Abstract
Genetically correlated traits do not evolve independently, and the covariances between traits affect the rate at which a population adapts to a specified selection regime. To measure the impact of genetic covariances on the rate of adaptation, we compare the rate fitness increases given the observed G matrix to the expected rate if all the covariances in the G matrix are set to zero. Using data from the literature, we estimate the effect of genetic covariances in real populations. We find no net tendency for covariances to constrain the rate of adaptation, though the quality and heterogeneity of the data limit the certainty of this result. There are some examples in which covariances strongly constrain the rate of adaptation but these are balanced by counter examples in which covariances facilitate the rate of adaptation; in many cases, covariances have little or no effect. We also discuss how our metric can be used to identify traits or suites of traits whose genetic covariances to other traits have a particularly large impact on the rate of adaptation.
Keywords: genetic covariances, genetic correlations, constraints, adaptation, phenotypic evolution
1. Introduction
It is well-known that traits do not evolve independently of one another. In evolutionary quantitative genetics, the genetic covariance serves as a measurable summary statistic that captures the effects of pleiotropy and linkage disequilibrium, and quantifies the extent to which the evolutionary response of one trait will be influenced by selection of another (Falconer & Mackay 1996; Lynch & Walsh 1998). Specifically, the change in the mean multivariate phenotype across a single generation is given by
| (1.1) |
where is a column vector of changes in the means of n traits (the T denotes transpose); G is the additive genetic (co)-variance matrix with diagonal elements representing genetic variances and off-diagonal elements representing genetic covariances (e.g. element Gii is the genetic variance of trait i and element Gij is the genetic covariance between traits i and j), and is a column vector of directional selection gradients (Lande 1979; Lande & Arnold 1983). Despite the availability of a robust theoretical framework describing the evolution of multiple quantitative traits, the extent to which—if at all—the covariance structure among traits constrains or facilitates multivariate evolution is unknown. Here, we suggest a simple and intuitively meaningful way to quantify the effects of genetic covariances on the rate of adaptation, and apply this metric to datasets from the literature.
The effects of covariances between traits on the response to artificial selection has long been appreciated by plant and animal breeders. For example, in index selection, multiple traits are combined via a weighting scheme into a single univariate trait (the index) which is itself selected upon (Falconer & Mackay 1996). The creation of a trait index specifically incorporates the covariances between the component traits, and weighting schemes can be designed to take advantage of these covariances and increase the response to selection of a preferred trait or set of traits (see Lin 1978; Falconer & Mackay 1996; B. Walsh & M. Lynch 2007, unpublished data, for overviews).
Using the Lande–Arnold framework (Lande 1979; Lande & Arnold 1983), the importance of genetic covariances in determining the response to natural selection can be illustrated by considering a simple two trait example, such that (1.1) can be written as
| (1.2a) |
| (1.2b) |
If z1 and z2 are under equally strong positive directional selection (β1=β2=β), and the two traits have equal amounts of genetic variance G11=G22=G, equation (1.2a) and (1.2b) simplifies to
| (1.3) |
where ρ12=G12/√(G11G22) is the genetic correlation between z1 and z2. If there is a maximally strong negative genetic correlation between these traits, ρ12=−1, then neither trait evolves despite there being genetic variance for both the traits. Examples such as this illustrate the potential of genetic correlations to constrain evolution. However, the example above is misleading in several respects. First, this example suggests that extremely strong genetic correlations (|ρ|≈1) are required to substantially constrain evolution. Given that empirically estimated genetic correlations are typically much less than ±1, one might be tempted to assume that genetic correlations do not pose a strong constraint on adaptation. However, correlations that are much smaller in magnitude than unity can prevent adaptation when there are more than two traits under selection (Dickerson 1955; Blows & Hoffmann 2005). Ultimately, the important quantity is not the genetic correlations per se but rather the amount of genetic variation in multivariate space in the direction of selection. Even if there is genetic variation in all n traits, this variation may be structured (as described by the co-variances) such that there is no variation in certain directions in multivariate space (i.e. the rank of G is less than n; Hine & Blows 2006). A complete lack of genetic variance in one or more directions can occur even when all genetic correlations are less than unity in magnitude when n>2. If one of these directions with no genetic variation matches the direction of selection (β), then there will be no evolution. In such a case, we can say that genetic correlations provide an absolute constraint to adaptation.
A second way in which our toy example is misleading is that it focuses on a case where no evolution occurs at all. The genetic correlation must be at its maximum negative value, ρ12=−1, to completely constrain evolution, as this is the only case in which there would be no genetic variation in the direction of selection. While a strict definition of the term ‘constraint’ might only apply to situations in which genetic correlations lead to no evolutionary response, this seems an overly stringent definition (Houle 1991). Any other value for the genetic correlation (e.g. ρ12=−0.5) will still allow for some adaptation—a fact noted by several investigators who have applied artificial selection to genetically correlated traits and still obtained a response to selection. These results come from agricultural systems (poultry: Nordskog 1977), model systems (mice: Cockrem 1959; Rutledge et al. 1973) and evolutionary and ecological systems (flour and dung beetles, butterflies, and short-lived plants: Bell & Burris 1973; Stanton & Young 1994; Emlen 1996; Beldade et al. 2002; Conner 2003; Frankino et al. 2005). As such, the relevant comparison is not whether any adaptation can occur, but how much occurs relative to a scenario with a different covariance structure. For example, when the genetic correlation between two traits is moderate (e.g. ρ12=−0.5), there is only half as much genetic variance in the direction of selection as there would be if there was no genetic correlation. Consequently, both traits evolve only half as much as with ρ12=−0.5 as they would if ρ12=0. Clearly, the genetic correlation constrains the rate of adaptation relative to what would be expected if the traits did not genetically covary. This idea forms the premise for our metric of constraint described below.
A third way in which the toy example is misleading is that it focuses on a particular genetic correlation in combination with a particular pattern of selection. Specifically, when ρ12=−1, there is no variation in the direction of selection given by β={β, β} and evolution is completely prevented. However, this same genetic correlation would have a very different effect if selection was in a different direction. For example, if β={β, −β}, both traits not only evolve but do so twice as fast with ρ12=−1 as they would if there was no genetic correlation. With this alternative pattern of selection, the genetic correlation increases, rather than reduces, the rate of adaptation. In much of the literature, there has been a focus on the role of genetic correlations as a source of constraint (Clark 1987; Arnold 1992) but this emphasis can be deceptive without the appropriate caveats. Any given pattern of genetic correlations among a set of traits can either constrain or facilitate adaptation depending on the pattern of selection (Lande 1979).
