Abstract
Eukaryotes typically have three multi-subunit enzymes that decode the nuclear genome into RNA, namely DNA-dependent RNA polymerases I, II and III. Remarkably, higher plants have five multi-subunit nuclear RNA polymerases: the ubiquitous Pol I, II and III, which are essential for viability, plus two non-essential polymerases, Pol IVa and Pol IVb that specialize in small RNA-mediated gene silencing pathways. RNA-directed DNA methylation of endogenous repetitive elements, silencing of transgenes, regulation of flowering time genes, inducible regulation of adjacent gene pairs, and spreading of mobile silencing signals are examples of phenomena that require Pol IVa and/or Pol IVb. Although biochemical details concerning Pol IV enzymatic activities are lacking, genetic evidence suggests several alternative models for how Pol IV might function.
RNA polymerases IVa and IVb: non-essential polymerases devoted to gene silencing
In all eukaryotes, DNA-dependent RNA polymerases (abbreviated RNAP, or Pol) I, II, and III transcribe essential genes that include rRNAs, mRNAs and tRNAs (see glossary for abbreviations used in the article). Pol I, II and III are complicated enzymes with 12–17 subunits, including structural and functional homologs of the five bacterial RNAP subunits [1]. The largest and second-largest RNAP subunits, the homologs of bacterial β’ and β, interact to form the DNA entry and RNA exit channels as well as the catalytic center for RNA synthesis [2] (Figure 1a).
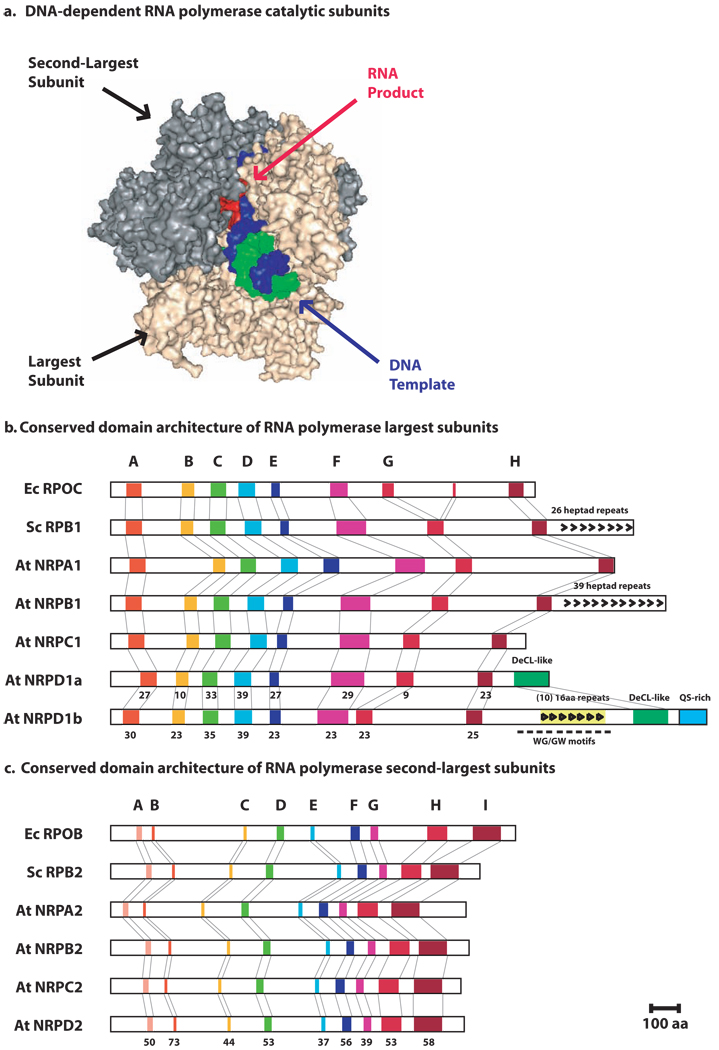
Figure 1.
Catalytic subunits of DNA-dependent RNA polymerases
(a) The largest and second-largest subunits form the catalytic center. The image is a surface rendering generated using the crystal coordinates for a yeast Pol II elongation complex determined by Westover, Bushnell and Kornberg (PDB:1R9T). Only the two largest Pol II subunits are shown. The DNA template strand is shown in blue, the non-template strand in green and the nascent RNA in red.
(b) Domain structures of RNAP largest subunits. E.coli (Ec RPOC) and yeast Pol II largest subunits (Sc RPB1) are compared to the Arabidopsis largest subunits for Pol I (At NRPA1), Pol II (At NRPB1), Pol III (At NRPC1), Pol IVa (At NRPD1a) and Pol IVb (At NRPD1b). Positions of conserved domains A–H are highlighted. Numbers below Pol IV domains are % identities to corresponding Arabidopsis Pol II subunit domains. CTDs of yeast and Arabidopsis Pol II largest subunits have 26 or 39 copies, respectively, of a seven amino acid (heptad) repeat. The domain with similarity to the Defective Chloroplasts and Leaves gene (DeCL domain), present in the CTDs of the Pol IVa and Pol IVb largest subunits, is shown in green. The CTD of NRPD1b also includes a region rich in WG/GW motifs, overlapping ten imperfect 16 amino acid repeats, and a domain composed of alternating glutamines and serines (QS-rich domain).
