tails than those with shorter tails. However, the tail length is ultimately constrained by the amount of energy and/or protein available to the bird, and hence by environmental quality. Measurable consequences of these phenomena in a natural population might be a negative skew in the distribution of tail lengths, reflecting a constraint on maximum tail size, and a difference in maximum tail size and the extent of this skew between populations experiencing benign and harsh conditions.
For the surviving individuals, we calculated the Pearson product–moment correlation coefficient r between the genetic quality ai and the trait size Si, which represents the reliability of the phenotype as an indicator of genetic quality in the population. When the environment has little or no effect on trait size, we would expect a strong positive correlation. As the effects of environmental stochasticity become more important, the prediction is that the correlation will weaken. Under certain conditions, the correlation may be so weak that the relationship is not significantly positive, implying that the trait is of no use to females in assessing the genetic quality. The value of r we report in the results is the mean correlation coefficient from 1000 replicate simulations. We also report the p value resulting from a sign test with the null hypothesis that the mean of r for a particular set of parameter values is not different from zero. Visual inspection of scatter plots under different combinations of parameter values suggested that it was reasonable to assume that the relationship between ai and Si was linear.
(b) Simulation results: effects of environmental and genetic heterogeneity
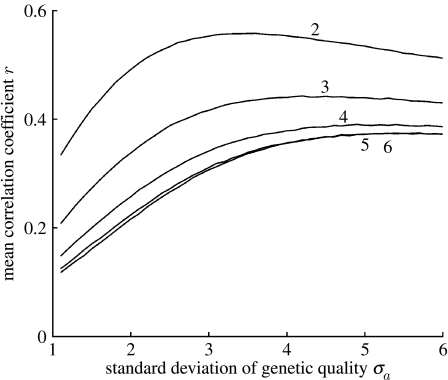
We explored the effect of varying the values of several parameters on the correlation between genetic quality and trait size, but we were most interested in the effect of varying the relative impact that environmental harshness has on sexual trait size. Initially, we explored the effect of altering the standard deviations of quality and environment, σa and σh, with all other parameters at default values (table 1). Simulations showed that as the heterogeneity of the environment increases, the positive correlation between genetic quality and trait size is disrupted (figure 1). As genetic variation in quality tends towards zero (σa=0), the correlation also tends towards zero. As σa increases, a positive relationship between genetic quality and trait size becomes increasingly likely, except at very high levels of genetic variation, where the relationship is weakened again, as a result of too much noise.
Figure 1.
Mean correlation coefficients r for the relationship between the genetic quality ai and the trait size Si in adult populations surviving mortality during development from 1000 simulations using a model with a multiplicative exponential function for Si (equation (2.2)), and a range of values for the standard deviation of genetic quality σa and standard deviation in environmental harshness σh. All other parameters values are shown in table 1. Numbers next to lines indicate values of σh.
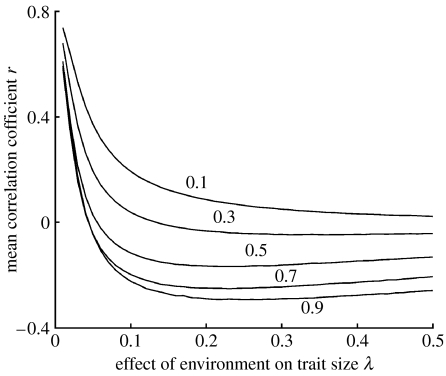
We next varied the effect of the environment on both survival β and trait size λ, keeping all other parameters at default values (table 1). Increasing both of these parameters made the correlation less positive (figure 2). When β is very low, high values of λ merely disrupt the positive correlation between genetic quality and trait size, but, when β is larger, large values of λ cause the correlation to be negative. In other words, females searching for a mate in this situation would be faced with a population of surviving adult males in which the higher quality individuals had smaller sexual traits.
Figure 2.
Mean correlation coefficients r for the relationship between the genetic quality ai and the trait size Si in adult populations surviving mortality during development from 1000 simulations using a model with a multiplicative exponential function for Si (equation (2.2)), and a range of values for the effects of the environment on survival β and trait size λ. Numbers next to lines indicate values of β.
In the version of the model for which we presented these results, trait size was assumed to be a deterministic function of λ and δ. We ran simulations that incorporated stochastic trait development, by treating the deterministic functions as the mean values, and randomly allocating trait size according to a standard deviation σs (table 1). These simulations produced qualitatively identical results (data not shown): the magnitude of all correlations was reduced, as might be expected by introducing extra stochasticity to the system.