Moreover, the toy example focuses too much attention on the genetic correlation, given that genetic correlations alone do not dictate the pattern of genetic variance in phenotypic space; genetic variances are also important. For example, in the toy example with equal variances (G11=G22), a genetic correlation of ρ12=−1 causes all the genetic variance to lie in a dimension orthogonal to the chosen direction of selection, β={β, β}. However, if trait variances were unequal and ρ12=−1, then all the variation would still lie only in one dimension (i.e. G has a single non-zero eigenvector), but this dimension would not be orthogonal to selection. Accordingly, a correlation of ρ12=−1 would not completely constrain evolution. As shown in equations (1.2a) and (1.2b), it is the genetic covariance that affects the response to selection. The effect of the genetic correlations on evolution is always dependent on trait variances because correlations only enter the equation through covariances (e.g. ). One can consider the effect of non-independence between traits on evolution by focusing directly on genetic covariances rather than the correlations.
While the concepts above are well-known to those familiar with quantitative genetics theory, the extent to which genetic covariances affect the evolution of real populations is unknown, in part because of the lack of a useful metric for quantifying these effects. Here, we present a simple metric for quantifying the extent to which genetic covariances affect the rate of adaptation. The impact of genetic covariances is measured by comparing the rate of adaptation with the observed G matrix to the rate of adaptation if all traits were genetically independent (i.e. all covariances set to zero). This simple approach measures the effect of genetic correlations while automatically accounting for all of the issues listed above (e.g. direction of selection relative to patterns of correlation, differences among traits in the amount of genetic variance).
Our metric is not intended to be a summary statistic of constraint in general. Obviously, a crucial aspect of constraint is the amount of genetic variation; adaptation will be constrained if traits have little or no genetic variation. Our metric does not address whether individual traits have particularly high or low levels of genetic variation (Mousseau & Roff 1987; Houle 1992; Hansen & Houle 2008). Rather, our metric addresses a specific question that focuses on genetic covariances: given the observed levels of genetic variance, how much does the covariance structure among these traits affect the rate of adaptation? We apply this metric to the data from the literature on real populations. We find some examples in which covariances strongly constrain adaptation and others where correlations facilitate adaptation. However, in many cases, the effect of covariances is small and the average effect is close to zero.
2. Material and methods
(a) Quantifying constraint
If there are no genetic correlations (or if selection acts only on one trait), then evolutionary change in trait i is simply the product of the genetic variance in that trait (Gii) and the selection gradient (βi)
| (2.1) |
Genetic covariances alter the response to selection from the simple univariate prediction shown in equation (2.1) to that given by equation (1.1). Genetic correlations are of interest because of their ability to change evolutionary trajectories relative to the patterns expected by considering each trait individually (as in equation (2.1)). Consequently, we suggest that the simplest way to quantify the effect of genetic correlations is to calculate the rate of adaptation (given the observed pattern of genetic covariances) relative to the rate of adaptation expected if there were no correlations.
For our purposes, we define the rate of adaptation as the rate of increase of fitness of the mean phenotype. Assuming fitness can be approximated by a quadratic function, the fitness of an individual with phenotype z={z1, z2, …, zn}T is
| (2.2) |
where γ is the matrix of quadratic selection gradients (Lande & Arnold 1983). The change in the fitness of the mean phenotype is given by
| (2.3a) |
Making the usual assumption that phenotypes are measured as deviations from the mean (i.e. ), then equation (2.3a) becomes
| (2.3b) |
To quantify the effect of covariances on rate of adaptation, we measure the ratio
| (2.4) |
is calculated from equations (2.3a) and (2.3b) using the value of given by equation (1.1) and therefore represents the rate of adaptation with the observed pattern of genetic covariance. is also calculated from equations (2.3a) and (2.3b) but using the value of given by equation (2.1) or equivalently by equation (1.1) but setting all off-diagonal elements in G to zero—this is the predicted rate of adaptation with the observed level of genetic variance in each trait but assuming all traits were genetically independent (ρij=0). In other words, R measures the rate of adaptation with covariances relative to the expected rate without them. Importantly, this metric R accurately quantifies evolutionary constraints in situations in which one or more of the traits are under weak or no selection but other traits are under strong selection. In principle, bootstrapping the underlying datasets for G and phenotypic selection parameter estimates (β and γ) could be used to generate confidence intervals on R.
The metric R is intuitively simple and biologically meaningful. If R=0, then adaptation is completely stalled by genetic correlations—there is an absolute constraint. If R=0.5, then the covariance structure causes the fitness of the mean phenotype to increase only 50 per cent as quickly as expected if the traits were genetically independent. If R=2, then genetic covariances accelerate evolution such that adaptation occurs twice as fast as expected under genetic independence. In some contexts, we use
L<0 implies that covariances constrain the rate of adaptation, whereas L>0 implies the opposite. L is helpful because it is easier to compare cases where covariances constrain versus facilitate the rate of adaptation. Consider two cases, the first in which covariances reduce the rate of adaptation by a factor of two (R=1/2) and the second in which covariances increase the rate of adaptation by a factor of two (R=2). It is easier to see that these situations are equal but opposite by comparing their L-values, which are −0.0301 and +0.0301, respectively.
Our metric focuses on the change in fitness of the mean phenotype . In principle, a better alternative would be to measure the change in mean fitness . However, calculation of the change in the mean fitness requires a prediction of not only the change in the mean phenotype (equation (2.1)) but also a prediction of how G will change. There is no consensus on how G changes and theory indicates that its evolution is quite sensitive to the underlying genetic architecture (Turelli 1988; Arnold 1992). To avoid these practical difficulties, we focus on rather than . The two approaches will be very similar provided that the curvature in the fitness surface is weak.
