Abstract
AIM: To investigate the roles and mechanism of signal transducer and activator of transcription 3 (STAT3) in invasion of human colon cancer cells by RNA interference.
METHODS: Small interfering RNA (siRNA) targeting Signal transducer and activator of transcription 3 (STAT3) was transfected into HT29 colon cancer cells. STAT3 protein level and DNA-binding activity of STAT3 was evaluated by western blotting and electrophoretic mobility shift assay (EMSA), respectively. We studied the anchorage-independent growth using colony formation in soft agar, and invasion using the boyden chamber model, anoikis using DNA fragmentation assay and terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL), respectively. Western blot assay was used to observe the protein expression of Bcl-xL and survivin in colon cancer HT29 cells.
RESULTS: RNA interference (RNAi) mediated by siRNA leads to suppression of STAT3 expression in colon cancer cell lines. Suppression of STAT3 expression by siRNA could inhibit anchorage-independent growth, and invasion ability, and induces anoikis in the colon cancer cell line HT29. It has been shown that knockdown of STAT3 expression by siRNA results in a reduction in expression of Bcl-xL and survivin in HT29 cells.
CONCLUSION: These results suggest that STAT3 siRNA can inhibit the invasion ability of colon cancer cells through inducing anoikis, which antiapoptotic genes survivin and Bcl-xL contribute to regulation of anoikis. These studies indicate STAT3 siRNA could be a useful therapeutic tool for the treatment of colon cancer.
Keywords: Colon cancer, Invasion, Signal transducer and activator of transcription 3, Anoikis
INTRODUCTION
Signal transducers and activator of transcription, STATs, are a family of transcription factors that transmit signals from cell surface receptors directly to the nucleus[1]. Activation of all the STAT proteins is caused by phosphorylation of a single tyrosine residue that leads to dimerization via an intermolecular SH2 phosphotyrosine interaction[2–5]. The dimerized STATs then translocate to the nucleus where they regulate gene expression by binding directly to high affinity DNA binding sites or by associating with other transcription factors[6–11]. They play a critical role in mediating cytokine and growth factor signaling involved in cell growth, differentiation and survival[12–14]. Among the seven members of the mammalian STAT family, STAT3 has been the most strongly implicated in oncogenesis[15].
Some studies showed that constitutively activated STAT3 is found in a wide variety of human tumors including multiple myelomas, breast, ovarian, prostate, and head and neck tumors[16–18]. Inhibition of STAT3 signaling with either dominant negative or antisense oligonucleotides against STAT3 suppresses the transformation process in some tumors[19]. Recent studies have shown that treatment of tumor cells with inhibitors of STAT signaling results in decreased cell viability and induction of apoptosis[20]. Together these findings demonstrate that STAT3 signaling plays a critical role in both the transformation process and tumor progression in some types of cancer.
Recent reports showed that STAT3 has a significant association with tumor invasion and metastasis of a few cancers[21–25]. In renal cell carcinoma, the positive rate of the expression of p-STAT3 correlated well with the depth of tumor invasion and with metastasis[21]. In colorectal adenocarcinoma, p-STAT3 protein was significantly correlated with the depth of tumour invasion, venous invasion, lymph node metastasis, and increasing stages of the Dukes' classification[25]. In addition, blockade of activated STAT3 via ectopic expression of dominant-negative STAT3 significantly could suppress angiogenesis, tumor growth, and metastasis in pancreatic cancer[26].Together these findings demonstrate that STAT3 activation might be a new potential target for therapy of human cancer metastasis. Recently it has been shown that STAT3 is constitutively activated in colon cancer[25], however, the role and mechanism of STAT3 signaling in metastasis of colon cancer remains elusive.