(c) Simulation results: effects of the shape of the function determining trait size
In order to explore the robustness of our results under different assumptions about the shape of the relationship between the genetic quality, environmental harshness and trait size, we employed eight functional forms for the relationship between ai, hi and Si. The functions were of four categories—linear, exponential, proportional and power—representing a broad range of simple forms that have asymptotes. Within each category, the interaction between the effects of genetic quality and environmental harshness could be either additive or multiplicative. All the functions we used gave rise to a weak GEI, where the effect of ai on Si was reduced at high values of hi; the functions used are shown in figure 3. In each case, we explored the effect on the correlation between the genetic quality ai and the trait size Si of two varying parameters: β, the effect of the environment on survival; and λ, the effect of the environment on trait size. We varied β between 0.1 and 0.9 while keeping α, the effect of genetic quality on survival, fixed at 0.5. Hence, the environment could have a greater (β>0.5) or smaller (β<0.5) effect on survival than genetic quality.
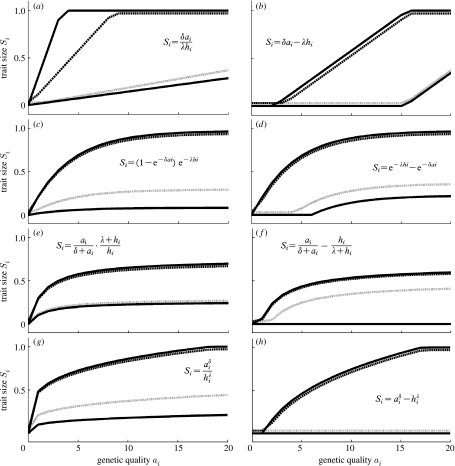
Figure 3.
Functional forms relating individual genetic quality ai and environmental harshness hi to trait size Si: (a,b) linear function, (c,d) exponential function, (e,f) proportional function and (g,h) power function. The effects of ai and hi are either (a,c,e,g) multiplicative or (b,d,f,h) additive. In each figure, the behaviour of the function concerned is displayed in four scenarios. The two solid lines show the relationship between ai and Si when the effect of the environment on trait size λ is at the extremes of the range that was explored (ranges given in table 2). The grey line shows the function at the value of λ for which the correlation across individuals between the genetic quality ai and the trait size Si in the population surviving development is the most negative. The broken line shows the function at the value of λ for which the correlation is the most positive (values shown in table 2). In all cases, environmental harshness hi=5, and the effect of genetic quality on trait size δ is as shown in table 2. All other parameter values are as shown in table 1.
We chose values for δ, the effect of genetic quality on trait size, which determined that, in a benign environment, the trait size Si for individuals of the highest genetic quality was approximately four times Si for individuals of the lowest genetic quality (full details of values used for this and other parameters that were varied are given in table 2). We employed values for λ, the effect of environment on trait size, which maximized the amount of parameter space explored. For the linear forms of the function determining Si, we assumed a maximum trait size of 1, and chose a range for λ that varied from negligible effects to dominant effects of the environment. For the exponential functional forms, we explored a range for λ that encompassed all increasing decelerating curves that did not approach an asymptote at very low values of Si. For the proportional functional forms, we varied λ up to the mean of ai, so that the shape of the slope varied between the immediately saturating and an almost straight line. For the power functional forms, we explored the whole range of increasing decelerating functions.
Table 2.
Mean (from 1000 simulations) minimum and maximum correlation coefficients r (all s.e.m.<0.001; not shown) for the observed correlation between trait size Si and genetic quality ai in populations composed of adults that have survived mortality during development. (Sign tests showed that all mean correlation coefficients differed significantly from zero (p≪0.001 in all cases). Results are given for eight different simulation models, each incorporating a different function to describe the effect of genetic quality and environmental harshness on trait size in individuals during development. Four functional types were employed, each with either additive (+) or multiplicative (×) effects of genetic quality and environmental harshness. The functions are shown in full in figure 3. For all models, the parameters controlling the effect of environment on survival β and trait size λ were varied systematically (range for β: 0.1–0.9; range for λ given in table). All other parameters were held at the default values shown in table 1, except for that controlling the effect of genetic quality on trait size δ, for which values are shown in this table and were chosen to maximize the impact of λ on r.)