(b) Assessing constraint from empirical datasets
As mentioned above, basic quantitative genetics theory makes no prediction about whether genetic covariances should tend to facilitate or constrain the rate of adaptation. However, there are several secondary ideas that are relevant. If there is constant directional selection, then genetic variation might eventually be exhausted in the selected direction (Lande 1982; Charnov 1989; Charlesworth 1990). The remaining variation will primarily reside in subspaces orthogonal to selection. In other words, there may be variation in all of the traits but the genetic correlations constrain further adaptation, i.e. R<1.
Alternatively, consistent long-term selection, particularly if there is stabilizing selection in dimensions orthogonal to the vector of directional selection, may drive the evolution of the covariance structure itself to match the selection surface (Cheverud 1982, 1984; Wagner 1989), such that correlations should tend to facilitate the rate of adaptation, i.e. R>1.
A third alternative is based on condition-dependence. If most traits experiencing directional selection are condition-dependent, then they will all be correlated with an underlying latent variable, ‘condition’. As others have argued (Houle 1991; Rowe & Houle 1996), the mutational target size for condition is very large, allowing considerable variation for condition despite it experiencing persistent directional selection. Because the key traits are condition-dependent, the correlations among them will align with the direction of selection. Again, R>1 is predicted (though little or no adaptation actually occurs because deleterious mutation counteracts the response to selection each generation).
To assess whether covariances tend to constrain or facilitate the rate of adaptation, we surveyed the literature for systems in which we could calculate R (see the electronic supplementary material for details). Ideally, a system would have both a good measure of the genetic (co)variance matrix, G, as well as good estimates of the selection parameters, β and γ. Unfortunately, good estimates of either genetics or selection are extremely difficult to obtain and we found few systems with both. To increase the number of systems we could use, we accepted the phenotypic matrix of (co)variance, P, as a substitute for G. Of course, the relative size of elements of P can differ from the corresponding elements of G (Willis et al. 1991). However, in many cases, the shape of P will be a reasonable reflection of the shape of G (Cheverud 1988; Roff 1995; Waitt & Levin 1998).
In some cases, estimates of heritabilities (but not genetic correlations) were available in addition to P. In these cases, we adjusted the matrix for different levels of genetic variability among the traits by multiplying the element in row i and column j of P by . When no heritabilities were available, we assumed a heritability of h2=0.5 (Roff 1997). When all traits are assigned the same heritability, the actual value of the heritability assumed will have little effect on the measure of R unless nonlinear selection gradients are strong.
We used studies reporting variance standardized selection gradients (Lande & Arnold 1983), or variance standardized selection gradients estimated with breeding values or family means, which are unbiased by environmental covariances between traits and fitness (Rausher 1992; Stinchcombe et al. 2002). (Because we were using variance standardized selection gradients, we ensured that our covariance matrices were also for variance standardized traits.) Many studies of selection either do not attempt to estimate nonlinear selection gradients, γ, or only estimate the diagonal elements of this matrix. We typically used what was available and assumed other elements to be zero. Some studies reported different measures of fitness for analysing fitness and we chose the one we thought best. A number of studies measured selection at multiple sites or in multiple years. In such cases, we calculated R for each selection estimate separately and used the geometric average value of R. Throughout the process of collating the data, we were required to make a number of judgement calls in regard to specific papers. In the electronic supplementary material, we provide additional notes on how each individual study used here was handled.
We were able to compile 45 datasets for which we could calculate R (see the electronic supplementary material). This included a wide range of plant and animal species. Most of these studies estimated selection in the field, although a few involved studies of selection in a laboratory or greenhouse setting intended to mimic some aspect of nature. A variety of fitness measures were used across these studies including viability, fecundity and mating success. A wide variety of traits were considered. Most were morphological, though there were some life-history traits too.
As explained above, we used the best available data for each study. This led to a fair degree of heterogeneity in the type of data used. In some cases G was available but in other cases P was used as proxy for G; in some cases nonlinear selection gradients were available but in other cases they were partially or completely absent. In order to have a more homogeneous dataset, we repeated our analysis using a ‘lowest common denominator’ (LCD) approach in which we used P as proxy for G for all datasets and only considered linear selection gradients. There were 11 studies that were excluded from this analysis because no P matrix data were available. The results using the best-available data and the LCD data were similar.
3. Results
(a) Do observed covariances affect the rate of adaptation?
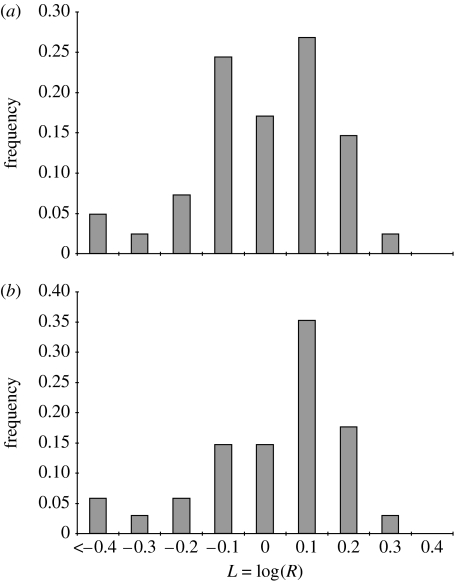
The distribution of observed L-values is shown in figure 1. There is a clear mode corresponding to the values close to L=0 (i.e. R=1). In many of datasets we examined, the covariances did not alter the rate of adaptation by more than 10 per cent in either direction. However, there were some examples with much larger effects. In 12/45 (27%) of studies, the patterns of covariance reduced the rate of adaptation by more than 30 per cent. In 7/45 (16%) of studies, the patterns of covariance increased the rate of adaptation by more than 30 per cent. The geometric mean value of R over all of the studies is (i.e. ), indicating that covariance patterns, on average, reduce the rate of adaptation by 11 per cent. Using the LCD data (figure 1b), ().
Figure 1.
Frequency histogram of L-values for surveyed studies where L=log(R). R measures the rate of adaptation relative to the rate expected if all traits were genetically independent. Negative and positive values for log(R) indicate cases where genetic covariances constrain and facilitate the rate of adaptation, respectively. (a) Distribution based on best available data (n=45). The mean of this distribution is (i.e. ) but this is not significantly different from . (b) Distribution based on data using the lowest common denominator (LCD) (see text for details; n=34). For this, distribution is (i.e. ). See the electronic supplementary material for a list of surveyed studies.