Colorectal cancer is the third most common malignant neoplasm worldwide[27] and the second leading cause of death due to cancer[28]. Despite recent advances in diagnostic and therapeutic measures, the prognosis of colorectal cancer patients with distant metastasis still remains poor. Liver metastasis is a major cause of morbidity and mortality in patients with colon cancer. Colorectal liver metastasis is associated with a very poor prognosis; most patients die within 2 years of diagnosis despite the availability of numerous therapies. To improve the choice of therapeutic strategy, it is critical that the mechanism of invasion and metastasis of colon cancer be clarified. Here we will show that knockdown of STAT3 expression by siRNA inhibit invasion ability and reduces anoikis resistance in colon cancer cells.
MATERIALS AND METHODS
Human colon cancer cell lines
The human colon cancer cell lines HT29, SW620 and SW480 were obtained from the Institute of Cell Biology, Shanghai, China. All cancer cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum (FCS). These studies were carried out with Medical Ethical Committee approval of Jiangsu University.
STAT3 siRNA and siRNA transfections
A double stranded siRNA oligonucleotide against STAT3 (5’-AACAUCUGCCUAGAUCGGCUAdTdT-3’; 3’-dTdTGUAGACGGAUCUAGCCGAU-5’) was designed and synthesized by Dharmacon Research, Inc. Oligofectamine (Invitrogen, Inc.). In brief, 1 d prior to transfection, cancer cells were seeded, without antibiotics, into a 24 well plate, 1 × 105 cells/well, corresponding to a density of 60%-70% at the time of transfection. There were the following groups: siRNA groups: cells transfected with siRNA at different doses; control groups: cells treated with oligofectamine alone without siRNA. All transfections were performed in triplicate for each time point. At different times after the beginning of the transfection period, all cells were harvested and the following assays were performed.
Western blot analyses
HT29 cells were harvested and lysed in a buffer containing 10 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mmol/L EDTA (pH 8.0), 2 mmol/L phenylmethylsulfonyl fluoride, 2 μg/mL aprotinin, 2 μg/mL leupeptin, and 1 mmol/L Na3VO4. For western blot analyses, 30 μg of total extracted proteins were applied per lane before SDS-PAGE. Following transfer to nitrocellulose membranes, protein expression levels were detected using polyclonal anti-STAT3 (Santa Cruz, CA), anti-phospho-STAT3, anti-Bcl-xL and anti-survivin (Alpha Diagnostics International, TX). The expression of β-actin (Sigma-Aldrich, MO) was used as a normalization control for protein loading.
Electrophoretic mobility shift assay (EMSA) for STAT3-DNA binding activity
The STAT3-DNA binding activity was assessed by EMSA using the nuclear extract from the three cell lines. The sense strand that binds activated STAT3 protein was 5'-TCGACATTTCCCGTAAATC-3'. Double-stranded oligonucleotide was end-labeled with [γ-32P] ATP using a T4 polynucleotide kinase according to the manufacturer's instruction. The final concentration of probe was 1.75 pmol/L. The labeled probes were then purified by G-25 spin columns. One microliter of 32P-labeled STAT3 oligonucleotide was added to each reaction. For STAT3 specific tests, a 150-fold unlabeled STAT3 probe was applied as a competitor. The final volume of reaction was 20 μL, including 10 μg of nuclear extract and 5 × binding buffer. The reactions were placed on ice for 30 min. The 45 g/L nondenaturing acrylamide gel was pre-run in 1 × TBE buffer at 25 mA for 60 min. After loading of the samples, the gel was run at room temperature in 1 × TBE buffer at 25 mA for 90 min. The gel was dried on a gel dryer, then exposed to X-ray film overnight at-80°C with intensifying screen. The protein-DNA complex was detected by autoradiography. The Quantity one software was used to analyze the scanned EMSA gel bands.
Anchorage-independent growth assay
For the anchorage-independent growth experiments, HT29 cells (8 × 103 cells/well) were seeded in 0.3% Difco Bactoagar (Difco, MI) supplemented with complete culture medium. This suspension was layered over 0.5 mL of 0.8% agar-medium base layer in 24 multiwell cluster dishes (Becton Dickinson, Italy). After 15 d, the colonies were stained with nitroblue tetrazolium, and colonies larger than 50 μm were acquired with a micro-Scopeman camera system (Moritex Europe Ltd, Italy) and analyzed with an Image-Pro Plus (Media Cybernetics, MD) computer program.
