Abstract
Purpose
To study an automatic repositioning method to reduce variability in longitudinal MRSI exams based on a priori image registration. Longitudinal proton magnetic resonance spectroscopic imaging (1H MRSI) exams to study the effects of disease or treatment are becoming increasingly common. However, one source of variability in such exams arises from imperfect re-localization of the MRSI grid in the follow-up exams.
Materials and Methods
Six healthy subjects were each scanned three times during the course of one day. In each follow-up exam, a manually placed MRSI grid was acquired in addition to the automatically repositioned MRSI grid. Then coefficients of variance between baseline and follow-up scans were calculated for N-acetylaspartate, creatine and choline. In addition, the overall MRSI grid overlap and individual voxel overlaps were also calculated for both the visually and automatically repositioned voxels.
Results
Streamlined workflow, reduced variability of metabolite concentration measurements and increased voxel overlaps are noted when this automatic repositioning procedure is compared to the visual MRSI grid repositioning approach.
Conclusion
Our results suggest that this approach is able to improve reproducibility in longitudinal MRS exams.
Keywords: automatic repositioning, prospective realignment, Magnetic Resonance Spectroscopy (MRS), brain, MR Spectroscopic Imaging (MRSI)
Longitudinal proton magnetic resonance spectroscopic imaging (1H MRSI) studies following disease progression or response to treatment are becoming more common. In vivo magnetic resonance spectroscopy (MRS) of the brain is routinely integrated into many clinical protocols to evaluate patients with strokes, oxygen deprivation, epilepsy, multiple sclerosis, Alzheimer’s, various brain tumors, and other brain disorders. (1–5). Two-dimensional (2D) MRSI is required in most clinical studies in order to obtain metabolic information of the entire lesion – possibly revealing new tumor growth that is not yet visible on a regular MRI. Furthermore, the progress of lactate and the neuronal marker N-Acetylaspartate (NAA) concentrations exhibit valuable information in stroke patients. In such studies, reproducible measurements of metabolite concentrations are of paramount importance to ensure maximum sensitivity in detecting the effect of the disease or treatment. One of the factors that can significantly influence the reproducibility of metabolite concentration measurements is the accuracy of repositioning the MRSI grids.
A posteriori image registration and MRSI data re-sampling method has recently been shown to significantly decrease scan-to-scan variability (6). However, using this approach, correction cannot be performed out of plane for single slice MRSI data sets or for the edge voxels of the MRSI grid if a rotation is observed between the baseline and follow-up MRSI grids.
Here, we present the results of a study designed to minimize scan-to-scan variability by using a priori image registration, followed by the acquisition of MRSI data sets with identical coverage and orientation as the baseline scans. Our approach to reproducibly repositioning MRSI grids consisted of an automatic registration algorithm and a tailored pulse sequence that allowed us to acquire images from the same position and orientation as the baseline scan. This sequence only added approximately one minute to the protocol. To assess the quality of our method, we measured the coefficients of variance (CVs) of the brain metabolite quantities between baseline exams and two follow-up exams. In addition, we quantified the voxel overlap and displacement errors between baseline scans and the follow-up scans.
MATERIALS AND METHODS
Subjects
For this study, six volunteers (two females) were each scanned three times within a 12 hour period. Between the baseline and two follow-up scans, the subjects were removed from the scanner. The protocol was approved by the Institutional Human Research Review Committee and informed consent was obtained from the participants before the study.
Registration Algorithm
Images were aligned based on the mutual information metric, introduced by two independent groups (7,8) and later refined by Mattes (9). We used an automatic registration algorithm and a tailored pulse sequence that allowed us to acquire MR images from the same position and orientation as the baseline scan. The scanner does not allow the rotation of the spectroscopy voxel in the prescription, instead a 3D fast spoiled gradient echo (FSPGR) sequence was modified to accept the three translations in left-right (LR), anterior-posterior (AP) and superior-inferior (SI) and the three rotations (around x, y and z) that were output from the registration algorithm. In addition, the registration algorithm calculated the prescription slice and the new RAS (Right Anterior Superior) locations for the follow-up 2D MRSI when prescribed from the new triple oblique 3D FSPGR image. Details of the registration algorithm can be found in Hancu et al. (10). The algorithm converges rapidly within 12 seconds on a modern workstation with approximately 1 mm accuracy.