| minimum correlation r | maximum correlation r | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| figure 3 panel | function | δ | λ range | β | λ | r | β | λ | r | |
| (a) | linear | × | 0.075 | 0.05–1.05 | 0.9 | 0.81 | −0.115 | 0.1 | 0.13 | 0.480 |
| (b) | linear | + | 0.075 | 0.034–0.23 | 0.9 | 0.23 | 0.102 | 0.1 | 0.034 | 0.919 |
| (c) | exponential | × | 0.25 | 0.006–0.5 | 0.9 | 0.246 | −0.296 | 0.1 | 0.006 | 0.767 |
| (d) | exponential | + | 0.25 | 0.006–0.5 | 0.9 | 0.204 | −0.192 | 0.1 | 0.006 | 0.766 |
| (e) | proportion | × | 1.5 | 0.2–10 | 0.9 | 9.2 | −0.078 | 0.1 | 0.2 | 0.284 |
| (f) | proportion | + | 1.5 | 0.2–10 | 0.9 | 4.6 | −0.153 | 0.1 | 9.6 | 0.395 |
| (g) | power | × | 0.25 | 0.02–1 | 0.9 | 0.54 | −0.102 | 0.1 | 0.02 | 0.966 |
| (h) | power | + | 0.25 | 0.02–1 | 0.9 | 0.84 | −0.111 | 0.1 | 0.02 | 0.978 |
We found significant negative correlations between genetic quality and trait size at certain combinations of parameter values (table 2) using seven of the eight functional forms shown in figure 3. In all cases, the correlation was most negative when the effect of the environment on survival β was the highest (0.9), and the most positive when β was the lowest (0.1). This is because decreasing β increases the relative importance of the effect of individual genetic quality on survival: when β is low, survivors tend to be individuals of the highest genetic quality.
Considering the effect of environment on trait size λ, the most positive correlations occurred when λ was at its smallest (table 2). This is because when the effect of the environment is negligible, trait size accurately reflects genotype. The most negative correlations occurred when λ was of intermediate magnitude; when λ is very high, there is too much noise and the correlation tends towards zero. In essence, only when the relative effect of genetic quality on trait size is sufficiently strong can any correlation occur. A negative correlation occurs when the high-quality individuals survive but mostly have small traits, and the low-quality individuals either have large traits (in benign environments) or do not survive.
3. Discussion
Our results show that the expected positive relationship between individual genetic quality and sexual trait size can be disrupted and even reversed by environmental effects on phenotypic expression and mortality during development. The relationship becomes negative only if the relative effect of genetic quality on trait size is smaller in more benign environments than in more harsh environments. In other words, the relationship between genetic quality and trait size is strongly affected by GEIs. Thus, our results help to explain why the evolution of (usually female) mate choice is sometimes made less likely by GEIs in the presence of environmental heterogeneity (Kokko & Heubel 2008). In this context, the inconsistent observed relationships between female preference and trait size (Andersson 1994; Griffith et al. 1999; Qvarnström 1999) are perhaps less puzzling than they have sometimes been considered to be.
Strong GEIs, such as those that result in ecological crossover, have been identified as being likely to disrupt the reliability of signals of male quality (Greenfield & Rodriguez 2004). In our model, we assume only a weak GEI (see for example, Hunt et al. 2004): superior genotypes are always superior, but the size of the selective advantage varies between environments. Kokko & Heubel (2008) have suggested that weak GEIs can produce similar patterns to those generated by ecological crossover and hence be detrimental to the evolution of female preference. Our results confirm that the weak GEIs are sometimes sufficient to completely disrupt any correlation between genetic quality and signal/trait size. This conclusion appears to be robust in the face of different modelling approaches. Kokko & Heubel's (2008) model considered only two types of environment, with only two alleles determining survival, in a deterministic framework. We modelled a large population of individuals, each with a different genotype and each experiencing a unique developmental environment, in a stochastic framework.
In this modelling exercise, we assumed that the individuals that have experienced different developmental environments come together to form well-mixed breeding populations. We made this assumption because our original objective was to explore the impact of stochastic micro-environmental variation on the reliability of sexual signals. Any well-mixed population of adults will contain individuals with varying experiences of developmental stress. For example, some individuals may have been unlucky enough to be exposed to serious diseases during development; some may have hatched or been born in particularly inclement weather; others may have suffered from reduced parental care after the death of a mother or father. In these circumstances, our results show that the effect of weak GEIs are generally disruptive of the relationship between genetic quality and trait size, and hence probably detrimental to the evolution of female preference. We did not consider, however, the variation among breeding populations or subpopulations that might result from environmental heterogeneity on a larger spatial or temporal scale. Kokko & Heubel (2008) have shown that such heterogeneity can actually promote the evolution of female preference if GEIs operate on individuals who develop in distinct environments and mix to only a limited extent when they become reproductively active.