Cell invasion assay
Cell invasion assay was performed using Boyden chambers and 8 μm pore size polyvinylpyrrolidone-free polycarbonate filters coated with 25 μg/filter Matrigel (Beckton Dickinson). After transfection for 48 h, Sub-confluent HT29 control or HT29 STAT3 cell lines were harvested by a mild trypsinization, washed twice with Cellect medium and counted. Cells (2 × 105 viable cells/sample) were allowed to invade Matrigel toward 10% FBS/Cellect at 37°C, 5% CO2 for 3 h. At the end of the assay, cells on the lower surface of the filter were fixed in ethanol, stained with hematoxylin, and 10 random fields/filter were counted at 200 × magnification. Data represent the average of three experiments, all performed in triplicate.
Induction of anoikis
To prevent cell adhesion, 6-well plates were coated with a solution of polyhydroxyethylmethacrylate (poly-HEMA,Sigma-Aldrich), dissolved at 10 mg/mL in ethanol[29,30]. To coat 6-well plates, 3 mL of poly-HEMA solution was added to each well. Plates were kept at 37°C for at least 3 d until the solvent had completely evaporated. To induce anoikis, 48 h after transfection, cells were harvested and moved to plates coated with poly-HEMA. 1 × 106 resuspended cells were cultured in DMEM medium containing 15% FCS for 12 h on poly-HEMA-coated dishes at 37°C and 5% CO2. Subsequently, cells were gently recovered and submitted to apoptosis detection assays.
Terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay
The TUNEL assay was performed according to a previous report[31]. Apoptosis index (AI) determination: apoptosis cells/total cell × 100%.
DNA fragmentation assay
For demonstration of internucleosomal DNA fragmentation, after cells were plated in 6-well plates coated with poly-HEMA for different times, the cells were harvested, washed with PBS solution at 4°C, and suspended in lysis buffer (10 mmol/L Tris-HCl pH 7.5, 10 mmol/L EDTA, and 0.2% Triton × 100). After incubation for 15 min at 4°C, samples were centrifuged at 13 000 × g for 10 min at 4°C. The supernatant containing the fragmented DNA was precipitated with NaCl 0.5 mol/L and 1 volume of isopropanol for at least 1 h at -70°C. Samples were centrifuged at 13 000 × g for 10 min at 4°C, and the pellet was washed once with 70% ethanol, and air-dried. The precipitates were dissolved in 10 μL TE-RNase (0.1 mg/mL) and incubated at 37°C for 30 min. Finally, the samples were electrophoresed through a 1% agarose gel.
Statistical analysis
Statistical significance of results was evaluated by SSPS10.0 software. All results with P values ≤ 0.05 were considered significant.
RESULTS
Effects of siRNA on the expression and DNA binding activity of STAT3 in the colon cancer cell line HT29
To inhibit STAT3 expression in colon cancer cells, we used the RNAi method adapted for mammalian cell culture by Elbashir et al[32]. A number of 21 bp double stranded RNAs to human STAT3 were synthesized and tested for their ability to knockdown STAT3 expression in colon cancer HT29 cells. The human colon cancer cell line HT29, harboring activated STAT3, was transfected with STAT3 siRNA using oligofectamine. Twenty-four and 72 h after transfection, the protein levels and DNA-binding activities of STAT3 were measured by western blot and EMSA, respectively. Transfection of cells with STAT3 siRNA (Figure 1A) resulted in a highly significant and reproducible decrease in STAT3 expression levels as judged by western blotting (Figure 1B). Figure 1A shows that STAT3 siRNA diminished STAT3 protein compared with control groups, accompanied by a significant decrease in STAT3 DNA-binding activity (Figure 1B). STAT3 knockdown by siRNA was found to be time dependent with the maximum effect achieved at 48-72 h of siRNA treatment (Data not shown).
Figure 1.