Acquisition Protocol
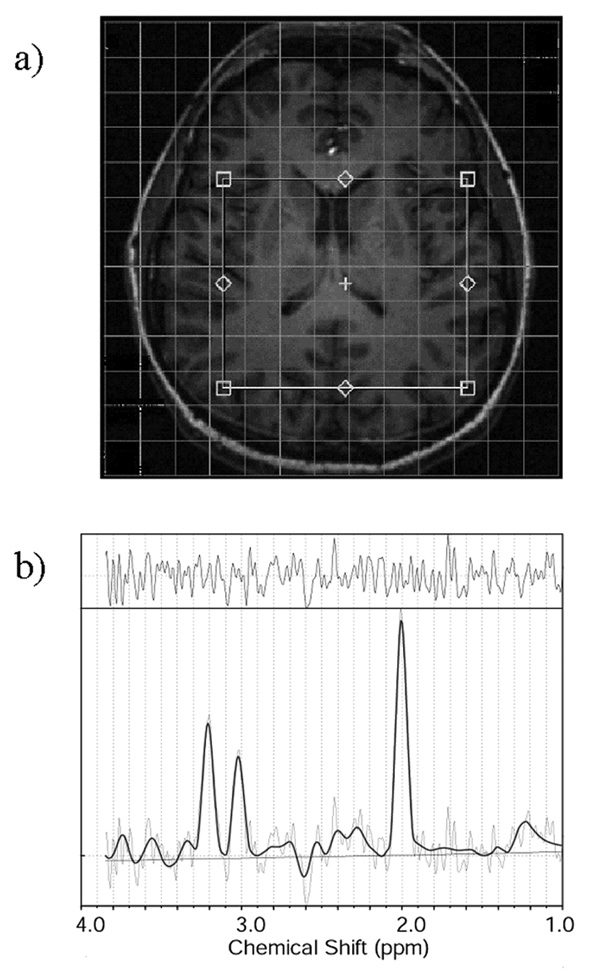
All experiments were done on a 3Tesla whole-body GE scanner (GE Healthcare, Milwaukee, WI, USA). The baseline scan for each subject included a sagittal T1 weighted localizer and a half brain axial (3D FSPGR) sequence with a resolution of 192×192 over FOV of 24cm. A volume consisting of 36 slices was acquired in 1.25min. 2D MRSI spectra were acquired over ~6.5min. using a PRESS sequence with CHESS water suppression. The MRSI sequence was acquired with a symmetric 220mm FOV, 16×16 phase-encoding steps, 10mm slice thickness, and TE/TR 135/1500. The volume of interest (VOI) was graphically prescribed over an axial image that included the caudate nucleus, putamen, internal capsule, the corpus callosum, the lateral ventricles and a variety of cortical structures resulting in approximately 6 × 5 voxels, as shown in Fig. 2a.
Figure 2.
Figure 2a: Typical location for the positioning of the MRSI grid. The volume of interest (VOI) was graphically prescribed over an axial image that included the caudate nucleus, putamen, internal capsule, the corpus callosum, the lateral ventricles and a variety of cortical structures, including the frontal operculum, the parietal cortex and the insula, resulting in approximately 6 × 5 voxels.
Figure 2b: Typical spectrum from a homogeneous brain region. The spectra included in the analysis had line widths between 4 Hz (from homogeneous white matter) and 12 Hz (from inhomogeneous regions surrounding the caudate nucleus and the putamen). Spectral data, along with fitted line (bold), fitted baseline and the residuals of the fit are displayed between 1 and 3.85 ppm.
To evaluate the registration algorithm, two MRSI data sets were acquired in the follow-up scans (with their order being randomized): one was placed on the axial FSPGR, using visual inspection of the baseline MRSI grid location for repositioning precision, and the second one was repositioned on a triple oblique slice, using the positioning information offered by the registration algorithm.
Data Analysis
Voxel Overlap Calculations
Overall MRSI grid overlap and individual voxel overlaps were also calculated for both the visually and automatically repositioned voxels using a procedure described elsewhere (10). Voxel overlap was calculated using the registration algorithm, hereby the voxel overlap was defined as the percentage of the baseline voxel volume encompassed by the follow-up voxel. In addition, displacements for the LR, AP, and SI coordinate of the MRSI grids and the offsets in the angles of rotation along x, y, and z with respect to the ideal voxel orientation was calculated. Subsequently, the average voxel overlap, the displacement and the rotation of both follow-up exams was calculated.