(c) Domain structures of RNAP second-largest subunits. E.coli (Ec RPOB) and yeast Pol II subunits (Sc RPB2) are compared to the Arabidopsis second-largest subunits for Pol I (At NRPA2) , Pol II (At NRPB2), Pol III (At NRPC2) or Pol IV (At NRPD2). Positions of conserved domains A–I are highlighted. Numbers below Pol IV domains are % identities to the corresponding Arabidopsis Pol II subunit domains.
At present, the catalytic subunits homologous to those depicted in Figure 1a are the only known Pol IVa and Pol IVb subunits in Arabidopsis, which is the species we will discuss throughout this review. They were initially identified in the newly sequenced Arabidopsis genome by the first author (CSP), who found two genes for an atypical fourth class of polymerase largest subunit and two genes for an atypical class of second-largest subunit. Collaborator J. Eisen (The Institute for Genomic Research) showed these putative subunits to be founding members of novel plant-specific clades [3] (see also refs. [4–6]). Like Pol I, II and III subunits, the atypical subunits have been shown to be nuclear proteins [4 7 8], representing a new class of polymerase, designated nuclear RNA polymerase IV (Pol IV) [4 5].
NRPD1a is the largest subunit of Pol IVa [4 5] whereas NRPD1b is the largest subunit of Pol IVb [9 10] (subunit nomenclature is discussed in Box 1). Both Pol IV largest subunits have C-terminal domains (CTDs) that share similarity with the Defective Chloroplasts and Leaves gene (abbreviated DeCL in this article) that is required for 4.5S rRNA processing in chloroplasts [11] (Fig. 1b). The CTD of NRPD1b also includes ten imperfect 16 amino acid repeats within a tryptophan and glycine (WG/GW)-rich region. A glutamine and serine (Q/S)-rich domain is present at the distal end of the CTD (Figure 1b). Whereas the WG/GW motifs are proposed to mediate Argonaute protein interactions [8 12], the significance of the DeCL and Q/S domains is unknown. However, the DCL and Q/S domains may facilitate additional molecular interactions in analogy to the CTD of the Pol II largest subunit, which mediates multiple interactions governing such processes as transcriptional activation by enhancers, transcription elongation and several mRNA processing steps [13–15]. Both Pol IVa and Pol IVb have an NRPD2 subunit that is encoded by the same gene, NRPD2a [4 5 9 10]. NRPD1a and NRPD1b each co-immunoprecipitate and colocalize with NRPD2 [7], but the alternative largest subunits do not immunoprecipitate one another, indicating that Pol IVa and Pol IVb are distinct physical entities.
Box 1- Pol IV subunit nomenclature
Nomenclature for Pol IV subunit genes derives from naming systems used in other eukaryotic model systems (such as the yeast, Saccharomyces cerevisiae) in which RNA polymerase I, II, and III are designated RPA, RPB, and RPC, respectively. In Arabidopsis, an N, for “nuclear,” was added ( e.g. NRPA, NRPB etc.) to polymerase subunit gene names in order to circumvent nomenclature conflicts with unrelated genes. The resulting gene names were registered with The Arabidopsis Information Resource by joint request of the David Baulcombe and Craig Pikaard labs. Largest subunits that are homologs of bacterial β′ are designated, by convention, with the number 1, such that the unique Arabidopsis genes NRPA1, NRPB1 and NRPC1 encode the largest subunits of pol I, II, and III, respectively. Likewise, the genes encoding the second-largest subunits of Arabidopsis pol I, II, and III are designated NRPA2, NRPB2, and NRPC2. Based on this naming scheme, the two related, but distinct Pol IV largest subunits were designated NRPD1a and NRPD1b. Likewise, the two Pol IV second-largest subunit genes are designated NRPD2a and NRPD2b. Only NRPD2a is functional in the Col-O ecotype of Arabidopsis that has been studied to date [4 5 9 10]. Therefore, NRPD2a can be referred to simply as NRPD2. In other plant species, there can be multiple functional genes for both the largest and second-largest subunits of Pol IV.
The full subunit compositions of Pol IVa and Pol IVb are not known, nor are their templates or enzymatic products. However, a flurry of studies in the past three years have shown that Pol IVa and (to a lesser extent) Pol IVb are key to a number of RNA-mediated gene silencing phenomena. These pathways, and the roles of Pol IV in them, are the focus of our review.
Roles of Pol IVa and Pol IVb in the RNA-directed DNA methylation pathway
Arabidopsis has four Dicer endonucleases (DCLs), six single-subunit RNA-dependent RNA polymerases (RDRs) and ten Argonaute proteins (AGOs) that participate in miRNA and siRNA-mediated transcriptional or post-transcriptional silencing [16–19]. In the RNA-directed DNA methylation (RdDM) pathway for transcriptional gene silencing [20–23], double-stranded RNAs generated with the involvement of RDR2 are cleaved by DCL3 and resulting siRNAs are loaded into AGO4- and/or AGO6-RISC complexes that mediate the de novo methylation of cytosines within DNA sequences complementary to the siRNAs [22 24–28]. Realization that Pol IVa and Pol IVb are players in the RdDM pathway came from a combination of genetic screens [5 10] and reverse-genetic analyses [4 9]. Silencing Defective (sde) mutants identified in screens for the derepression of a silenced transgene locus led to the identification of sde4 as an allele of NRPD1a [5]. A subsequent test to determine if one of the atypical second-largest subunit (NRPD2) genes might partner with NRPD1a revealed that insertional mutants of NRPD2a, like sde4/nrpd1a mutants, disrupted the silencing pathway, coincident with the disappearance of 24 nt siRNAs and loss of cytosine methylation at corresponding loci [5]. Our laboratory initially focused on NRPD2, showing that it was not redundant with the equivalent Pol I, II or III subunits and did not co-purify with Pol I, II or III [4]. However, NRPD2 was found to localize within the nucleus and to affect the coalescence of heterochromatic sequences into chromocenters [4]. Heterochromatic DNA is typically heavily methylated, and loss of cytosine methylation occurred at a subset of heterochromatic loci in nrpd2 as well as in nrpd1a mutants [4]. Collectively, the initial studies of NRPD1a and NRPD2 pointed to the existence of Pol IVa.