R provides some insight to the relationship between G and β with respect to the rate of adaptation. As discussed above, basic quantitative genetics makes no prediction with respect to this relationship but subsequent evolutionary perspectives predict particular types of relationships between G and β to evolve such that R will tend to be negative or positive.
We tested whether properties of the observed distribution of R values (mean and variance) differ from that expected when there is no relationship between G and β using the following approach. We paired each of our observed G-matrices (or P-based approximations of G) with randomly generated β vectors and calculated a pseudo- value. Each random β was scaled to be the same length as the corresponding observed β. We repeated this process 5000 times to generate a distribution of pseudo- values. (For simplicity, we always used the observed γ values in creating these pseudo- values because γ values were missing or incomplete in most studies and γ will tend to be much less important than β with respect to R unless nonlinear selection is quite strong. We also performed all of these analyses assuming no nonlinear selection and results were similar.) Our observed was not significantly different from that distribution of pseudo- values; p=0.83. We observed a qualitatively similar result using the LCD data; p=0.88.
The small mean effect of covariances ( or ) occurs because large positive values of L are balanced by large negative values rather than because all values are close to zero. We tested whether the observed variance in L was different from what would be expected by chance. Our observed variance in L was not significantly different from expected when our observed G matrices were paired with randomly generated β vectors rather than our observed β vectors (p<0.24; p<0.35 for LCD data). In summary, neither the mean nor the variance of our observed distribution was different from expected by chance.
(b) The potential for covariances to affect the rate of adaptation
While there is little evidence that the observed covariances affect the rate of adaptation, they do not address whether the observed covariances or selection gradients have the potential to affect the rate of adaptation (i.e. R≠1). Two necessary (but not sufficient) conditions for correlational structure to affect adaptation are that: (i) traits must be correlated and (ii) selection must act upon multiple traits. Below, we describe and verify the usefulness of an approach for quantitatively assessing these conditions, and then apply this approach to the data gathered from our literature survey.
It is possible for either selection or the covariances between traits to be structured in a way that there is little potential for covariances to affect the rate of adaptation. For example, if selection is primarily on a single trait (or on a low proportion of traits), then all (or most) covariances will be of little importance (i.e. most possible sets of correlations would have little effect on R with such selection). Conversely, when the strength of selection is more evenly distributed across traits then there should be high potential for covariances to affect the rate of adaptation. Thus, we would expect that the potential for a specified pattern of directional selection to be affected by the pattern of covariances to depend on the evenness in selection across traits. Evenness of β can be calculated analogously to the Shannon–Wiener index for species diversity and evenness
| (3.1) |
where is the magnitude of directional selection on trait i relative to the total amount of directional selection on all traits. For the calculations involving Eβ (and Eλ described below), selection estimates and G were scaled such that the traits were measured on scale where the genetic variances of all traits were unity.
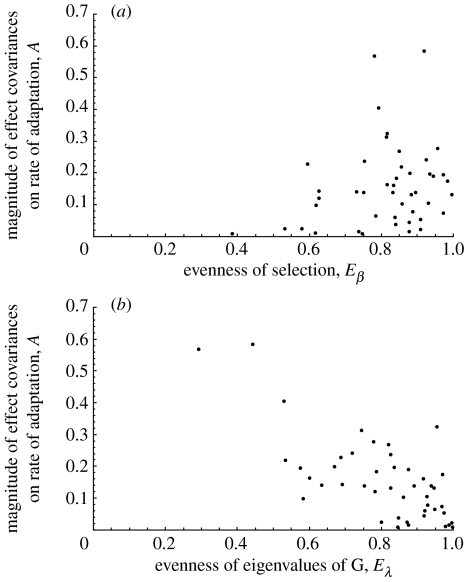
The evenness of β, Eβ, should be positively correlated to A where A=|L|=|log(R)|. (Recall that if covariances have a strong effect on the rate of adaptation, R≠1 and L≠0, and |L| will be larger for cases in which correlations have a strong effect on the rate of adaptation, regardless of the direction of that effect.) As expected, there is a positive correlation between Eβ and A, ρ=0.24 (figure 2a).
Figure 2.
Assessing the potential for covariances to affect the rate of adaptation. (a) The evenness of selection, Eβ, is plotted against A=|log(R)|. The correlation between Eβ and A is positive (ρ=0.24), indicating that when selection is more evenly distributed across traits, covariances are more likely to affect the rate of adaptation. (b) The evenness of the eigenvalues of the G matrix, Eλ, is plotted A. The negative correlation (ρ=−0.71) confirms that the rate of adaptation is more likely to be altered in cases where correlations cause genetic variation to be more abundant in some directions than others (Eλ≪1). See text for details.
The second necessary condition for covariance structure to affect the rate of adaptation is that traits must covary. If traits are very weakly correlated, then there would be little potential for these correlations to affect the rate of adaptation, regardless of the nature of selection. We can get some sense of the overall strength of correlational structure by examining the eigenvalues of the genetic correlation matrix (i.e. G with all traits scaled to have unit variances). When traits are scaled such that the genetic variance in each trait is unity, the n eigenvalues (λ) for an n-dimensional G matrix will all be equal if there are no genetic correlations. (The vector of eigenvalues is represented by λ.) If traits are correlated, then there will be more variation in some multivariate directions than in others, and the eigenvalues will differ. As we have done with β, we can measure the evenness of eigenvalues as
| (3.2) |
where . Low values of Eλ occur when correlations exist such that there is more variance in some directions of multivariate space than others, while high values of Eλ suggest that genetic variance is evenly distributed among the measured traits. As expected, there is a negative correlation between Eλ and A, ρ=−0.71 (figure 2b).
The results above confirm that the evenness of β and G (Eβ and Eλ) are useful measures of the potential for a given β or a given G to affect R. We assessed whether our datasets had less potential to show values of R different from unity than expected by chance by evaluating the mean evenness of βs. The mean evenness of βs in our datasets was . This value was not significantly different from the mean evenness value obtained from randomly generated βs (p<0.74, i.e. pseudo values were calculated from 5000 sets of randomly generated βs of equal dimensionality to the original datasets; the frequency of these pseudo-values that were larger than our observed value of was 1850/5000). In principle, one could perform an analogous test on but it is unclear what biological model could be used for generating the random G matrices required to create the null distribution.