The results allow us to make some predictions about the situations in which male traits should be particularly unreliable indicators of quality to females. These predictions make possible several empirical tests of the model. First, we expect that sexual traits will be less correlated with genetic quality when mortality during development is common. We might look for negative correlations in species where most individuals do not survive to maturity (e.g. insects), or in harsh seasons (e.g. Wilson et al. 2006). Second, the correlation should be less strong when the developmental environment is highly heterogeneous. This would be expected, for example, in species with dispersing embryos, eggs or juvenile stages, and where parental care is absent or minimal. In species with male parental care, sons may only be able to develop large traits if a benign environment is provided by parents. Third, quality and trait size should be less correlated when trait heritability is low relative to environmental effects. For example, where food availability is relatively stochastic and more a result of good fortune than foraging skill, environmental effects may dominate expression of traits. Finally, in all the above situations, in which GEIs have the greatest potential to erode the information value of male traits, we would expect the evolution of female choice to be inhibited (Kokko & Heubel 2008).
Our model is very simple and therefore involves several simplifying assumptions. First, we have assumed that there is zero covariance between the genetic quality and environmental harshness: higher quality genotypes do not generally experience higher quality environments. It is not difficult to envisage a situation where this assumption may not hold, such as where parental quality plays a role in determining the environment into which offspring are born, or when parental effort determines food availability. It would thus be interesting to explore the impact of the relaxation of this assumption on the role of GEIs in the development of phenotypic traits. Second, we have assumed that genetic quality determines both survival and trait size. That is, we assume that, in the absence of environmental heterogeneity, traits are perfect indicators of the ability to survive development. Reduction in the genetic covariance between trait size and survival could have interesting consequences for predictions of models of this type. For example, if poor males can increase their reproductive success by investing proportionally more in trait size, the reliability of traits as indicators of genetic quality would be further reduced.
Our model highlights the importance of developmental selection in the evolution of traits. We assume that a significant proportion of individuals die before maturity. Evidence indicates that this is a robust assumption: in most species studied, mortality rate is the highest in neonates (e.g. Wilson et al. 2006). Such empirical results are likely to be underestimates, since mortality very early in development is easily missed. In the context of our study, even mortality of eggs or embryos in the womb has the potential to affect the relationship between the genetic quality and the trait size. Our results show that all such mortality has the potential to play a role in the erosion of the information content of traits that might otherwise have usefulness as honest indicators of male quality. Intuitively, we can see that this is likely to have an important impact on the evolution of female mate choice. Although further work is needed to explore the consequences fully, recent evidence is indeed that GEIs of the sort described by our model make mate choice less likely to evolve (Kokko & Heubel 2008). However, we can speculate that, in many cases, selection for trait size might proceed very slowly when the correlation between the genetic quality and the trait size is weakened in this way. Indeed, since our results show that the usefulness of a trait as an indicator of genetic quality decreases with variance in the population, our mechanism might provide a ‘brake’ on the erosion of genetic variance. Furthermore, in cases of extreme environmental heterogeneity or stochasticity in developmental selection, selection may be unable to occur at all. As a result, females might be reduced to selecting for direct benefits only, and hence the role of indirect benefits in mate choice may have been overstated in some mating systems.
Acknowledgments
We thank John Brookfield, Francis Gilbert, Jan Lindström, Alan McElligott and Graeme Ruxton for stimulating discussion and insight, Hanna Kokko and three anonymous referees for critically reading the manuscript, and Markus Owen for mathematical advice. A.D.H. was supported by NERC grant no. NE/E016626/1 awarded to Graeme Ruxton. This work is dedicated to the memory of Prof. Chris Barnard.