A genetic screen for mutations causing the derepression of a reporter gene silenced by RdDM identified DRD1, a member of the SWI2/SNF2 chromatin remodeling protein family [29] as well as DRD2 and DRD3, which turned out to be NRPD2a and NRPD1b, respectively [10]. The realization that the NRPD1b gene had been mistakenly annotated as two genes [4 5 10] also led to a reverse-genetic examination of cytosine methylation and siRNA phenotypes in nrpd1b insertional mutants [9]. Collectively, these independent studies revealed the existence of Pol IVb and showed that siRNAs eliminated in Pol IVa mutants [4 5] are not abolished in Pol IVb mutants [9 10] despite similar losses of cytosine methylation [9 10]. These observations, based on a small number of loci, indicated that Pol IVa and Pol IVb act at different steps in the RdDM pathway, with Pol IVa acting upstream of siRNA production and Pol IVb functioning at a later step in the pathway, mostly downstream of siRNA production [10]. Recent genome-wide analyses of small RNA populations have shown that there are at least 4600 Arabidopsis loci that give rise to small RNAs, with 94% of them being dependent on Pol IVa [30]. Pol IVb plays little, if any role in siRNA abundance at approximately one-third of these loci, has intermediate effects at another one-third of the loci, and is absolutely required for siRNA production at one-third of the Pol IVa-dependent loci [30]. However, there are no definitive examples of siRNAs that are dependent on Pol IVb only, and do not require Pol IVa. These results are consistent with the hypothesis that Pol IVa acts upstream of siRNA production. The role of Pol IVb in siRNA production is less clear, and could be indirect. A positive feedback relationship exists between heterochromatin formation and continued siRNA production such that Pol IVb's role in facilitating RdDM may explain Pol IVb's influence on siRNA abundance, as has been depicted in circular models for the RdDM pathway [7 8].
Localization of proteins involved in RdDM have provided insight into the pathway [7 8 31 32]. Pol IVa, Pol IVb and DRD1 colocalize with chromosomal loci that are both sources and targets of abundant siRNAs, suggesting an involvement in the generation of siRNA precursors or the targeting of siRNA-directed chromatin modifications [7]. AGO4 and DRM2, the primary de novo DNA methyltransferase, also colocalize at source/target loci in some nuclei [32]. RNA-FISH combined with protein immunolocalization showed that siRNAs colocalize with RDR2, DCL3, AGO4 and NRPD1b within a nucleolar compartment interpreted to be an siRNA processing center [7]. The processing center includes several molecular markers of Cajal bodies [8], which are dynamic compartments important for assembling ribonucleoprotein complexes involved in pre-mRNA splicing, pre-rRNA processing, RNA methylation and pseudouridylation, telomerase assembly and histone mRNA 3’ end formation [33 34]. Formation of siRNA-RISC complexes is consistent with the overall theme of assembling ribonucleoprotein complexes within Cajal bodies [8 33–35]. Recent evidence suggests that miRNA processing in plants may also occur within nucleolus associated Cajal body-like entities that include the spliceosomal proteins SmB and SmD3, which are found in both Cajal bodies and spliceosomes, but that lack the canonical Cajal body protein, coilin [36]. Other groups have suggested that these miRNA processing centers are not Cajal bodies due to the absence of coilin [37 38]. However, Drosophila lacks coilin yet has functional Cajal bodies [39]. The observations can be reconciled by the hypothesis that there are multiple sub-classes of Cajal bodies, some of which have coilin and some of which do not [34 35 39].
Because Pol IVa colocalizes with loci that give rise to abundant 24 nt siRNAs and because loss of NRPD1a function causes all other known components of the RdDM pathway to mislocalize, Pol IV is thought to act at an initial step of the pathway, upstream of RDR2 [7]. CLSY1, a SWI-SNF family protein (like DRD1) co-localizes with RDR2 at the inner perimeter of the nucleolus and in clsy1 mutants, RDR2 localization is severely disrupted [40]. Pol IVa localization is also affected, albeit to a lesser degree [40], suggesting that CLSY1 functions at the interface between Pol IVa and RDR2, presumably facilitating the generation of dsRNAs that are diced by DCL3 and loaded into AGO4 effector complexes [16 17 26 41] within the nucleolar siRNA processing center [7 8]. NRPD1b colocalizes with AGO4 both within the processing center [7 8] and at target loci [32], interacting with AGO4 via the CTD [8 12]. Current models suggest that siRNA-AGO4-Pol IVb effector complexes then locate their targets by virtue of siRNA-target base-pairing interactions [7 8]. Pol IVb, DRD1 and DRM2 are then thought to collaborate in the siRISC-directed DNA methylation process via an unknown mechanism [21]. DNA methylation then appears to feed back on the production of siRNAs, such that siRNAs are depleted in drm mutants at some loci [4 7 41] and in ddm1 (decrease in DNA methylation 1) or met1 (cytosine methyltransferase 1) mutants at other loci [42]. Therefore, it is possible that Pol IVa preferentially transcribes methylated DNA [4] or aberrant RNAs generated from methylated loci [7 43 44] as a means of perpetuating the repression cycle.