It should be noted that estimated values of Eλ will be downwardly biased because sampling error causes estimated eigenvalues to be overdispersed. This bias will be small for phenotypic correlation matrices estimated from moderate sample sizes but could be quite large for genetic correlation because of the high sampling variance. In general, evenness values should be used only as a heuristic for thinking about the potential of a G matrix to cause constraint and should be regarded with caution given the bias in their estimation.
Alternatively, the potential for a specified G matrix to affect the rate of adaptation can be assessed in a different way that does not rely on the evenness of eigenvalues. Matrix theory shows that the ratio xTAx/xTBx must lie between the minimum and maximum eigenvalues of B−1A, where x is a vector and A and B are matrices (Gantmakher 1960, p. 318). If selection is linear, we can write our ratio of interest as R=βTGβ/βTG*β, where G* is G with the covariances set to zero. Thus, R must lie between the minimum and maximum eigenvalues of M, where M=(G*)−1G. (Note that we can also express this matrix as M=σ−1Cσ where σ is a diagonal matrix containing the square roots of the genetic variances (i.e. genetic standard deviations) and C is the genetic correlation matrix. Thus, the elements of this matrix are , where ρij is the genetic correlation between traits i and j.) The bounds for R given by the range of eigenvalues of M give the minimum and maximum potential effect of covariances on the rate of adaptation for any possible pattern of linear selection on a G matrix of interest.
4. Discussion
Evolutionary biologists have been interested in genetic covariances because they can alter the response to selection from the simple univariate prediction. A genetic correlation provides a pathway that allows the evolution of one trait to be influenced by selection acting on another trait. Such correlations can either facilitate or restrain the rate of adaptation. Our primary goal was to provide a metric for quantifying the net effect of all the correlations among a set of traits, holding the trait variances constant.
For any pair of traits, zi and zj, it is simple to determine whether the correlation between them will have a positive or negative effect on the rate of adaptation. If the correlation is of the same (opposite) sign as the product of the directional selection gradients, βi×βj, the effect of the correlation will be positive (negative). Using this approach, one can consider the qualitative effect of each correlation individually (Etterson & Shaw 2001). However, this approach does not provide a quantitative measure of the importance of the correlation. Nor does it allow the joint effects of all correlations among a set of traits to be assessed simultaneously.
When n traits are under consideration, there will be n(n−1)/2 genetic correlations. Some of these may facilitate the rate of adaptation and while others may constrain it not all of them will affect the rate of adaptation equally. Correlations that are large in magnitude or involve traits that are under strong selection, or both, will be more important than correlations that are weak in magnitude and/or involve traits that are only weakly selected. The metric R provides the net effect of all correlations, appropriately weighting important ones more so than unimportant ones. R measures effect of the correlations in terms of fitness, allowing for comparisons to be made between different populations or different species.
Although R quantifies the net effect of all correlations, it is also possible to slightly modify this approach to identify individual traits whose correlations to other traits are particularly important in affecting the rate of adaptation. For example, suppose we are interested in assessing the extent to which trait j alters the rate of adaptation through its genetic correlations with other traits. This can be accomplished by calculating , where is the change in the fitness of the mean phenotype assuming that all genetic covariances involving trait j are zero. Specifically, is calculated from equations (2.3a) and (2.3b) with but with all elements Gij set to zero for i≠j. Rj is the ratio of the rate of adaptation with the observed correlations relative to the predicted rate of adaptation if trait j was genetically independent from all other traits.
For example, Caruso (2004) measured selection and G on seven floral traits in Lobelia siphilitica. Using the data from the CERA population in 2000, we find that R=0.41, i.e. the predicted rate of adaptation is only 41 per cent of what it would be in the absence of any genetic correlations. Using the method above to calculate Rj, we can ask how each trait individually affects the rate of adaptation through its correlations with other traits. For the traits: (i) lobe length, (ii) lobe width, (iii) corolla length, (iv) corolla width, (v) stigma–nectary distance, (vi) stigma exsertion, and (vii) flower number, the corresponding values are R1=1.00, R2=1.01, R3=0.59, R4=0.96, R5=0.34, R6=1.35, R7=0.68. These results reveal that correlations involving lobe length, lobe width or corolla width have little effect on the rate of adaptation under this selection regime. However, the predicted rate of adaptation is reduced relative to what it would be if corolla length, stigma–nectary distance or flower number were genetically independent of other traits (i.e. correlations involving any of these three traits tend to constrain the rate of adaptation). By contrast, correlations involving stigma exsertion tend to facilitate the rate of adaptation.
One could extend this approach to examine the effect of correlations between sets of traits by calculating after setting all covariances between traits from different trait suites to zero. For example, consider a hypothetical study involving four traits. The first trait suite (e.g. floral traits) comprise traits 1 and 2, whereas traits 3 and 4 belong to a separate trait suite (e.g. vegetative traits). To examine the effect of correlations between floral and vegetative trait suites one would set G13=G14=G23=G24=0 to do the calculation. Analysis of data in this way may serve as a useful approach to study the phenotypic integration and modularity (Cheverud 1982; Arnold 1992).
In different contexts, other authors have used other measures to consider how correlations affect evolution. Schluter (1996) measured the angle between the major axis of genetic variation gmax (the leading eigenvector of the G) and major axis of divergence among species within lineages having undergone adaptive radiations. He found that this angle was smaller than expected by chance, leading him to infer that patterns of correlations bias patterns of long-term evolution (ca 4 Myr). McGuigan et al. (2005) applied a related but more advanced approach with a set of fish populations that had repeatedly adapted to lake and stream environments. They found that the divergence of traits that were likely to be under strong selection showed less evidence of being affected by correlations than traits that were less likely to be targets of strong selection.