Spectroscopy Data Processing and Quantification
MRS raw data was processed offline using SAGE spectral analysis program (GE Healthcare, Milwaukee, WI) for preprocessing. Preprocessing included 1.25Hz spectral apodization, internal water referencing and spectral zerofilling before Fourier transformation. Quantities of the brain metabolites NAA, creatine (Cr), and choline (Cho) were determined using LCModel 6.0–1 (11). Fits were performed between 1.00 and 3.85ppm. Voxels with poor signal-to-noise ratio and those located within CSF-spaces were excluded from the data analysis.
Statistical data analysis
All statistical data analyses described below were performed using Statistical Analysis System (SAS) software (version 8; SAS, Cary, NC). First, displacement errors for the LR, AP, and SI coordinated for the MRSI grid and the offsets in the angles of rotation along x, y, and z with respect to ideal voxel orientation were averaged for the two follow-up exams for the manual and automatic repositioning method, respectively. Hereby, the absolute values had been averaged since offsets are expected in either direction. To evaluate the displacement errors and the rotation offsets in the angles non-parametric Wilcoxon Signed Rank tests were performed since a normal distribution of absolute displacement error and rotational offsets cannot be expected. The percent overlap was calculated for all four corner voxels, the average of these corner voxels, the center voxels and the overall MRSI grid. Student paired t-tests were conducted to examine mean differences in the percent overlap for the two repositioning methods.
For both the visual and automatic repositioning method, intra subject CVs were calculated for Cr, Cho, NAA, Cho/Cr and NAA/Cr by means of CV =SD/mean·100 where both the standard deviation, SD, and the mean are calculated for each voxel out of the 3 measurements. We chose to normalize the standard deviations by their mean value because we expect the variability to scale with the absolute value of the metabolite. Subsequently, Wilcoxon Signed Rank tests were used comparing the median CVs across all voxels for each subject. Note that an assumption has been made that CVs from different voxels in one subject are independent of each other. We also performed Wilcoxon Signed Rank tests and one-tailed signed t-test comparing the median CVs across all voxels and all 6 subjects.