A role for Pol IV in flowering
Although non-essential for viability, Pol IVa and Pol IVb nonetheless play roles in development, affecting flowering time in the context of the RdDM pathway. Under short-day conditions, flowering in nrpd1a and nrpd1b mutants is significantly delayed, as is also the case in rdr2, dcl3, ago4 and drm mutants [9 45]. The flowering time regulators FCA and FPA were identified in screens for mutants disrupting RNA-directed gene silencing, and appear to be players in the RdDM pathway that act at some, but not all, loci [46]. At least two flowering genes, FWA and FLC , appear to be targets of silencing via Pol IV-dependent siRNA pathways [45 47 48].
The role of Pol IV in abiotic and biotic stress-inducible siRNA production
Pol IV plays an important role in the production of nat-siRNAs derived from natural antisense transcripts [49–53]. These siRNAs are generated from dsRNAs derived from the overlapping 3′ ends of convergently transcribed gene pairs. Transcription of one member of the gene pair is constitutively expressed but the other is inducible, as in the case of the SRO5 and P5CDH gene pair. Salt stress induces SRO5 expression such that its transcript can anneal with the P5CDH mRNA to form a region of dsRNA. In a process involving Pol IVa, RDR6, SGS3 and DCL2, a 24 nt nat-siRNA is produced which is thought to guide the cleavage of P5CDH transcripts, setting the phase for generation of additional DCL1-dependent 21 nt siRNAs [49]. The resulting down-regulation of P5CDH results in increased proline synthesis, a physiological response that helps confer salt tolerance.
Pathogen-inducible siRNAs provide two examples of additional means for generating nat-siRNAs [54 55]. In the first, infection of Arabidopsis with Pseudomonas syringae generates a 22 nt nat-siRNA in a pathway that requires Pol IVa, RDR6 and SGS3, similar to the salt stress-induced nat-siRNA discussed above except that DCL2 is not involved; instead, DCL1, HYL1 and HEN1 are required for siRNA production in the pathogen response. The end-result is the down-regulation of PPRL, a negative regulator of pathogen resistance. More recently, a 39–41nt RNA was also shown to be induced upon Pseudomonas syringae infection [54]. This so-called lsiRNA (long siRNA) matches the overlapping region of the SRRLK and AtRAP gene pair and specifically downregulates AtRAP, another negative regulator of the pathogen defense response, in a pathway requiring Pol IVa and Pol IVb, DCL1, HYL1, HEN1, HST1 (HASTY1), RDR6, DCL4, AGO7, and SDE3 (SILENCING DEFECTIVE 3). Most of these proteins (DCL1, HYL1, HEN1, HST1, RDR6, DCL4, AGO7) are also players in the so-called trans-acting siRNA pathway in which a miRNA-mediated cleavage of a specific target mRNA initiates the subsequent production of siRNAs from the cleaved mRNA [56–59]. Resulting siRNAs then target additional mRNAs for cleavage, thereby amplifying the signal in a regulatory cascade. Whether a similar regulatory cascade occurs upon bacterial infection, and where Pol IVa and Pol IVb fit within such a pathway, is not yet clear.
Roles of Pol IV in the spread of silencing
Pol IVa is required for both short-range spreading of RNA silencing cell-to-cell via plasmadesmata and long-range silencing via the phloem [60 61]. Two independent screens revealed a requirement for Pol IVa and RDR2 in short-range spreading of silencing [40 62] in addition to DCL4 [40 63], DCL1, HEN1 and AGO1 [62]. By contrast HYL1, DCL3, AGO4, RDR6 [40 62 63], Pol IVb (NRPD1b) and DRD1 [40] are all dispensable. Although both 24- and 21-nt transgene-specific siRNAs are produced, the DCL4-dependent 21-nt siRNAs are believed to be the primary short-range mobile signals [40 62 63]. However, longer siRNAs can suffice when overproduced in mutants of DRB4, a dsRNA binding protein that partners with DCL4 in the production of 21 nt siRNAs [62].
In Pol IVa mutants, silencing is impaired even in the phloem cells where the silencing signal is initiated, suggesting that Pol IVa acts at an initiating step in the process that ultimately gives rise to the mobile silencing signal(s). Interestingly, the spread of silencing can be dramatically enhanced in dcl3 and ago4 mutants [40], coincident with increased 21 nt siRNA production and loss of 24 nt siRNAs. A possibility is that Pol IVa/RDR2-dependent dsRNA substrates can be channeled into either 24 nt or 21 nt siRNA production, with the 21 nt siRNAs acting as the primary short-range mobile signals.
An ability to distinguish between production and perception of silencing signals has come from a study in which wild-type or mutant rootstocks or scions (shoots) were grafted to one another and monitored for long-distance silencing of a GFP transgene [64]. Pol IVa (NRPD1a), RDR2, DCL3, AGO4 and RDR6 are all required for the scion to respond to a silencing signal derived from a dsRNA hairpin expressed in the rootstock [65]. However, none of these proteins are required to generate the mobile signal. Interestingly, RDR6 is required for the perception of the long-distance signal [65] but is dispensable for short-range silencing [40 62]. Pol IVb (NRPD1b) is dispensable for both short and long-distance silencing, consistent with the hypothesis that Pol IVb functions in chromatin modification rather than RNA production.