A: Expression of STAT3 protein in HT29 cells; B: STAT3 DNA-binding activity by EMSA in HT29 cells. 1: Control; 2, 3, 4: siRNA 2, 4, 8 nmol/L.
STAT3 RNAi inhibits anchorage-independent growth in colon cancer cells
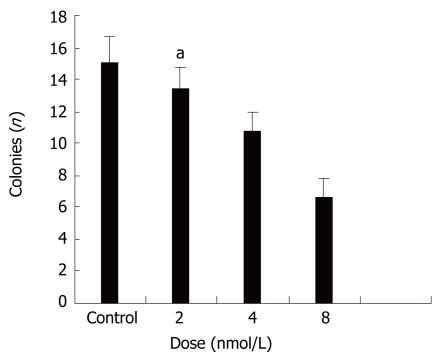
Next we evaluated the biological effects of STAT3 suppression in colon cancer HT29 cells by using several different types of assays. Colony formation in soft agar is a property closely associated with malignancy[33]. Figure 2 shows that treatment with STAT3 siRNA induced significant anchorage-independent growth inhibition in a dose-dependent manner.
Figure 2.
Effects of STAT3 siRNA on anchorage-independent growth of HT29 cells. It shows that treatment with STAT3 siRNA inhibit anchorage-independent growth in a dose-dependent manner. aP < 0.05 between siRNA 8 nmol/L and control groups.
STAT3 RNAi suppresses invasion in colon cancer cells
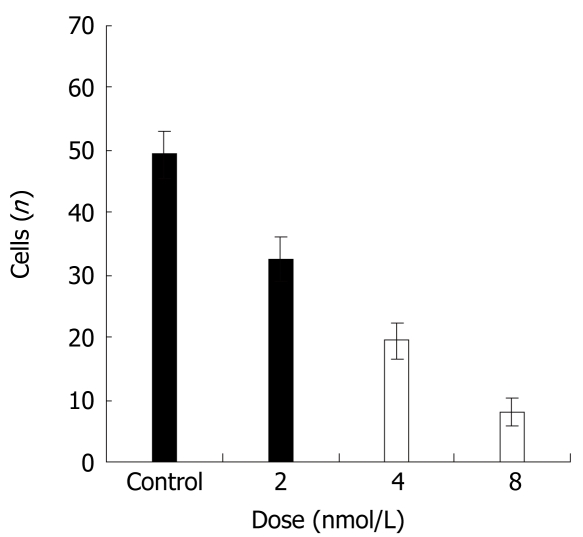
Next, we assessed whether STAT3 suppression might decrease the ability of cancer cells to invade the extracellular matrix. HT29 cancer cells were subjected to cell invasion assays, by using Boyden chambers and 8 μm pore size filters coated with Matrigel. Compared with the control group, HT29 cells transfected with siRNA exhibited a scarce ability to cross Matrigel (Figure 3). This difference was statistically significant as compared with control groups (P < 0.01).
Figure 3.
Effects of STAT3 suppression on the invasion ability of HT29 cells.
Effects of STAT3 suppression on anoikis
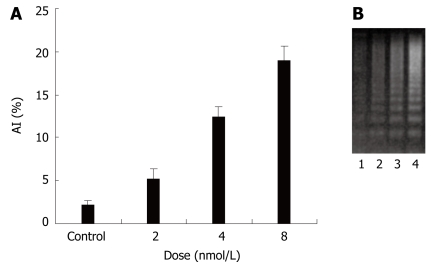
To explore the molecular mechanisms of invasion of colon cancer, anoikis was studied. To investigate the effect of STAT3 siRNA on anoikis, HT29 cells were subjected to plates seeded in poly-HEMA. After transfection for 48 h, all cells were removed to plates seeded in poly-HEMA for 12 h. TUNEL and DNA ladder were performed to evaluate the anoikis of HT29 cells. There was little apoptosis in all cancer cells in attached culture. When in a suspended culture, there was significant apoptosis in cancer cells treated with STAT3 siRNA. A significantly higher percentage of apoptotic index was observed in STAT3 siRNA treated cells than in only oligofectamine treated cells (Figure 4A). Consistent with this observation, agarose electrophoresis analysis of HT29 cancer cell extracts showed that there was DNA ladder in cells treated with STAT3 siRNA (Figure 4B). Together these data indicate that siRNA-mediated suppression of the STAT3 gene reduces resistance to anoikis of HT29 cells.