RESULTS
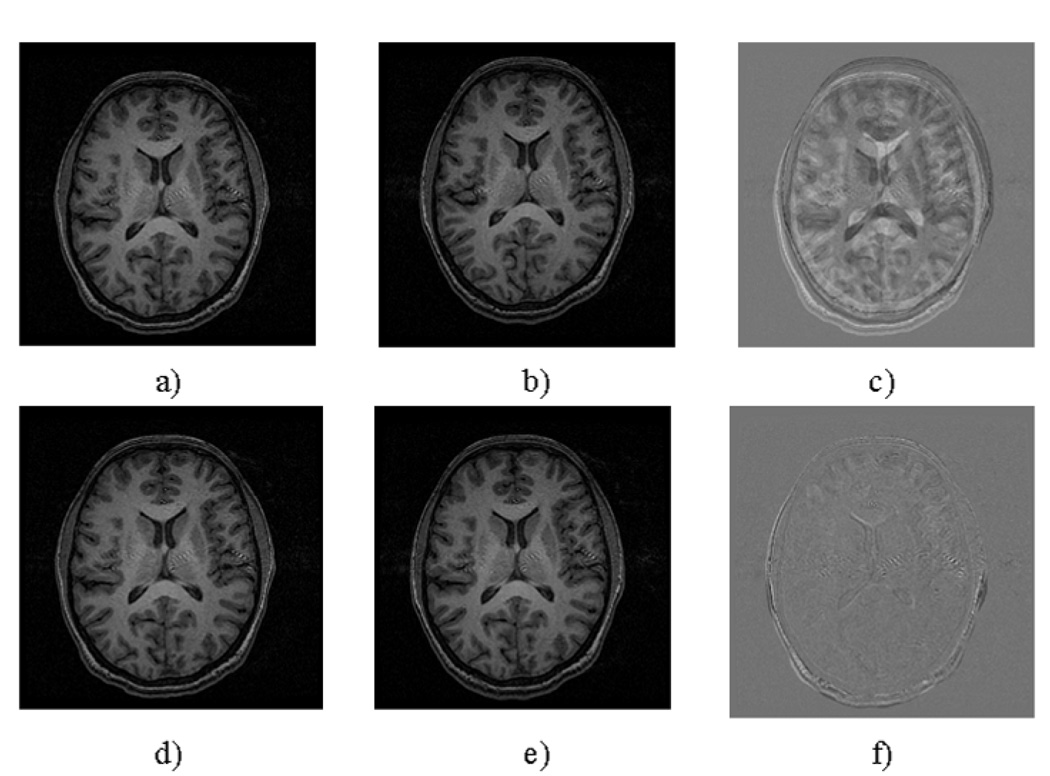
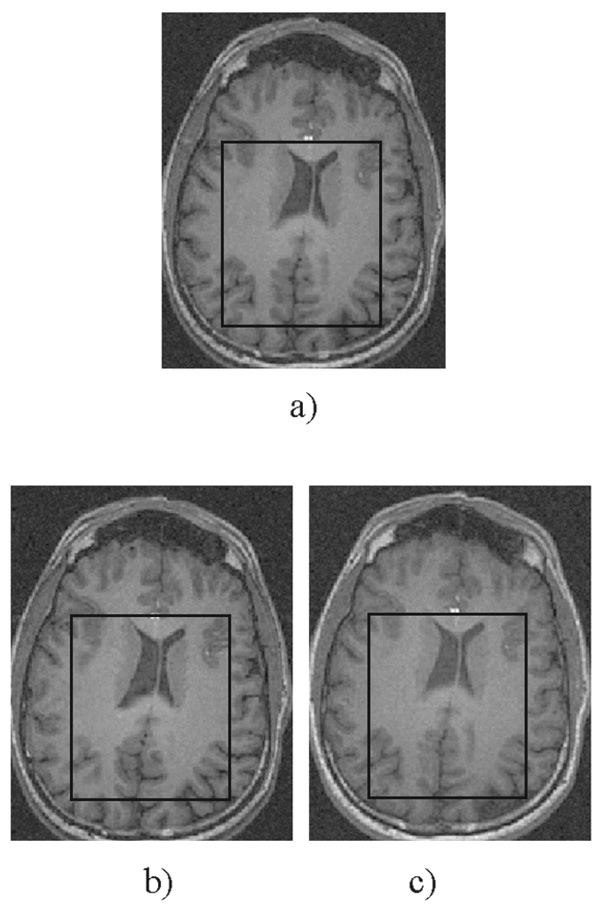
Typical performance of the registration procedure is presented in Fig. 1. Differences in head positioning and/or brain coverage in the two exams (translations and rotations) are easily visible through the brain structure in the difference image. Figure 2a presents the typical location for the positioning of the MRSI grid, and Fig. 2b a typical spectrum from a homogeneous brain region. The spectra included in the analysis had line widths between 4Hz (from homogeneous white matter) and 12Hz (from inhomogeneous regions surrounding the caudate nucleus and the putamen). Despite the great effort to visually reposition the MRSI grid on the same location, it is obvious from Fig. 3 that the automatic repositioning method resulted in a better overlap with the baseline scan.
Figure 1.
Axial slices from the brain of a normal volunteer a) Baseline scan b) Same slice numbers in a manually placed follow-up scan c) Pixel-by-pixel difference between the baseline (a) and follow-up scans (b) d) The same baseline slice e) algorithm derived reacquisition of the same plane f) Pixel-by-pixel difference between the baseline scan (d) and automatically prescribed reacquisition (e).
Figure 3.
Typical voxel grid location in the (a) baseline exam including the axial image and the VOI of the MRSI grid, (b) follow-up exam using manual repositioning and (c) follow-up exam using the automatic repositioning method. On the manually positioning MRSI grid, the follow-up MRSI grid location was closely co-registered in relation to the ventricles, however, the parietal cortex exhibits a deviating location.
The displacement errors for the LR, AP, and SI coordinates and the sum of the displacement and averages are shown in Table 1. The average offsets in the rotation angles and the means were also calculated. Wilcoxon Signed Rank tests across the six volunteers revealed significant improvements for the rotation offsets
Table 1.