Blows et al. (2004) examined the relationship between G and selection by measuring the angle between β and the projection of the ‘major’ subspace of G onto β, and by comparing the angles between the axes of major subspaces of G and γ. As with our metric, this approach is intended to give a sense of the extent to which patterns of genetic variation constrain or facilitate evolution. This approach can be useful when applied to systems where the biology is well understood and the results are carefully interpreted. However, it has several limitations. First, the resulting metrics are not simple to understand or interpret. Second, the choice of principal component axes used to define the subspaces is somewhat arbitrary, and not all of the principal component axes can be used (Blows et al. 2004). Third, it is possible for this approach to give misleading results when only one or a few of the correlated traits are under selection unless results are interpreted with great caution. For example, if arm and leg length are strongly genetically correlated but only arm length is under selection, there will be a large angle between β and the gmax (the one-dimensional subspace of G). A large angle is interpreted as a mismatch between selection and G. However, in this example, the correlation between the traits is meaningless with respect to the rate of adaptation because only arm length is under selection.
Our metric R is intuitively meaningful and can be easily calculated and interpreted. This is because R measures the effects of correlations in terms of fitness. However, this property also limits the usefulness of R in some respects. In the arm/leg length example above, we would find that R=1 (or L=0), correctly indicating that the correlation between arm and leg length has no effect on the rate of increase in fitness. However, this correlation is clearly of great importance with respect to understanding the evolution of leg length. In this example, leg length, a neutral character evolves because it is genetically correlated to a selected trait (arm length). Because R measures effects with respect to fitness, it does not describe how correlations affect the phenotypic evolution of individual traits. For this reason, we advocate that R be used in addition to, rather than instead of, other ways of understanding how correlations affect evolution.
Some particularly interesting approaches have been recently described by Hansen & Houle (2008). Building from earlier work (Houle 1992; Schluter 1996; Hansen et al. 2003), these authors have suggested several measures for quantifying constraint and evolutionary potential in multidimensional space. Evolvability is a standardized measure of the amount of genetic variation in the direction of selection, β. Conditional evolvability gives the amount of genetic variation in this direction assuming that movement in any other direction is strongly prohibited by stabilizing selection, i.e. it is the amount of variation along β that is independent of variation in other directions.
These measures quantify the combined effects of trait variances as well as the correlations, and in this sense provide a more complete picture of genetic constraint. By contrast, our goal was to isolate the effect of genetic covariances. The simple logic of our metric could be applied to their elegant measures to determine how correlations affect conditional evolvability. This can be easily accomplished by calculating the ratio of the evolvability (or conditional evolvability) when the genetic correlations are set to zero to the actual evolvability (or conditional evolvability) using the observed G. In fact, calculating this ratio for evolvability will be equivalent to the ratio R discussed here if there is no nonlinear selection.
One important limitation of our approach is that the value of R depends on how traits are defined. This dependency on trait definitions is an inherent part of our question: how do the genetic correlations among a specified set of traits affect the rate of adaptation? If traits are redefined along an alternative set of axes in the same multidimensional space (i.e. an orthogonal transformation to a new coordinate system), then there will be a new set of genetic correlations among these newly defined traits. The value of R can be re-calculated to measure how these correlations affect the rate of adaptation, but the new value of R will be different from the original since the new R refers to a different set of correlations. While we can measure R with respect to any defined set of traits, is it meaningful to do so?
A reasonable argument can be made that the way in which traits are defined is somewhat arbitrary and that natural selection may view traits in a very different way than humans do (see the target review by Blows (2007) and ensuing commentaries for further discussion of trait identification in studies of multivariate evolution). Even though selection may perceive them differently, the traits chosen for study usually are significant to those biologists measuring them. Often, something is known about the function of a trait or a trait is meaningful to how the observer perceives the study organism, e.g. arm and leg length have more meaning than two alternate traits defined by orthogonal linear combinations of these traits. Moreover, some aspects of organisms need to be measured or chosen for measurement by biologists in the first place before any coordinate or orthogonal transformations can take place, suggesting that escaping human perceptual biases is likely to be quite difficult or impossible. Biologists identify measurable traits and are often interested in how the relationships between these measurable traits affect evolution. Our metric helps to understand this relationship better. Our metric is not designed to show selection views on an organism in multivariate space. (It should be noted that the evolutionary hypotheses discussed previously for the average value of R should be reasonably robust to how traits are defined. For example, if genetic variation is exhausted in the direction of selection, genetic correlations should be constraining regardless of the coordinate system used.)
In addition to the metric R that requires information on both selection and G, we have proposed alternative perspectives from which the potential for correlations to affect the rate of adaptation can be assessed. First, we can ask whether the rate of adaptation under a specified pattern of selection is likely to be sensitive to correlations by measuring the evenness of selection. If the evenness of selection is high across a set of traits, then there will be high potential for a random set of correlations among these traits to affect the rate of adaptation. This metric depends only on selection and thus can be measured even in the absence of any information on G.
We can also ask whether the covariance structure is such that covariances are likely to affect the rate of adaptation. This can be assessed by the evenness of the eigenvalues of the G. When the evenness of eigenvalues is low, it means that correlations cause genetic variation to be reduced in some directions. Consequently, there is potential for covariances to affect the rate of adaptation if selection occurs in these directions. Measuring potential from only this perspective requires an estimate of G. However, estimates of the evenness of eigenvalues should be regarded with caution because of the downward bias in their estimation. Alternatively, we can avoid relying on the estimates of the evenness of the eigenvalues of G and instead determine the minimum and maximum possible values of R for a given G from the range of eigenvalues of M=σ−1Cσ.
Our survey of the literature revealed a distribution of L-values centred close to zero. Most datasets were only weakly affected by correlations. There were examples of datasets where correlations were fairly strongly constraining the rate of adaptation but these were balanced by datasets where correlations were facilitating the rate of adaptation. Neither the mean nor variance of L was significantly different from that expected by chance.
