Abstract
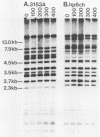
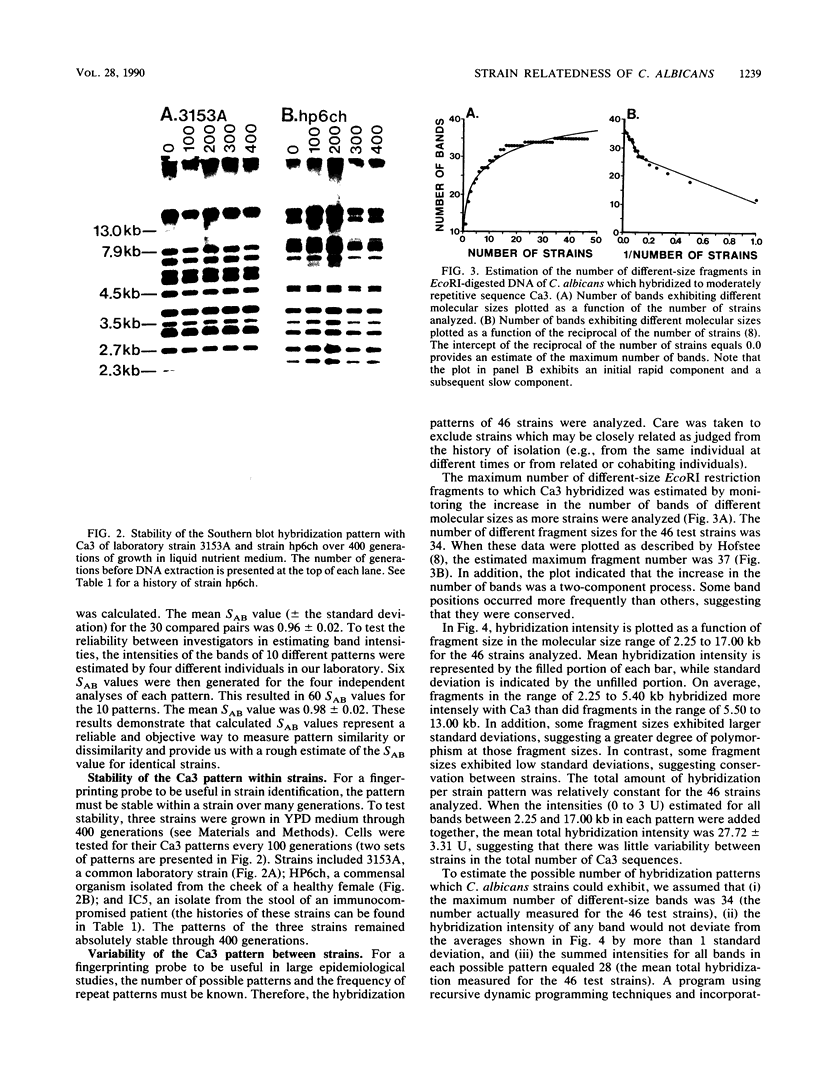
When used to probe EcoRI-digested Candida albicans DNA, the moderately repetitive sequence Ca3 generated a Southern blot hybridization pattern which included 15 to 25 bands, depending upon the strain. The pattern was stable through 400 generations in each of three independent strains but variable between most of 46 unrelated tester strains, making it a very effective probe for discrimination between strains. Computer-assisted methods (Dendron) were developed for storage of Ca3 patterns in data files, calculation of similarity (SAB) values between strains based upon band positions and intensities, and generation of histograms and dendrograms based on SAB values for all strains or any subset of strains in large epidemiological studies. In testing the effectiveness of the system, it was found that (i) multiple isolates from different body locations of the same healthy individual could represent either the same strain or different strains, (ii) isolates from oral lesions of a husband and wife represented the same strain, (iii) strains isolated from the mouths of 10 healthy individuals on the same day and in the same geographical location were as dissimilar on average as the 46 unrelated tester strains, and (iv) strains isolated from seven immunocompromised patients hospitalized over a 2.5-month period in the same hospital were highly similar, indicating nosocomial origin. The apparent effectiveness of these fingerprinting methods and the Dendron program suggests that interlaboratory procedures for fingerprinting should be standardized and all patterns should be analyzed and stored in a common and accessible data base for broad epidemiological analysis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler J. E., Glee P. M., Horn H. L. Candida albicans- and Candida stellatoidea-specific DNA fragment. J Clin Microbiol. 1988 Sep;26(9):1720–1724. doi: 10.1128/jcm.26.9.1720-1724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox B. C., Mobley H. L., Wade J. C. The use of a DNA probe for epidemiological studies of candidiasis in immunocompromised hosts. J Infect Dis. 1989 Mar;159(3):488–494. doi: 10.1093/infdis/159.3.488. [DOI] [PubMed] [Google Scholar]
- HASENCLEVER H. F., MITCHELL W. O. Antigenic studies of Candida. I. Observation of two antigenic groups in Candida albicans. J Bacteriol. 1961 Oct;82:570–573. doi: 10.1128/jb.82.4.570-573.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HASENCLEVER H. F., MITCHELL W. O., LOEWE J. Antigenic studies of Candida. II. Antigenic relation of Candida albicans group A and group B to Candida stellatoidea and Candida tropicalis. J Bacteriol. 1961 Oct;82:574–577. doi: 10.1128/jb.82.4.574-577.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFSTEE B. H. Non-inverted versus inverted plots in enzyme kinetics. Nature. 1959 Oct 24;184:1296–1298. doi: 10.1038/1841296b0. [DOI] [PubMed] [Google Scholar]
- Hunter P. R., Fraser C. A., Mackenzie D. W. Morphotype markers of virulence in human candidal infections. J Med Microbiol. 1989 Feb;28(2):85–91. doi: 10.1099/00222615-28-2-85. [DOI] [PubMed] [Google Scholar]