Average displacement errors and average offsets in the rotation angles
| Volunteer | dLR (mm) | dAP (mm) | dSI (mm) | d (mm) | rx (°) | ry (°) | rz (°) | rmean | |
|---|---|---|---|---|---|---|---|---|---|
| Visual Repositioning | 1 | 0.14 | 0.10 | 0.08 | 0.02 | 1.05 | 1.11 | 6.48 | 7.71 |
| 2 | 2.87 | 1.77 | 2.12 | 0.51 | 0.78 | 1.70 | 3.73 | 5.17 | |
| 3 | 8.59 | 4.52 | 7.00 | 1.33 | 1.56 | 2.52 | 4.78 | 7.60 | |
| 4 | 2.01 | 0.39 | 0.98 | 1.66 | 1.84 | 0.59 | 0.68 | 0.28 | |
| 5 | 4.08 | 1.37 | 2.29 | 3.08 | 2.95 | 0.67 | 0.25 | 1.91 | |
| 6 | 2.59 | 2.19 | 0.56 | 1.24 | 1.96 | 0.40 | 0.29 | 0.74 | |
| Average | 3.38 | 1.73 | 2.17 | 1.31 | 1.69 | 1.17 | 2.70 | 3.90 | |
| Automatic Repositioning | 1 | 1.17 | 0.39 | 1.10 | 0.13 | 0.22 | 0.33 | 0.37 | 0.46 |
| 2 | 0.75 | 0.15 | 0.68 | 0.27 | 0.30 | 0.14 | 0.11 | 0.26 | |
| 3 | 0.93 | 0.09 | 0.92 | 0.04 | 0.27 | 0.09 | 0.15 | 0.16 | |
| 4 | 0.98 | 0.03 | 0.72 | 0.66 | 1.09 | 0.15 | 0.23 | 0.12 | |
| 5 | 1.22 | 0.07 | 1.15 | 0.29 | 0.21 | 0.08 | 0.04 | 0.10 | |
| 6 | 0.69 | 0.12 | 0.61 | 0.28 | 0.88 | 0.41 | 0.13 | 0.28 | |
| Average | 0.96 | 0.14 | 0.86 | 0.28 | 0.49 | 0.20 | 0.17 | 0.23 | |
| Wilcoxon p value | 0.0625 | 0.22 | 0.0625 | 0.0625 | 0.0313 | 0.0625 | 0.0313 | 0.0313 | |
Table 2 presents the percent voxel overlap between the baseline scans and two follow-up scans for the visual and automatic repositioning procedure of all four corners of the MRSI grid, the average percent voxel overlap, the center voxel, and the whole grid overlap. The analysis revealed that the whole grid overlaps were generally high for both repositioning procedures, but the automatic repositioning methods exhibited a 94.9% overlap between baseline scan and follow-up exams compared to only 81.7% overlap that was achieved using the visual repositioning method. The overlap for corner voxels showed a wide range of variability when using the visual repositioning. Student’s paired t-tests revealed that the automatic repositioning method has significantly better percent overlaps compared to visual repositioning.
Table 2.
Average percentage voxel overlap
| Volunteer | Corner voxel lowerleft |
Corner voxel upper left |
Corner voxel lower right |
Corner voxel upper right |
Average Corner voxel |
Center voxel |
Overall MRSI grid |
|
|---|---|---|---|---|---|---|---|---|
|
% Overlap Visual Repositioning |
1 | 97.2 | 97.0 | 97.2 | 97.0 | 97.1 | 97.4 | 99.2 |
| 2 | 39.7 | 51.5 | 59.0 | 55.5 | 51.4 | 65.6 | 86.9 | |
| 3 | 11.1 | 14.3 | 31.5 | 22.2 | 19.7 | 21.3 | 73.6 | |
| 4 | 80.4 | 68.7 | 73.3 | 57.5 | 69.9 | 73.7 | 82.1 | |
| 5 | 58.9 | 30.4 | 64.8 | 36.4 | 47.6 | 49.2 | 66.0 | |
| 6 | 68.6 | 59.7 | 65.6 | 55.3 | 62.3 | 68.2 | 82.7 | |
| Average | 59.3 | 53.6 | 65.2 | 54.0 | 58.0 | 62.6 | 81.7 | |
| % Overlap Automatic Repositioning | 1 | 81.8 | 85.0 | 84.4 | 86.8 | 84.5 | 85.6 | 96.3 |
| 2 | 89.3 | 90.6 | 88.5 | 88.1 | 89.1 | 90.2 | 95.8 | |
| 3 | 87.2 | 86.9 | 88.3 | 88.8 | 87.8 | 89.4 | 96.3 | |
| 4 | 77.0 | 86.9 | 80.0 | 91.4 | 83.8 | 86.1 | 91.7 | |
| 5 | 84.7 | 85.2 | 85.3 | 86.8 | 85.5 | 86.6 | 94.9 | |
| 6 | 85.8 | 87.2 | 91.1 | 83.5 | 86.9 | 90.5 | 94.7 | |
| Average | 84.3 | 86.9 | 86.2 | 87.6 | 86.2 | 88.1 | 94.9 | |
|
Paired t-test p value |