As stated above, basic quantitative genetics theory makes no prediction about whether correlations should tend to constrain or facilitate the rate of adaptation. However, there are several secondary sets of ideas that do make predictions in this regard (e.g. exhaustion of variation in the direction of selection, long-term shaping of mutational/developmental pathways to facilitate the generation of variation along selected directions, condition-dependence of strongly-selected traits). In light of these hypotheses, there are at least three different ways to view our result that correlations seem to have little or no effect on average. First, it is possible that none of the three hypotheses are important. Second, all three hypotheses may be important in individual cases but none of them are sufficiently common or strong to generate a clear signal across all the datasets. The different hypotheses were suggested in the context of different types of traits (e.g. morphological traits versus life-history traits and/or sexually selected traits) but our dataset is not large enough to meaningfully partition these studies into different trait types for separate analyses. Third, our data may be too noisy to perform a reasonable test. As described above and in the electronic supplementary material, our data are far from perfect. Both estimates of selection and G are notorious for their large confidence intervals. Moreover, we had to make a number of ‘approximations’ (e.g. using P as an approximation to G) to obtain a reasonable number of studies for examination. Even with all these levels of uncertainty, the estimates we used for selection and G should be closer to the true values than randomly chosen estimates would be. If there was a strong tendency for R to differ from unity, we should have been able to detect it through the noise. However, we have no way to assess the power of our test; it is possible that there is a fairly strong effect of correlations on average but our data are too noisy to detect it. Regardless, we could not know whether a sufficiently strong effect existed without attempting to look for it.
Genetic correlations have intrigued evolutionary biologists since it became well-understood that correlations affect how traits respond to multivariate selection. Genetic correlations became the perspective by which evolutionary quantitative geneticists considered constraint. While some have focused on the idea of absolute constraint, it is more useful to view constraint quantitatively rather than qualitatively. The metric R does so with respect to fitness. Our literature survey reveals examples with strong and weak constraint (R<1) as well as cases of negative constraint, i.e. patterns of correlations that facilitate the rate of adaptation (R>1). Though the average effect of correlations was not significantly different from expected by chance, the available data are noisy and we urge caution in interpreting our results. Nonetheless, we can say that our results provide no support for the emphasis on constraint that seems to permeate the literature on genetic correlations.
Acknowledgments
We thank W. G. Hill, D. Houle, T. Hansen and an anonymous reviewer for their several helpful comments. T. Hansen pointed out the connection between our metric R and classic matrix theory results for the min and max value for a ratio of quadratic forms. We thank L. Rowe for helpful discussion and E. Drummond for assistance with searching the literature. This work was supported by Discovery grants from the Natural Sciences and Engineering Research Council of Canada (A.F.A. and J.R.S.).
Supplementary Material
A description of how studies were chosen for the literature survey. We present a table that provides key information about each study included in the survey and how it was treated
References
- Arnold S.J. Constraints on phenotypic evolution. Am. Nat. 1992;140:S85–S107. doi: 10.1086/285398. doi:10.1086/285398 [DOI] [PubMed] [Google Scholar]
- Beldade P., Koops K., Brakefield P.M. Developmental constraints versus flexibility in morphological evolution. Nature. 2002;416:844–847. doi: 10.1038/416844a. doi:10.1038/416844a [DOI] [PubMed] [Google Scholar]
- Bell A.E., Burris M.J. Simultaneous selection for two correlated traits in Tribolium. Genet. Res. 1973;21:29–46. [Google Scholar]
- Blows M.W. A tale of two matrices: multivariate approaches in evolutionary biology. J. Evol. Biol. 2007;20:1–8. doi: 10.1111/j.1420-9101.2006.01164.x. doi:10.1111/j.1420-9101.2006.01164.x [DOI] [PubMed] [Google Scholar]
- Blows M.W., Hoffmann A.A. A reassessment of genetic limits to evolutionary change. Ecology. 2005;86:1371–1384. doi:10.1890/04-1209 [Google Scholar]
- Blows M.W., Chenoweth S.F., Hine E. Orientation of the genetic variance–covariance matrix and the fitness surface for multiple male sexually selected traits. Am. Nat. 2004;163:329–340. doi: 10.1086/381941. doi:10.1086/381941 [DOI] [PubMed] [Google Scholar]
- Caruso C.M. The quantitative genetics of floral trait variation in Lobelia: potential constraints on adaptive evolution. Evolution. 2004;58:732–740. doi: 10.1111/j.0014-3820.2004.tb00406.x. doi:10.1554/03-329 [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Optimization models, quantitative genetics, and mutation. Evolution. 1990;44:520–538. doi: 10.1111/j.1558-5646.1990.tb05936.x. doi:10.2307/2409433 [DOI] [PubMed] [Google Scholar]
- Charnov E.L. Phenotypic evolution under Fisher's fundamental theorem of natural selection. Heredity. 1989;62:113–116. doi: 10.1038/hdy.1989.15. doi:10.1038/hdy.1989.15 [DOI] [PubMed] [Google Scholar]
- Cheverud J.M. Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution. 1982;36:499–516. doi: 10.1111/j.1558-5646.1982.tb05070.x. doi:10.2307/2408096 [DOI] [PubMed] [Google Scholar]
- Cheverud J.M. Quantitative genetics and developmental constraints on evolution by selection. J. Theor. Biol. 1984;110:155–171. doi: 10.1016/s0022-5193(84)80050-8. doi:10.1016/S0022-5193(84)80050-8 [DOI] [PubMed] [Google Scholar]
- Cheverud J.M. A comparison of genetic and phenotypic correlations. Evolution. 1988;42:958–968. doi: 10.1111/j.1558-5646.1988.tb02514.x. doi:10.2307/2408911 [DOI] [PubMed] [Google Scholar]
- Clark A.G. Genetic correlations: the quantitative genetics of evolutionary constraints. In: Loeschcke V., editor. Genetic constraints on adaptive evolution. Springer; New York, NY: 1987. pp. 25–45. [Google Scholar]
- Cockrem F. Selection for relationships opposite to those predicted by the genetic correlation between two traits in the house mouse (Mus musculus) Nature. 1959;183:342–343. doi: 10.1038/183342a0. doi:10.1038/183342a0 [DOI] [PubMed] [Google Scholar]
- Conner J.K. Artificial selection: a powerful tool for ecologists. Ecology. 2003;84:1650–1660. doi:10.1890/0012-9658(2003)084[1650:ASAPTF]2.0.CO;2 [Google Scholar]
- Dickerson G.E. Genetic slippage in response to selection for multiple objectives. Cold Spring Harb. Symp. Quant. Biol. 1955;20:213–224. doi: 10.1101/sqb.1955.020.01.020. [DOI] [PubMed] [Google Scholar]
- Emlen D.J. Artificial selection on horn length body size allometry in the horned beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae) Evolution. 1996;50:1219–1230. doi: 10.1111/j.1558-5646.1996.tb02362.x. doi:10.2307/2410662 [DOI] [PubMed] [Google Scholar]
- Etterson J.R., Shaw R.G. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. doi:10.1126/science.1063656 [DOI] [PubMed] [Google Scholar]
- Falconer D.S., Mackay F.C. Longman; Harlow, UK: 1996. Introduction to quantitative genetics. [Google Scholar]
- Frankino W.A., Zwaan B.J., Stern D.L., Brakefield P.M. Natural selection and developmental constraints in the evolution of allometries. Science. 2005;307:718–720. doi: 10.1126/science.1105409. doi:10.1126/science.1105409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantmakher, F. R. 1960 The theory of matrices, vol. 1. New York, NY: Chelsea Publishing.