Figure 4.
Effects of STAT3 siRNA on anoikis of human colon cancer cell line HT29. A: AI (Apoptosis index) induced with STAT3 siRNA in HT29 cells; B: DNA ladder by STAT3 siRNA in HT29 cells. 1: Control; 2, 3, 4: siRNA, 2, 4, 8 nmol/L.
Knockdown of STAT3 expression down regulates survival genes
Recent data indicate that some genes, especially anti-apoptotic genes, contribute to regulation of anoikis[34].Constitutive activation of STAT3 induces the expression of a number of anti-apoptotic genes including Bcl-xL, a member of the Bcl-2-family of anti-apoptotic genes[20,35], and survivin, a member of the IAP, inhibitors of apoptotic proteins family[36]. Moreover, both of these genes are expressed in colon cancer[37,38]. In order to determine whether these two genes might be involved in the STAT3 mediated anoikis resistance in colon cancer, western blot analyses were performed to examine the expression levels of the two genes. Western blot analysis showed that Bcl-xL and survivin protein levels were drastically reduced upon treatment with STAT3 siRNA. Thus, these data show that STAT3 regulates anoikis of at least two distinct antiapoptotic genes in colon cancer HT29 cell (Figure 5).
Figure 5.
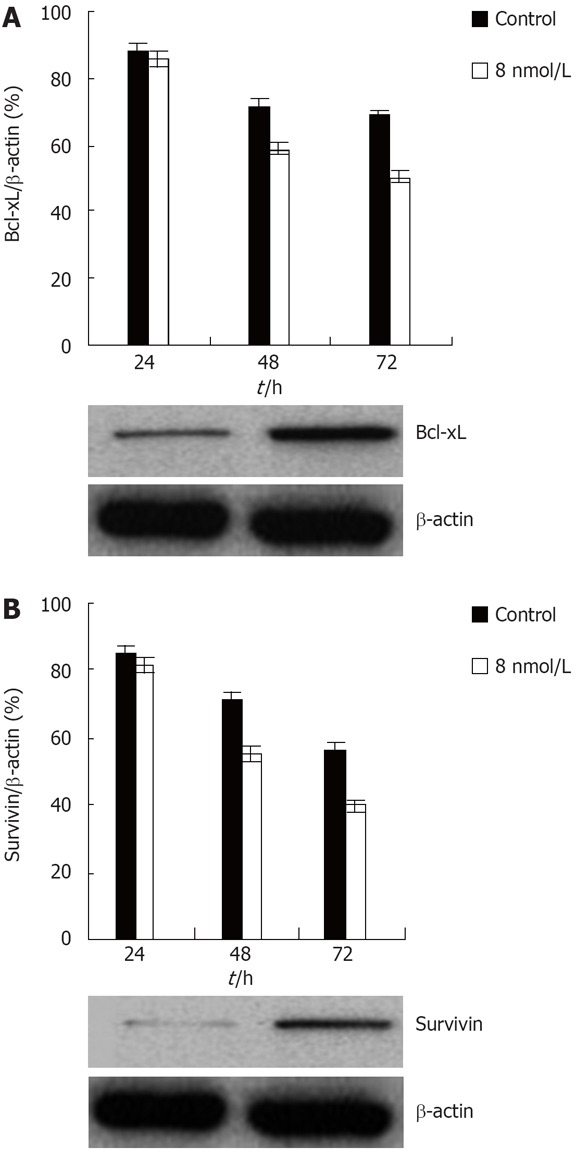
Knockdown of STAT3 expression downregulates Bcl-xL and survivin protein level in the HT29 human colon cancer cell line. A: Inhibition of transfection with STAT3 siRNA on Bcl-xL expression of colon cancer HT29 cell; B: Suppression of transfection with STAT3 siRNA on survivin expression of colon cancer HT29 cell.