The nature of the long-distance silencing signal is unknown, but dcl1-8 hypomorphs and dcl2,3,4 triple mutants defective for miRNA or siRNA production, respectively, continue to produce the mobile signal in roots, as do mutants for Pol IVa, Pol IVb, RDR2 and RDR6 [65]. Therefore, it seems unlikely that dicer-generated small RNAs are the long-distance signaling molecules. Instead, larger RNAs may serve as the mobile signal(s). An intriguing observation is that siRNAs produced in the scion upon reception of the silencing signal do not correspond to the ~2/3 of the GFP gene that was used as the hairpin trigger sequence; instead, the siRNAs neatly correspond to the ~1/3 of the GFP transgene located downstream (3′) of the trigger sequences [65]. It is not clear why this should be the case if siRNAs are the mobile signal. Antisense siRNAs could anneal anywhere throughout the first 2/3 of the target mRNA and might be expected to prime RDR activity in the upstream direction. Likewise, siRNA-directed cleavage of target mRNAs, which would render the 3′ target fragment uncapped, “aberrant” and a potential substrate for RDR6 [66] would generate a diverse set of cleaved fragments throughout the first 2/3 of the GFP target. Therefore, a possibility is that the dsRNA trigger molecule itself, or its component strands, might be the mobile signal(s), which is plausible given the evidence that intact mRNAs can traffic through phloem [67]. If the antisense strand of the dsRNA trigger were to anneal to the intact mRNA in the shoot such that only the 3′ portion of the GFP mRNA were to remain single-stranded, the resulting structure might somehow direct RDR6 and Pol IVa-dependent amplification of the single-stranded sequences 3′ of the trigger sequence.
Unsolved mysteries and future directions
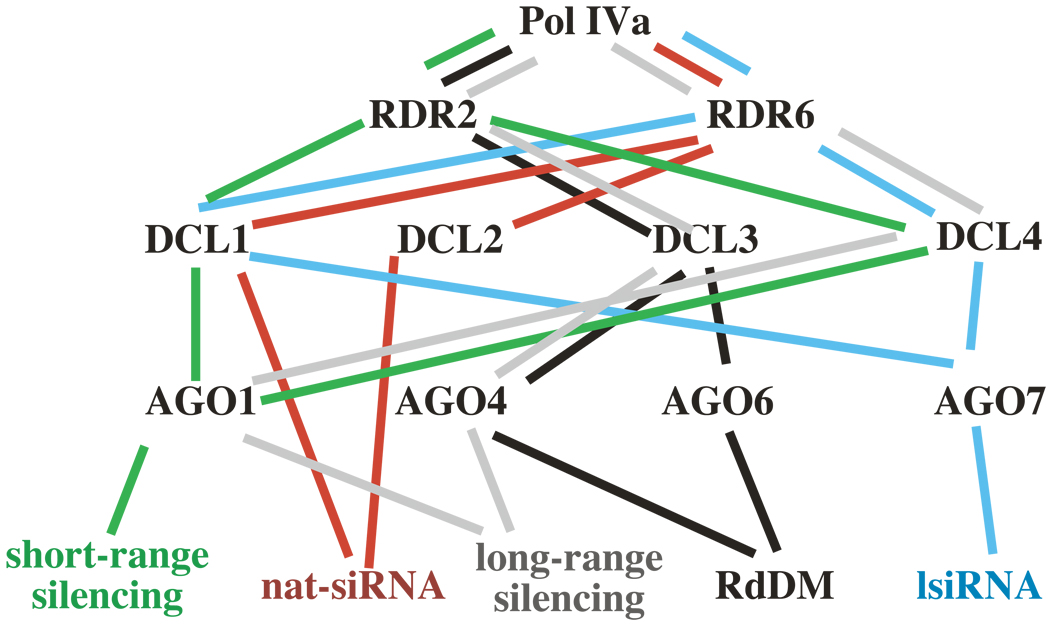
Pol IVa is integral to multiple RNA silencing pathways, including the RdDM pathway, the nat-siRNA and lsiRNA pathways, the short-range spread of silencing pathway and the pathway for the perception of long-distance silencing signals (Figure 2). Pol IVb is apparently less gregarious, acting primarily in the RdDM pathway [30], but also playing an undefined role in the lsiRNA pathway [54]. It seems probable that both Pol IVa and Pol IVb possess enzymatic activity given that the NRPD1a, NRPD1b and NRPD2 subunits possess the key conserved amino acids of the metal A and metal B sites found within the catalytic centers of other multi-subunit RNA polymerases [68 69]. But what do Pol IVa and Pol IVb transcribe, and what are their products? At present, we have no answer. In fact, our only biochemical clue is a negative result: that chromatographic fractions enriched for Pol IV lack the DNA-dependent RNA polymerase activity of RNA polymerases I, II and III in a conventional, promoter-independent transcription assay [70] using sheared double-stranded template DNA [4]. Based on this result, it seems likely that Pol IVa and Pol IVb may utilize very specific templates.
Figure 2.
A variety of proteins participate in Pol IVa-dependent silencing pathways. The figure shows a subset of the proteins that are involved in RdDM, nat-siRNA, lsiRNA, short-range silencing and long-distance silencing pathways. Proteins involved in the various pathways are linked by color-coded lines. The diagram does not imply the order of events, but illustrates the diversity of functional collaborations that are possible. Not all mutants have been tested in every pathway, therefore other potential connections may exist. However, the figure reflects the models provided by the authors of the studies discussed in the text.