- Magee B. B., D'Souza T. M., Magee P. T. Strain and species identification by restriction fragment length polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol. 1987 Apr;169(4):1639–1643. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T. Electrophoretic karyotypes and chromosome numbers in Candida species. J Gen Microbiol. 1987 Feb;133(2):425–430. doi: 10.1099/00221287-133-2-425. [DOI] [PubMed] [Google Scholar]
- McCreight M. C., Warnock D. W. Enhanced differentiation of isolates of Candida albicans using a modified resistogram method. Mykosen. 1982 Nov;25(11):589–598. doi: 10.1111/j.1439-0507.1982.tb01926.x. [DOI] [PubMed] [Google Scholar]
- Odds F. C., Abbott A. B. A simple system for the presumptive identification of Candida albicans and differentiation of strains within the species. Sabouraudia. 1980 Dec;18(4):301–317. [PubMed] [Google Scholar]
- Olivo P. D., McManus E. J., Riggsby W. S., Jones J. M. Mitochondrial DNA polymorphism in Candida albicans. J Infect Dis. 1987 Jul;156(1):214–215. doi: 10.1093/infdis/156.1.214. [DOI] [PubMed] [Google Scholar]
- Polonelli L., Archibusacci C., Sestito M., Morace G. Killer system: a simple method for differentiating Candida albicans strains. J Clin Microbiol. 1983 May;17(5):774–780. doi: 10.1128/jcm.17.5.774-780.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rikkerink E. H., Magee B. B., Magee P. T. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988 Feb;170(2):895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Stevens D. A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987 Apr;25(4):675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B., Buffo J., Soll D. R. High-frequency switching of colony morphology in Candida albicans. Science. 1985 Nov 8;230(4726):666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- Snell R. G., Hermans I. F., Wilkins R. J., Corner B. E. Chromosomal variations in Candida albicans. Nucleic Acids Res. 1987 Apr 24;15(8):3625–3625. doi: 10.1093/nar/15.8.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Galask R., Isley S., Rao T. V., Stone D., Hicks J., Schmid J., Mac K., Hanna C. Switching of Candida albicans during successive episodes of recurrent vaginitis. J Clin Microbiol. 1989 Apr;27(4):681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Langtimm C. J., McDowell J., Hicks J., Galask R. High-frequency switching in Candida strains isolated from vaginitis patients. J Clin Microbiol. 1987 Sep;25(9):1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D. R., Staebell M., Langtimm C., Pfaller M., Hicks J., Rao T. V. Multiple Candida strains in the course of a single systemic infection. J Clin Microbiol. 1988 Aug;26(8):1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey S. B., Mori M., Pfaller M. A., Woolson R. F., Wenzel R. P. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch Intern Med. 1988 Dec;148(12):2642–2645. doi: 10.1001/archinte.148.12.2642. [DOI] [PubMed] [Google Scholar]
- Wills J. W., Troutman W. B., Riggsby W. S. Circular mitochondrial genome of Candida albicans contains a large inverted duplication. J Bacteriol. 1985 Oct;164(1):7–13. doi: 10.1128/jb.164.1.7-13.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]