DISCUSSION
STAT3 is activated by a number of cytokines and growth factors and has diverse functions during embryogenesis and early development[12–14]. Some studies showed that there is persistent activation of STAT3 in a number of human solid tumors including colon cancer[16–18]. Elevated STAT3 activity has been shown to render cells resistant to apoptosis, and inhibition of STAT3 signaling in a number of tumor cell lines with some ways causing a decrease in cell viability and subsequent apoptosis[16,17]. Lots of studies have demonstrated that the transformation process induced by diverse oncogenic protein tyrosine kinases is dependent on STAT3 activation[18,39,40]. Recently it has been shown that STAT3 is constitutively activated in colon cancer. However, the relationships of STAT3 with invasion and metastasis in colon cancer cells remain unclear.
In order to determine the role of STAT3 in colon cancer directly, we have used RNAi to specifically knock down the expression of STAT3, studied the effects of anchorage-independent growth and invasion in the colon cancer cell line HT29. SiRNAs are short oligonucleotides of 21-23 nucleotides in length that can be used in vitro to produce sequence specific gene silencing of mammalian cells[32]. It has been shown that siRNAs can be used effectively in vivo to suppress gene expression in adult mice[41,42]. SiRNAs can be directly introduced into the CNS to reduce endogenous gene expression[43]. Transfection of colon cancer HT29 cells with STAT3 siRNA resulted in colonies forming membranes with reduced biochemical changes indicative of invasion. In addition, consistent with the results from HT29 cancer cell, SW480 and SW620 cancer cells transfectd with the STAT3 siRNA led to a decrease of both invasion and anchorage-independent growth. These results are similar to reports previously observed with AG490 inhibiting STAT3 in pancreatic cancer cells[34]. All data suggest that STAT3 down-regulation by RNAi inhibit metastasis of human cancer cell in vitro.
The metastasis and invasion of cancer cells were involved in a lot of mechanisms. Anoikis resistance was thought to be high. Anoikis is a peculiar form of apoptosis that is induced by disruption of the interactions between epithelial cells and extracellular matrix[29]. Induction of apoptosis upon loss of anchorage has been termed anoikis (Greek for homelessness)[30]. Anoikis can be considered as a safety program for maintaining normal cell and tissue homeostasis, which prevents survival and reattachment of detached cells to new matrices at inadequate locations. Anoikis has been suggested to act as a physiological barrier to metastasis; resistance to anoikis may allow survival of cancer cells during systemic circulation, thereby facilitating secondary tumour formation in distant organs. Gaining anoikis resistance or anchorage-independent survival is a hallmark of oncogenic transformation. For these experiments, cells are usually seeded in tissue culture dishes that are coated with poly-HEMA, which does not allow cells to attach by preventing matrix deposition. Tumor cells are usually resistant to anoikis[30]. Our results here showed that, in attached culture, neither control cells nor transfected cells exhibited significant apoptosis, suggesting HT29 cells are resistant to anoikis. When seeded in poly-HEMA plates, compared to the control group, there were significant signs of apoptosis in cancer cells treated with STAT3 siRNA. It was shown that clear DNA ladder, the augmented the apoptosis index. These results suggest that STAT3 siRNA induces anoikis of HT29 colon cancer cells.
One mechanism by which STAT3 participates in tumorigenesis is by inhibiting apoptosis through the induction of anti-apoptotic genes such as Bcl-2, Bcl-xL, and Mcl-1[18]. STAT3 responsive elements are found in the promoter region of these genes, suggesting that they are directly regulated by STAT3[16,36]. Interestingly, some reports showed that some anti-apoptotic genes regulate anoikis. Biochemical studies have shown that Bcl-xL, a member of the Bcl-2 family of proteins, induce anoikis of some cancer cells[34,45], and we have found survivin gene is overexpressed in gastric cancer and survivin siRNA reduced the resistance of anoikis of gastric cancer cells (data not shown). In the present study, we have found that both Bcl-xL and survivin are expressed in HT29 cells. Treating HT29 cells with STAT3 siRNA significantly reduces expression levels of both of these genes. These findings suggest that induction of Bcl-xL and/or survivin gene by constitutively activated STAT3 contributes to regulating anoikis of colon cancer cells.