0.13 | 0.039 | 0.078 | 0.024 | 0.050 | 0.035 | 0.066 | |
Table 3 displays the intra subject median CVs calculated for the aforementioned metabolites. On average, 23 voxels per volunteer were included in the data analysis. In addition, the average median CV over all the volunteers was computed. Wilcoxon Signed Rank tests between the CV obtained with manual and automatic repositioning showed statistically significant improvement in some subjects for some metabolites (** with p<0.05, * with p<0.06). In addition, the p values were calculated when including all voxels across all subjects using Wilcoxon Signed Rank tests and signed t-tests. Since all subjects were scanned within 1 to 3 hours, no variations in metabolite concentrations and relaxation times are expected between the baseline scans and the two follow-up exams. The calculated CV for the metabolite concentrations and ratios are a good measure to assess the effect of the two repositioning methods. Also, during this short time period instrument stability should be comparable for both re-localization methods. We verified that the receiver gains did not change between scan, and the transmitter gains were constant within less than 1% variability. The manual re-localization procedure in our study displayed a wide range of CVs – e.g. up to 30% variance for NAA in subject 1. The variability is considerably lower when the automatic repositioning method is used - the maximum CV is only 17.4%. Except for Cho/Cr, all median standard deviations are lower when the automatic repositioning was performed.
Table 3.
Coefficients of variance for metabolite concentrations and ratios
| Volunteer | # of voxels |
Cr | Cho | NAA | Cho/Cr | NAA/Cr | |
|---|---|---|---|---|---|---|---|
| Median CVs Visual Repositioning | 1 | 21 | 30.2** | 28.1** | 37.0** | 13.5 | 13.1 |
| 2 | 25 | 13.85 | 11.07* | 7.67 | 10.65 | 14.06 | |
| 3 | 22 | 12.68** | 17.82 | 14.22 | 12.82 | 15.54 | |
| 4 | 26 | 10.66 | 9.19 | 9.22 | 10.16 | 13.88* | |
| 5 | 17 | 12.86 | 11.62 | 5.87 | 14.42 | 17.43 | |
| 6 | 24 | 13.08 | 12.44 | 7.66 | 14.17 | 11.43 | |
| Average | 23 | 15.5 | 15.0 | 13.6 | 12.6 | 14.2 | |
| Median CVs Automatic Repositioning | 1 | 21 | 15.1** | 14.9** | 17.4** | 11.8 | 12.3 |
| 2 | 25 | 13.38 | 9.63* | 5.94 | 14.02 | 10.70 | |
| 3 | 22 | 10.77** | 16.77 | 14.53 | 13.37 | 12.78 | |
| 4 | 26 | 9.60 | 10.01 | 6.45 | 10.25 | 11.48* | |
| 5 | 17 | 10.83 | 9.46 | 5.74 | 17.36 | 11.69 | |
| 6 | 24 | 12.79 | 12.80 | 7.90 | 16.00 | 10.47 | |
| Average | 23 | 12.1 | 12.3 | 9.7 | 13.8 | 11.6 | |
| Wilcoxon p value | 135 | 0.007 | 0.05 | 0.002 | 0.09 | 0.004 | |
| Signp value | 135 | 0.05 | 0.13 | 0.04 | 0.9 | 0.05 | |
P<0.05,
P<0.06
DISCUSSION
We have presented a method to reproducibly align MRSI grids. We compared the overlap of the MRSI excitation volume and the overlap volume of individual voxels measurements between the visual and automatic repositioning method. Our results indicate that the percent overlaps of the entire MRSI VOI were improved when the automatic repositioning method was used. Displacements and rotation errors were considerably decreased when the automatic repositioning methods was used for the re-localization of the VOI of the MRSI grid.