A distinct possibility is that Pol IVa transcribes RNA [7 43 44]. Pol IVa is mislocalized by RNAse treatment of nuclei, but not by DNase treatment, whereas RNA polymerase II shows the opposite nuclease sensitivities [7]. Moreover, there is precedent for DNA-dependent RNA polymerases transcribing RNA. Hepatitis Delta Virus (HDV) and plant viroid RNAs are replicated by Pol II transcription [71 72]. Likewise, E. coli RNAP is regulated by binding 6S RNA, which is transcribed in order to be released [73].
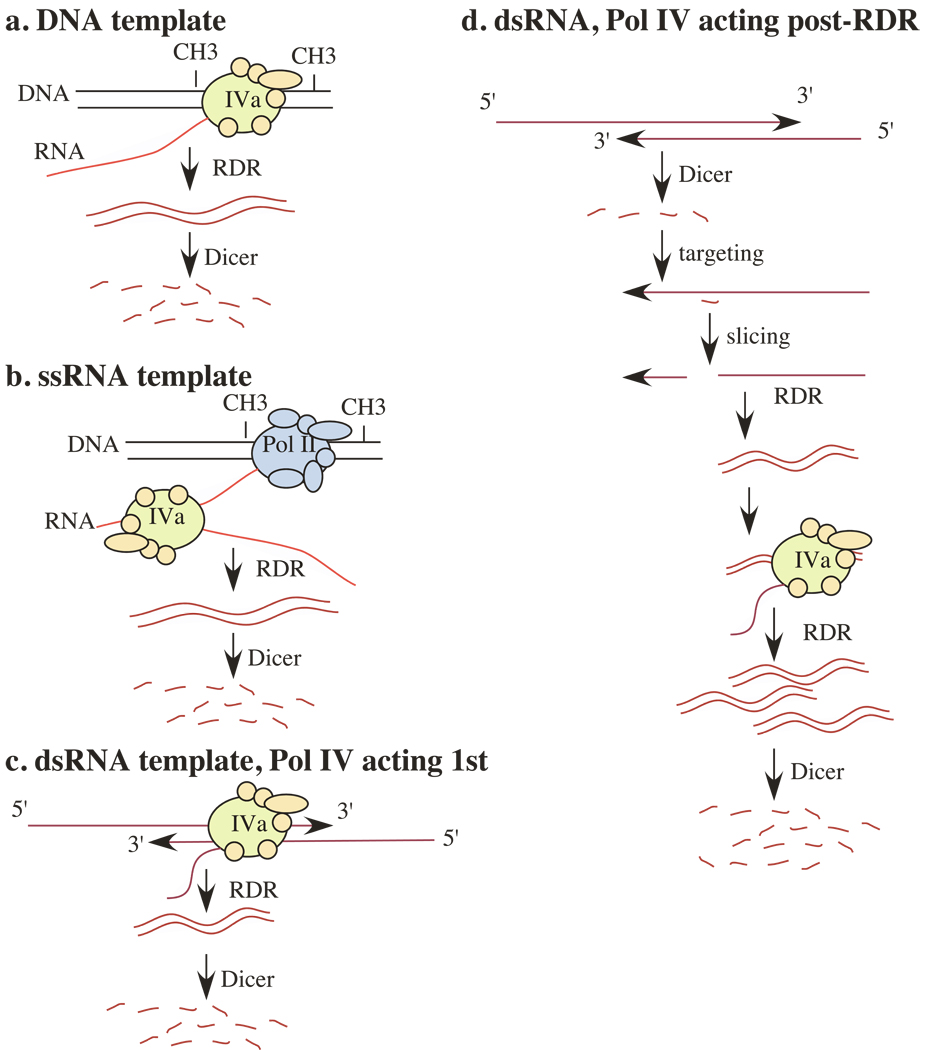
Previous models for the RdDM pathway have suggested that Pol IVa transcribes methylated DNA or transcripts of methylated loci, with resulting Pol IVa transcripts being amplified or made double-stranded by RDR2 (Figures 3a and b). However, in the nat-siRNA and lsiRNA pathways, regions of dsRNA are apparently generated via Pol II transcription of overlapping gene pairs and these transcripts persist in nrpd1a mutants, suggesting that there is no need for Pol IVa in initial dsRNA formation. Likewise, Pol IVa plays roles in short-range spreading of silencing triggered by dsRNA hairpin trigger sequences, as well as longdistance silencing likely to involve annealing of a mobile RNA to target mRNAs, thereby forming dsRNA. In each of these cases, there is no obvious need for Pol IVa in the initial generation of dsRNAs.
Figure 3.
Possible modes of Pol IVa function. Pol IVa may transcribe a specialized DNA template, such as methylated DNA (a) or single-stranded RNA transcripts derived from methylated DNA loci (b). Alternatively, Pol IVa may transcribe dsRNA generated from bidirectional transcripts, including transcripts of natural antisense gene pairs, or dsRNAs resulting from the annealing of long-distance mobile RNAs with target mRNAs (c and d). The model shown in D may account for the involvement of multiple dicers and multiple RDR inputs in the nat-siRNA and long-distance silencing pathways.
Pol IVa may use initial dsRNAs as templates, generating transcripts that are then made double-stranded by RDR2 or RDR6, one (or both) of which are involved in all known Pol IVa-dependent pathways (Figure 3c). Subsequent dicing, siRNA-mediated target slicing in trans, and RDR transcription of sliced templates might then amplify the initial signal and generate small RNAs beyond the region of initial transcript overlap. Alternatively, dicing of initial dsRNA regions may result in siRNAs that prime RDR on sliced or unsliced target RNAs, resulting in secondary dsRNAs that are then transcribed by Pol IVa and amplified by further RDR activity (Figure 3d). The model in Figure 3d would account for the involvement of more than one Dicer and more than one RDR-requiring step in the nat-siRNA and longdistance silencing pathways.