References
- Andersson M. Princeton University Press; Princeton, NJ: 1994. Sexual selection. [Google Scholar]
- Borgia G. Sexual selection and the evolution of mating systems. In: Blum M.S., Blum N.A., editors. Sexual selection and reproductive competition in insects. Academic Press; New York, NY: 1979. pp. 19–80. [Google Scholar]
- Danielson-François A.M., Kelly J.K., Greenfield M.D. Genotype×environment interaction for male attractiveness in an acoustic moth: evidence for plasticity and canalization. J. Evol. Biol. 2006;19:532–542. doi: 10.1111/j.1420-9101.2005.01006.x. doi:10.1111/j.1420-9101.2005.01006.x [DOI] [PubMed] [Google Scholar]
- Darwin C.R. John Murray; London, UK: 1871. The descent of man and selection in relation to sex. [Google Scholar]
- Ellner S., Hairston N.G. Role of overlapping generations in maintaining genetic variation in a fluctuating environment. Am. Nat. 1994;143:403–417. doi:10.1086/285610 [Google Scholar]
- Emlen D.J. Environmental control of horn length dimorphism in the beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae) Proc. R. Soc. B. 1994;256:131–136. doi:10.1098/rspb.1994.0060 [Google Scholar]
- Fisher R. Oxford University Press; Oxford, UK: 1930. The genetical theory of natural selection. [Google Scholar]
- Gillespie J.H., Turelli M. Genotype–environment interactions and the maintenance of polygenic variation. Genetics. 1989;121:129–138. doi: 10.1093/genetics/121.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield M.D., Rodriguez R.L. Genotype–environment interaction and the reliability of mating signals. Anim. Behav. 2004;68:1461–1468. doi:10.1016/j.anbehav.2004.01.014 [Google Scholar]
- Griffith S.C., Owens I.P.F., Burke T. Environmental determination of a sexually selected trait. Nature. 1999;400:358–360. doi:10.1038/22536 [Google Scholar]
- Hunt J., Bussiere L.F., Jennions M.D., Brooks R. What is genetic quality? Trends Ecol. Evol. 2004;19:329–333. doi: 10.1016/j.tree.2004.03.035. doi:10.1016/j.tree.2004.03.035 [DOI] [PubMed] [Google Scholar]
- Jennions M.D., Møller A.P., Petrie M. Sexually selected traits and adult survival: a meta-analysis. Q. Rev. Biol. 2001;76:3–36. doi: 10.1086/393743. doi:10.1086/393743 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Ryan M.J. The evolution of mating preferences and the paradox of the lek. Nature. 1991;350:33–38. doi:10.1038/350033a0 [Google Scholar]
- Knell R.J., Fruhauf N., Norris K.A. Conditional expression of a sexually selected trait in the stalk-eyed fly Diasemopsis aethiopica. Ecol. Entomol. 1999;24:323–328. doi:10.1046/j.1365-2311.1999.00200.x [Google Scholar]
- Kokko H., Heubel K. Condition-dependence, genotype-by-environment interactions and the lek paradox. Genetica. 2008;132:209–216. doi: 10.1007/s10709-007-9166-1. doi:10.1007/s10709-007-9166-1 [DOI] [PubMed] [Google Scholar]
- Kokko H., Jennions M.D., Brooks R. Unifying and testing models of sexual selections. Annu. Rev. Ecol. Evol. Syst. 2006;37:43–66. doi:10.1146/annurev.ecolsys.37.091305.110259 [Google Scholar]
- Moller A.P. Developmental selection against developmentally unstable offspring and sexual selection. J. Theor. Biol. 1997;185:415–422. doi:10.1006/jtbi.1996.0332 [Google Scholar]
- Qvarnström A. Genotype-by-environment interactions in the determination of the size of a secondary sexual character in the collared flycatcher (Ficedula albicollis) Evolution. 1999;53:1564–1572. doi: 10.1111/j.1558-5646.1999.tb05419.x. doi:10.2307/2640901 [DOI] [PubMed] [Google Scholar]
- Qvarnström A., Brommer J.E., Gustafsson L. Testing the genetics underlying the co-evolution of mate choice and ornament in the wild. Nature. 2006;441:84–86. doi: 10.1038/nature04564. doi:10.1038/nature04564 [DOI] [PubMed] [Google Scholar]
- Spencer K.A., Buchanan K.L., Goldsmith A.R., Catchpole C.K. Developmental stress, social rank and song complexity in the European starling (Sturnus vulgaris) Proc. R. Soc. B. 2004;271:S121–S123. doi: 10.1098/rsbl.2003.0122. doi:10.1098/rsbl.2003.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.J., Pemberton J.M., Pilkington J.G., Coltman D.W., Mifsud D.V., Clutton-Brock T.H., Kruuk L.E.B. Environmental coupling of selection and heritability limits evolution. PLoS Biol. 2006;4:1270–1275. doi: 10.1371/journal.pbio.0040216. doi:10.1371/journal.pbio.0040216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.J., Pemberton J.M., Pilkington J.G., Clutton-Brock T.H., Coltman D.W., Kruuk L.E.B. Quantitative genetics of growth and cryptic evolution of body size in an island population. Evol. Ecol. 2007;21:337–356. doi:10.1007/s10682-006-9106-z [Google Scholar]
- Zahavi A. Mate selection—a selection for a handicap. J. Theor. Biol. 1975;53:205–214. doi: 10.1016/0022-5193(75)90111-3. doi:10.1016/0022-5193(75)90111-3 [DOI] [PubMed] [Google Scholar]