It has to be noted that the data were acquired by one highly skilled operator, with voxels repositioned on high-resolution scouts, and with the baseline axial image and prescribed MRSI grid displayed on a screen next to the scanner. This is a best case scenario for the visual voxel placement procedure. In a clinical setting where longitudinal MRSI studies are being performed by multiple MR technicians at different levels of expertise and where baseline images may be not available, the percentage voxel overlap will exhibit higher levels of variance for the visual placement procedure.
We observed significant improvement in measurement reproducibility for Cho, Cr, NAA and NAA/Cr using the automatic procedure. Our data is consistent within the range of reproducibility measures presented in the literature (6,12–17). While intra individual CVs for single voxel MRS were reported to vary between 3.3–8.1% for absolute quantification (12), reproducibility of metabolite ratios obtained with MRSI was somewhat poorer. Tedeschi et al. obtained intra-individual CVs of 8.2–22.2% and 4.5–1.0% for NAA/Cr and Cho/Cr, respectively. Hereby the highest variabilities were found in the caudate and thalamus. The majority of voxels were kept for analysis, regardless of their position across brain regions with high iron contents (such as the putamen) or proximity to the skull. Those voxels in particular significantly increase overall variability.
Approaches to prospectively align MR images have been presented in the literature. Gedat et al. (18) used the three orthogonal localizer images of the initial examination as a reference for patient position. Subsequently, after a follow-up localizer and registration of these images to the initial localizer the method involves calculation of the transformation matrix and adaptation of the gradient control such that a follow-up imaging examination uses this transformation and gradient adaptation. Our results compare favorably with (18) who report 0.1mm with 1.5mm SD in translation and 0.2 degrees with 1.5 degrees SD in rotation. Our improved accuracy is likely due to the increased FOV image used for registration. The main difference in these two papers lays in the different focus. While the primary focus of Gedat et al. is the mechanism for registration using a 3-plane localizer, our goal was to evaluate the effects of repositioning on metabolite concentration.
Van der Kouwe et al. (19) described an automatic slice repositioning for imaging using a statistical human brain atlas to automatically align a localizer image. This method is limited to human brain imaging, and cannot been transformed to other body parts or a pathological brain. Recently, Wu et al. proposed that a fully deformable model, which combines piecewise linear registration for coarse alignment with demons algorithm for voxel level refinement is more accurate for co-localization of functional signal from different subjects (20).
Our approach of a rigid body registration is extremely fast and robust, results in reduced variability of metabolite concentration measurements, increased voxel overlaps and can be adapted to other rigid structures e.g. extremities or animals. We have not explicitly studied the effects on B1 or B0 inhomogeneities, however, we did not observe any problems related to B1 artifacts (10).
Future studies will be performed in which we test the performance of this re-location program when treatment in form of surgery or radiation has been performed on tumor patients to see whether the program is yet capable of overlaying these images correctly. Local changes in morphology should produce only minor disruption to the global transformation. However, even if the global registration is good, the grid would no longer cover the tumor in the same manner as underlying tumor morphology has changed. Thus, the subjects we intend to study will be for the purposes of post-surgical follow-up and should not experience significant morphological changes. Preliminary results indicate good performance of our co-registration procedure between baseline and follow-up images for post-surgical follow-up scans.
In conclusion, significantly increased volume overlap and better measurement reproducibility, along with improved exam workflow, were observed for the automatic repositioning procedure compared to a visual repositioning procedure, suggesting this approach as the future of longitudinal MRS exams.
ACKNOWLEDGEMENTS
Grant Support: This work was supported by grants from the NIH, including R01NS050041 and AI028691.
The authors would like to thank Mr. Jeffrey Bombardier for proofreading this manuscript.