Pol IVa appears to be dispensable in some dsRNA-initiated phenomena. For instance, in a screen for methylation-defective mutants based on the use of a dsRNA hairpin to trigger RNA-directed DNA methylation, nine alleles of NRPD1b, and twelve alleles of NRPD2a were recovered, but no alleles of NRPD1a or RDR2 were identified [10], suggesting that the production of dsRNA hairpins had bypassed a need for Pol IVa or RDR2. Similarly, deep sequencing of small RNA libraries has shown that >90% of all siRNAs are Pol IVa-dependent, mostly derived from transposable elements and tandem repeats [30 74]. Inverted repeats, however, can contribute to the siRNA pool by a Pol IVa-independent mechanism [74]. Because transcription of inverted repeats can produce hairpin dsRNAs on their own, their Pol IVa-independence fits with the idea that Pol IVa functions at other loci in the production of dsRNAs that then feed into siRNA production. Why some dsRNA hairpin-initiated silencing phenomena require Pol IVa, but others do not, is not clear. The strength of the promoters driving hairpin formation may be an important variable.
Pol IVb is even more of a mystery than Pol IVa. NRPD1b mostly appears to reinforce Pol IVa-dependent siRNA production [9 30] yet is required, in addition to Pol IVa, for RdDM [9 10 75]. One possibility is that Pol IVb binds to DNA and interacts with AGO4 via its CTD [8 12], facilitating siRNA-DNA base-pairing that recruits DRM2. Alternatively, siRNA-AGO4 complexes may anneal to Pol IVb transcripts, thereby recruiting DRM2 and/or histone modifying enzymes to the vicinity of the corresponding DNA, as in models for siRNA-mediated silencing in fission yeast, Schizosaccharomyces pombe. [76 77]. AGO4 can slice RNAs in an siRNA-guided process, providing evidence that AGO4-siRNA RISC complexes can interact with RNA transcripts [78]. Nonetheless, direct siRNA interactions with DNA cannot be ruled out.
Clearly, there is much that needs to be learned concerning the templates, products, subunit structures, and interacting partners of Pol IVa and Pol IVb. Development of in vitro assays will be invaluable for deciphering the functions of these enigmatic polymerases and is a major challenge for the future.
Acknowledgments
Our work is supported by NIH grants R01GM60380 and R01GM077590 and by the Monsanto Company-Washington University Plant Biology Research Agreement.
Glossary of Abbreviations
- AGO
ARGONAUTE, proteins in this family bind small RNAs including siRNAs and miRNAs and are capable of cleaving RNAs complementary to the small RNAs (slicing)
- CLSY1
CLASSY1, a putative chromatin remodeling protein involved in RNA-directed DNA methylation
- CTD
C-terminal domain
- DCL1
Arabidopsis DICER-LIKE 1, involved primarily in miRNA biogenesis
- DCL2
Arabidopsis DICER-LIKE 2; generates 22 nt siRNAs
- DCL3
Arabidopsis DICER-LIKE 3, involved in 24 nt siRNA biogenesis
- DCL4
Arabidopsis DICER-LIKE 4; generates 21 nt siRNAs
- DNA
Deoxyribonucleic acid
- RNA-FISH
RNA fluorescent in situ hybridization; a means for locating specific RNAs
- DRD1
DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1, a putative chromatin remodeling protein involved in RNA-directed DNA methylation
- DRM2
DOMAINS REARRANGED METHYLYTRANSFERASE 2, the primary Arabidopsis de novo DNA methyltransferase
- dsRNA
double-stranded RNA
- GFP
Green fluorescent protein, initially derived from jellyfish.
- HEN1
HUA ENHANCER 1; methylates the 2′ hydroxyl groups of siRNA and miRNA 3′ - terminal nucleotides
- HST1
HASTY1, an exportin 5 homolog implicated in nuclear export of miRNAs
- HYL1
HYPONASTIC LEAVES 1, a dsRNA-binding protein that interacts with DCL1
- lsiRNA
long siRNA of ~40 nt, as opposed to the predominant 21–24 nt size range
- mRNA
messenger RNA, encodes proteins
- miRNA
micro-RNA, small RNAs transcribed from dedicated genes, mediate mRNA cleavage or translational arrest
- nat-siRNA
siRNA derived from natural antisense transcripts derived from adjacent genes
- Pol I
DNA-DEPENDENT RNA POLYMERASE I; synthesizes the precursor for the three largest rRNAs
- Pol II
DNA-DEPENDENT RNA POLYMERASE II; transcribes most genes including mRNAs and miRNAs
- Pol III
DNA-DEPENDENT RNA POLYMERASE III; mostly transcribes 5S rRNA genes and tRNA genes
- Pol IVa
nuclear RNA polymerase IVa; includes the NRPD1a and NRPD2a subunits
- Pol IVb
nuclear RNA polymerase IVb; includes the NRPD1b and NRPD2a subunits
- RdDM
RNA-directed DNA methylation, one of several gene silencing pathways in the nucleus
- RDR2
RNA-DEPENDENT RNA POLYMERASE 2, required for the biogenesis of 24nt siRNAs in Arabidopsis in the RNA-directed DNA methylation pathway
- RDR6
RNA-DEPENDENT RNA POLYMERASE 6, involved in the tasiRNA, nat-siRNA, lsiRNA, transgene and viral silencing, and long-distance silencing pathways
- RISC
RNA-induced silencing complex, includes an ARGONAUTE protein and siRNA (siRISC) or miRNA (miRISC)
- RNA
Ribonucleic acid
- RNAP
DNA-dependent RNA polymerase
- RNP
ribonucleoprotein, a complex of RNA and proteins
- rRNA
ribososomal RNA; four rRNAs are present in ribosomes
- SDE3
SILENCING DEFECTIVE 3; a putative RNA helicase
- SGS3
SUPPRESSOR OF GENE SILENCING 3; a putative coiled-coil protein
- siRNA
small interfering RNA
REFERENCES CITED
- 1.Werner F. Structure and function of archaeal RNA polymerases. Mol Microbiol. 2007;65:1395–1404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- 2.Kettenberger H, et al. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 3.The-Arabidopsis-Genome-Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 4.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Herr AJ, et al. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 6.Luo J, Hall BD. A multistep process gave rise to RNA polymerase IV of land plants. J Mol Evol. 2007;64:101–112. doi: 10.1007/s00239-006-0093-z. [DOI] [PubMed] [Google Scholar]
- 7.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 8.Li CF, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 9.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanno T, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 11.Bellaoui M, et al. DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Mol Biol. 2003;53:531–543. doi: 10.1023/B:PLAN.0000019061.79773.06. [DOI] [PubMed] [Google Scholar]