- Hansen T.F., Houle D. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 2008;21:1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. doi:10.1111/j.1420-9101.2008.01573.x [DOI] [PubMed] [Google Scholar]
- Hansen T.F., Armbruster W.S., Carlson M.L., Pélabon C. Evolvability and genetic constraint in Dalechampia blossoms: genetic correlations and conditional evolvability. J. Exp. Zool. B. 2003;296:23–39. doi: 10.1002/jez.b.14. doi:10.1002/jez.b.14 [DOI] [PubMed] [Google Scholar]
- Hine E., Blows M.W. Determining the effective dimensionality of the genetic variance–covariance matrix. Genetics. 2006;173:1135–1144. doi: 10.1534/genetics.105.054627. doi:10.1534/genetics.105.054627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D. Genetic covariance of fitness correlates: what genetic correlations are made of and why it matters. Evolution. 1991;45:630–648. doi: 10.1111/j.1558-5646.1991.tb04334.x. doi:10.2307/2409916 [DOI] [PubMed] [Google Scholar]
- Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Quantitative genetic analysis of multivariate evolution, applied to brain : body allometry. Evolution. 1979;33:402–416. doi: 10.1111/j.1558-5646.1979.tb04694.x. doi:10.2307/2407630 [DOI] [PubMed] [Google Scholar]
- Lande R. A quantitative genetic theory of life history evolution. Ecology. 1982;63:607–615. doi:10.2307/1936778 [Google Scholar]
- Lande R., Arnold S.J. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. doi:10.2307/2408842 [DOI] [PubMed] [Google Scholar]
- Lin C.Y. Index selection for genetic improvement of quantitative characters. Theor. Appl. Genet. 1978;52:49–56. doi: 10.1007/BF00281316. [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B. Sinauer Associates, Inc; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- McGuigan K., Chenoweth S.F., Blows M.W. Phenotypic divergence along lines of genetic variance. Am. Nat. 2005;165:32–43. doi: 10.1086/426600. doi:10.1086/426600 [DOI] [PubMed] [Google Scholar]
- Mousseau T.A., Roff D.A. Natural selection and the heritability of fitness components. Heredity. 1987;59:181–197. doi: 10.1038/hdy.1987.113. doi:10.1038/hdy.1987.113 [DOI] [PubMed] [Google Scholar]
- Nordskog A.W. Success and failure of quantitative genetic theory in poultry. In: Pollak E., Kempthorne O., Bailey T.B. Jr, editors. Proc. Int. Conf. on Quantitative Genetics. Iowa State University Press; Ames, IA: 1977. pp. 47–51. [Google Scholar]
- Rausher M.D. The measurement of selection on quantitative traits: biases due to environmental covariances between traits and fitness. Evolution. 1992;46:616–626. doi: 10.1111/j.1558-5646.1992.tb02070.x. doi:10.2307/2409632 [DOI] [PubMed] [Google Scholar]
- Roff D.A. The estimation of genetic correlations from phenotypic correlations: a test of Cheverud's conjecture. Heredity. 1995;74:481–490. doi:10.1038/hdy.1995.68 [Google Scholar]
- Roff D.A. Chapman and Hall; Toronto, Canada: 1997. Evolutionary quantitative genetics. [Google Scholar]
- Rowe L., Houle D. The lek paradox and the capture of genetic variance by condition dependent traits. Proc. R. Soc. B. 1996;263:1415–1421. doi:10.1098/rspb.1996.0207 [Google Scholar]
- Rutledge J.J., Heisen E.J., Legates J.E. An experimental evaluation of genetic correlation. Genetics. 1973;75:709–726. doi: 10.1093/genetics/75.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. doi:10.2307/2410734 [DOI] [PubMed] [Google Scholar]
- Stanton M., Young H.J. Selecting for floral character associations in wild radish, Raphanus sativus L. J. Evol. Biol. 1994;7:271–285. doi:10.1046/j.1420-9101.1994.7030271.x [Google Scholar]
- Stinchcombe J.R., Rutter M.T., Burdick D.S., Tiffin P., Rausher M.D., Mauricio R. Testing for environmentally induced bias in phenotypic estimates of natural selection: theory and practice. Am. Nat. 2002;160:511–523. doi: 10.1086/342069. doi:10.1086/342069 [DOI] [PubMed] [Google Scholar]
- Turelli M. Phenotypic evolution, constant covariances, and the maintenance of additive variance. Evolution. 1988;42:1342–1347. doi: 10.1111/j.1558-5646.1988.tb04193.x. doi:10.2307/2409017 [DOI] [PubMed] [Google Scholar]
- Wagner G.P. Multivariate mutation–selection balance with constrained pleiotropic effects. Genetics. 1989;122:223–234. doi: 10.1093/genetics/122.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitt D.E., Levin D.A. Genetic and phenotypic correlations in plants: a botanical test of Cheverud's conjecture. Heredity. 1998;80:310–319. doi:10.1046/j.1365-2540.1998.00298.x [Google Scholar]
- Willis J.H., Coyne J.A., Kirkpatrick M. Can one predict the evolution of quantitative characters without genetics? Evolution. 1991;45:441–444. doi: 10.1111/j.1558-5646.1991.tb04418.x. doi:10.2307/2409678 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A description of how studies were chosen for the literature survey. We present a table that provides key information about each study included in the survey and how it was treated