REFERENCES
- 1.Ross B, Kreis R, Ernst T. Clinical tools for the 90s: magnetic resonance spectroscopy and metabolite imaging. Eur J Radiol. 1992;14:128–140. doi: 10.1016/0720-048x(92)90226-y. [DOI] [PubMed] [Google Scholar]
- 2.Cecil KM, Lenkinski RE. Proton MR spectroscopy in inflammatory and infectious brain disorders. Neuroimaging Clin N Am. 1998;8:863–880. [PubMed] [Google Scholar]
- 3.Nelson SJ, Vigneron DB, Dillon WP. Serial evaluation of patients with brain tumors using volume MRI and 3D 1H MRSI. NMR Biomed. 1999;12:123–138. doi: 10.1002/(sici)1099-1492(199905)12:3<123::aid-nbm541>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 4.Van Zijl PC, Barker PB. Magnetic resonance spectroscopy and spectroscopic imaging for the study of brain metabolism. Ann N Y Acad Sci. 1997;820:75–96. doi: 10.1111/j.1749-6632.1997.tb46190.x. [DOI] [PubMed] [Google Scholar]
- 5.Lee PL, Gonzalez RG. Magnetic resonance spectroscopy of brain tumors. Curr Opin Oncol. 2000;12:199–204. doi: 10.1097/00001622-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Chu W-J, Pan C, Pan JW, Hetherington HP. Reproducibility of 1H Spectroscopic Imaging of the Human Hippocampus. Kyoto, Japan: 2004. [Google Scholar]
- 7.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 8.Viola P, Wells WI. Mutual information: an approach for the registration of object models and images. Int J Comp Vis. 1997;24:137–154. [Google Scholar]
- 9.Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Trans Med Imaging. 2003;22:120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- 10.Hancu I, Blezek DJ, Dumoulin MC. Automatic repositioning of single voxels in longitudinal 1H MRS studies. NMR Biomed. 2005;18:352–361. doi: 10.1002/nbm.965. [DOI] [PubMed] [Google Scholar]
- 11.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 12.Brooks WM, Friedman SD, Stidley CA. Reproducibility of 1H-MRS in vivo. Magn Reson Med. 1999;41:193–197. doi: 10.1002/(sici)1522-2594(199901)41:1<193::aid-mrm27>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 13.Lee PL, Yiannoutsos CT, Ernst T, et al. A multi-center 1H MRS study of the AIDS dementia complex: validation and preliminary analysis. J Magn Reson Imaging. 2003;17:625–633. doi: 10.1002/jmri.10295. [DOI] [PubMed] [Google Scholar]
- 14.Rose SE, de Zubicaray GI, Wang D, et al. A 1H MRS study of probable Alzheimer's disease and normal aging: implications for longitudinal monitoring of dementia progression. Magn Reson Imaging. 1999;17:291–299. doi: 10.1016/s0730-725x(98)00168-4. [DOI] [PubMed] [Google Scholar]
- 15.Schirmer T, Auer DP. On the reliability of quantitative clinical magnetic resonance spectroscopy of the human brain. NMR Biomed. 2000;13:28–36. doi: 10.1002/(sici)1099-1492(200002)13:1<28::aid-nbm606>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Bertolino A, Callicott JH, Nawroz S, et al. Reproducibility of proton magnetic resonance spectroscopic imaging in patients with schizophrenia. Neuropsychopharmacology. 1998;18:1–9. doi: 10.1016/S0893-133X(97)00090-0. [DOI] [PubMed] [Google Scholar]
- 17.Jackson EF, Doyle TJ, Wolinsky JS, Narayana PA. Short TE hydrogen-1 spectroscopic MR imaging of normal human brain: reproducibility studies. J Magn Reson Imaging. 1994;4:545–551. doi: 10.1002/jmri.1880040406. [DOI] [PubMed] [Google Scholar]
- 18.Gedat E, Braun J, Sack I, Bernarding J. Prospective registration of human head magnetic resonance images for reproducible slice positioning using localizer images. J Magn Reson Imaging. 2004;20:581–587. doi: 10.1002/jmri.20153. [DOI] [PubMed] [Google Scholar]
- 19.van der Kouwe AJ, Benner T, Fischl B, et al. On-line automatic slice positioning for brain MR imaging. Neuroimage. 2005;27:222–230. doi: 10.1016/j.neuroimage.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Wu M, Carmichael O, Lopez-Garcia P, Carter CS, Aizenstein HJ. Quantitative comparison of AIR, SPM, and the fully deformable model for atlas-based segmentation of functional and structural MR images. Hum Brain Mapp. 2006 doi: 10.1002/hbm.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tedeschi G, Bertolino A, Campbell G, et al. Reproducibility of proton MR spectroscopic imaging findings. Am J Neuroradiol. 1996;17:1871–1879. [PMC free article] [PubMed] [Google Scholar]