Data presented in this paper are consistent with the growing body of evidence suggesting STAT3 may be an ideal therapeutic target in tumors including colon cancer. We are the first to report that STAT3 siRNA can suppress invasion of colon cancer through inducing anoikis. These results suggest that siRNA may become a useful clinical tool in the future. Since STAT3 signaling is important for the survival of a number of human tumors, STAT3 siRNA could become an effective therapeutic agent for STAT3 dependent tumors.
COMMENTS
Background
Some reports showed that STAT3 has a significant association with tumor invasion and metastasis of a few of cancers, however, the role and mechanism of STAT3 signaling in metastasis of colon cancer remain elusive.
Research frontiers
Induction of apoptosis upon loss of anchorage has been termed anoikis, and anoikis has been suggested to act as a physiological barrier to metastasis. Here we will show that knockdown of STAT3 expression by siRNA inhibit invasion ability by reducing the resistance anoikis in colon cancer cells. In addition, down-regulation of Bcl-xL and/or survivin by STAT3 siRNA might play an important role in inducing anoikis in human colon cancer cell.
Innovations and breakthroughs
The paperwork showed that that STAT3 siRNA can inhibit invasion ability of colon cancer cells through inducing anoikis, and suppression of Bcl-xL and/or survivin by STAT3 siRNA might contribute to regulation of anoikis of human colon cancer cell.
Applications
Here we will show that knockdown of STAT3 expression by siRNA inhibits invasion ability and reduces the resistance anoikis in colon cancer cells. The paper would help to clarify the mechanism of invasion and metastasis of colon cancer and improve the choice of therapeutic strategy.
Terminology
Anoikis (Greek for homelessness) is a peculiar form of apoptosis that is induced by disruption of the interactions between epithelial cells and extracellular matrix.
Peer review
This is an interesting paper investigating the in vitro effect and mechanisms of STAT3 siRNAs in a colorectal cancer (CRC) cell line. The main finding of the study was that STAT3 siRNA can inhibit the invasion ability of colon cancer cells through inducing anoikis, in addition antiapoptotic genes survivin and Bcl-xL may contribute to regulation of anoikis.
Supported by the Program of Science and Technology, Zhenjiang City, No. SH2006019
Peer reviewer: Peter L Lakatos, MD, PhD, Assistant Professor, 1st Department of Medicine, Semmelweis University, Koranyi S 2A, Budapest H1083, Hungary
S- Editor Zhu LH L- Editor Alpini GD E- Editor Ma WH
References
- 1.Ihle JN. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 2.Shuai K, Schindler C, Prezioso VR, Darnell JE Jr. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 3.Schindler C, Shuai K, Prezioso VR, Darnell JE Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 4.Shuai K, Stark GR, Kerr IM, Darnell JE Jr. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 5.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE Jr. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 6.Hayes TE, Kitchen AM, Cochran BH. Inducible binding of a factor to the c-fos regulatory region. Proc Natl Acad Sci USA. 1987;84:1272–1276. doi: 10.1073/pnas.84.5.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE Jr. Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE Jr, Levy DE. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 11.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Akira S. STAT family of transcription factors in cytokine-mediated biological responses. Cytokine Growth Factor Rev. 2000;11:199–207. doi: 10.1016/s1359-6101(00)00005-8. [DOI] [PubMed] [Google Scholar]