- 12.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shilatifard A. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim Biophys Acta. 2004;1677:79–86. doi: 10.1016/j.bbaexp.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 18.Herr AJ, Baulcombe DC. RNA silencing pathways in plants. Cold Spring Harb Symp Quant Biol. 2004;69:363–370. doi: 10.1101/sqb.2004.69.363. [DOI] [PubMed] [Google Scholar]
- 19.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 20.Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol. 2000;43:203–220. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- 21.Huettel B, et al. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Wassenegger M. The role of the RNAi machinery in heterochromatin formation. Cell. 2005;122:13–16. doi: 10.1016/j.cell.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- 24.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 25.Zilberman D, et al. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–652. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matzke M, et al. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harb Symp Quant Biol. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- 28.Zheng X, et al. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. Embo J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 30.Mosher RA, et al. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pikaard CS. Cell Biology of the Arabidopsis Nuclear siRNA Pathway for RNA-directed Chromatin Modification. Cold Spring Harb Symp Quant Biol. 2006;71:473–480. doi: 10.1101/sqb.2006.71.046. [DOI] [PubMed] [Google Scholar]
- 32.Li CF, et al. Dynamic Regulation of ARGONAUTE4 within Multiple Nuclear Bodies in Arabidopsis thaliana. PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 34.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Pontes O, Pikaard CS. siRNA and miRNA processing: new roles for Cajal bodies. Curr. Opin. Gen. Dev. 2008 doi: 10.1016/j.gde.2008.01.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujioka Y, et al. Location of a Possible miRNA Processing Site in SmD3/SmB Nuclear Bodies in Arabidopsis. Plant Cell Physiol. 2007;48:1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 37.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song L, et al. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JL, et al. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zilberman D, et al. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 42.Lippman Z, et al. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaughn MW, Martienssen RA. Finding the right template: RNA Pol IV, a plant-specific RNA polymerase. Mol Cell. 2005;17:754–756. doi: 10.1016/j.molcel.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Vaucheret H. RNA polymerase IV and transcriptional silencing. Nat Genet. 2005;37:659–660. doi: 10.1038/ng0705-659. [DOI] [PubMed] [Google Scholar]
- 45.Chan SW, et al. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 46.Baurle I, et al. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science. 2007;318:109–112. doi: 10.1126/science.1146565. [DOI] [PubMed] [Google Scholar]
- 47.Swiezewski S, et al. Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci U S A. 2007;104:3633–3638. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, et al. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borsani O, et al. Endogenous siRNAs derived from a pair of natural cisantisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henz SR, et al. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 2007;144:1247–1255. doi: 10.1104/pp.107.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jen CH, et al. Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 2005;6:R51. doi: 10.1186/gb-2005-6-6-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang XJ, et al. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin H, et al. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katiyar-Agarwal S, et al. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katiyar-Agarwal S, et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 57.Allen E, et al. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Peragine A, et al. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshikawa M, et al. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 61.Himber C, et al. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. Embo J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunoyer P, et al. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 63.Dunoyer P, et al. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 64.Palauqui JC, et al. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. Embo J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brosnan CA, et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Axtell MJ, et al. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 67.Kim M, et al. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science. 2001;293:287–289. doi: 10.1126/science.1059805. [DOI] [PubMed] [Google Scholar]
- 68.Lehmann E, et al. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 69.Cramer P. Recent structural studies of RNA polymerases II and III. Biochem Soc Trans. 2006;34:1058–1061. doi: 10.1042/BST0341058. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz LB, et al. Isolation and partial characterization of the multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in the mouse myeloma, MOPC 315. J Biol Chem. 1974;249:5889–5897. [PubMed] [Google Scholar]
- 71.Greco-Stewart VS, et al. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology. 2007;357:68–78. doi: 10.1016/j.virol.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 72.Ding B, Itaya A. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol Plant Microbe Interact. 2007;20:7–20. doi: 10.1094/MPMI-20-0007. [DOI] [PubMed] [Google Scholar]
- 73.Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X, et al. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. Embo J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buhler M, et al. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 77.Buhler M, et al. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 78.Qi Y, et al. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]