- 13.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19:2548–2556. doi: 10.1038/sj.onc.1203551. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 17.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 18.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 20.Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 21.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 22.Suiqing C, Min Z, Lirong C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J Dermatol. 2005;32:354–360. doi: 10.1111/j.1346-8138.2005.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu HJ, Moroi Y, Masuda T, Yasumoto S, Kokuba H, Imafuku S, Koga T, Tetsuya T, Tu YT, Aburatani H, et al. Expression of phosphorylated Stat3, cyclin D1 and Bcl-xL in extramammary Paget disease. Br J Dermatol. 2006;154:926–932. doi: 10.1111/j.1365-2133.2005.06951.x. [DOI] [PubMed] [Google Scholar]
- 24.Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, Xie K, Sawaya R, Huang S. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- 25.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Nagayasu T, Sekine I. Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J Clin Pathol. 2005;58:833–838. doi: 10.1136/jcp.2004.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 27.Shike M, Winawer SJ, Greenwald PH, Bloch A, Hill MJ, Swaroop SV. Primary prevention of colorectal cancer. The WHO Collaborating Centre for the Prevention of Colorectal Cancer. Bull World Health Organ. 1990;68:377–385. [PMC free article] [PubMed] [Google Scholar]
- 28.Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH, et al. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112:594–642. doi: 10.1053/gast.1997.v112.agast970594. [DOI] [PubMed] [Google Scholar]
- 29.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frisch SM, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 31.Fan Y, Zheng S, Xu ZF, Ding JY. Apoptosis induction with polo-like kinase-1 antisense phosphorothioate oligo-deoxynucleotide of colon cancer cell line SW480. World J Gastroenterol. 2005;11:4596–4599. doi: 10.3748/wjg.v11.i29.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 33.Boyd DD, Levine AE, Brattain DE, McKnight MK, Brattain MG. Comparison of growth requirements of two human intratumoral colon carcinoma cell lines in monolayer and soft agarose. Cancer Res. 1988;48:2469–2474. [PubMed] [Google Scholar]
- 34.Frankel A, Rosen K, Filmus J, Kerbel RS. Induction of anoikis and suppression of human ovarian tumor growth in vivo by down-regulation of Bcl-X(L) Cancer Res. 2001;61:4837–4841. [PubMed] [Google Scholar]
- 35.Zushi S, Shinomura Y, Kiyohara T, Miyazaki Y, Kondo S, Sugimachi M, Higashimoto Y, Kanayama S, Matsuzawa Y. STAT3 mediates the survival signal in oncogenic ras-transfected intestinal epithelial cells. Int J Cancer. 1998;78:326–330. doi: 10.1002/(SICI)1097-0215(19981029)78:3<326::AID-IJC12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 36.Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto Y, Hosotani R, Wada M, Lee JU, Koshiba T, Fujimoto K, Tsuji S, Nakajima S, Doi R, Kato M, et al. Immunohistochemical analysis of Bcl-2, Bax, Bcl-X, and Mcl-1 expression in pancreatic cancers. Oncology. 1999;56:73–82. doi: 10.1159/000011933. [DOI] [PubMed] [Google Scholar]
- 38.Sarela AI, Verbeke CS, Ramsdale J, Davies CL, Markham AF, Guillou PJ. Expression of survivin, a novel inhibitor of apoptosis and cell cycle regulatory protein, in pancreatic adenocarcinoma. Br J Cancer. 2002;86:886–892. doi: 10.1038/sj.bjc.6600133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE Jr. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999;59:5059–5063. [PubMed] [Google Scholar]
- 41.Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 42.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 43.Makimura H, Mizuno TM, Mastaitis JW, Agami R, Mobbs CV. Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC Neurosci. 2002;3:18. doi: 10.1186/1471-2202-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C, Cao J, Huang KJ, Zhang F, Jiang T, Zhu L, Qiu ZJ. Inhibition of STAT3 activity with AG490 decreases the invasion of human pancreatic cancer cells in vitro. Cancer Sci. 2006;97:1417–1423. doi: 10.1111/j.1349-7006.2006.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coll ML, Rosen K, Ladeda V, Filmus J. Increased Bcl-xL expression mediates v-Src-induced resistance to anoikis in intestinal epithelial cells. Oncogene. 2002;21:2908–2913. doi: 10.1038/sj.onc.1205388. [DOI] [PubMed] [Google Scholar]