Abstract
Differentiation of active from latent tuberculosis (TB) is a major challenge in the control of TB. In this study, PBMC from latent TB-infected subjects, TB patients, and tuberculin skin test-negative donors stimulated with the Mycobacterium tuberculosis (Mtb)-specific Ag, early secretory antigenic target 6, and mRNA for 45 immune-related genes was measured by quantitative real-time PCR. Univariate analysis showed significant differences in the expression of 10 genes (IFN-γ, FOXP3, IL-1α, IL-1β, IL-2, IL-6, IL-8, IL-12α, IL-12β, and IL-24) in PBMC from TB patients vs latent TB-infected subjects (p < 0.01). Multivariate logistic regression and classification and regression tree analyses revealed that expression of three genes, IL-8, FOXP3, and IL-12β, is predictive for TB vs latent Mtb infection. Thus, measurement of Ag-specific expression of these three genes may offer a specific and noninvasive means of differentiating between latent Mtb infection and TB.
It is estimated that approximately one-third of the world’s population is infected with Mycobacterium tuberculosis (Mtb),3 but only ~10% of those infected ever develop active tuberculosis (TB). Thus, ~90% of individuals with Mtb infection contain the infection in a latent state (latent TB infected (LTBI)), but all of those who have been infected are still at increased risk of developing disease. Central to the pathogenesis of TB is the inability of macrophages infected with Mtb to maintain containment of the organism and the failure of T cells to confer long-lasting protective immunity (1). Defects in either the IFN-γ or the IL-12 pathways render the host susceptible to TB (2). Compelling evidence suggests that different T cell populations, including CD4+, CD8+, double-negative, and γδ T cells, participate in protective immunity against TB (3), and all of these populations have been shown to secrete IFN-γ. However, IFN-γ alone is insufficient to protect against development of TB (1), suggesting a role for other immune mechanisms. A newly described subset of CD4+CD25high T lymphocytes, referred to as T regulatory cells (Tregs), has been shown to play a vital role in self-tolerance (4). Tregs specifically express the X-linked forkhead/winged helix transcription factor FOXP3 and suppress a variety of immune responses (5). Recent work suggests that Tregs may suppress antibacterial immune responses and contribute to the persistence of organisms such as Helicobacter pylori and Mtb in vivo (6, 7).
The tuberculin skin test (TST) has long been used to diagnose Mtb infection by assessing cell-mediated immunity as measured by delayed-type hypersensitivity response to purified protein derivative (PPD) (8). PPD is a mixture of poorly defined mycobacterial Ags, some of which are shared with several nontuberculous mycobacteria and with Mycobacterium bovis bacille Calmette Guérin (BCG), the current vaccine for TB. Thus, the TST has low specificity in populations with high BCG coverage or exposure to nontuberculous mycobacteria (9). Recently, alternatives to the TST that measure IFN-γ release from peripheral blood lymphocytes in response to challenge with Mtb Ags have been developed (10). Current versions of these assays use two recombinant Mtb Ags, early secretory antigenic target 6 (ESAT-6), and culture filtrate protein 10 (CFP-10). These proteins are encoded within the region of difference 1 of the Mtb genome that is deleted in all attenuated BCG vaccine strains and most nontuberculous mycobacteria (11). High ESAT-6-specific IFN-γ responses have been positively correlated with pathology (12). This is in contrast to clinical studies that have used ESAT-6-induced immune responses to identify those groups of putatively infected individuals (latent infection, healthy contacts), some of whom were found to express elevated levels of IL-4 splice variants that appear to correlate with control of infection in the long term (13, 14). During the past 5 years, a number of studies have compared the TST with the IFN-γ-based test for diagnosing Mtb infection, and it appears that the IFN-γ-based tests are more specific (10). However, neither test is able to distinguish LTBI from active TB.
In an examination of gene expression using quantitative realtime PCR (qPCR) in ESAT-6-activated PBMC, we found that expression of 10 of 45 immune-related genes was significantly different between LTBI individuals and those with active TB. Moreover, using classification and regression tree (CART) analysis and logistic regression to model the predictors for infection, mRNA levels of three genes, IL-8, FOXP3, and IL-12β, differentiated LTBI from active TB with a high degree of accuracy.
Materials and Methods
Participants
This investigation was approved by the institutional review boards of Stanford University and the University of California, San Francisco. Patients with active pulmonary TB were recruited from the TB Clinic at the San Francisco Department of Public Health/San Francisco General Hospital. To minimize the effect of anti-TB treatment on gene expression, only patients on standard anti-TB therapy for <3 wk were included in the initial studies. PBMC were obtained from 10 patients who had received no treatment, 8 patients treated for <1 wk, 6 patients treated for 1–2 wk, and 6 patients treated for 2–3 wk (Table I). Of the 30 TB patients, 24 were culture positive. The remainder were culture negative, but were classified with clinically active TB based on the American Thoracic Society and Centers for Disease Control and Prevention criteria (15), including symptoms and chest radiograph consistent with TB and clinical and/or radiographic improvement in association with treatment. Retrospective analysis revealed no significant differences in gene expression between culture positive and negative TB patients; therefore, they were grouped for the results shown here. Additionally, 42 TB patients who had been treated for >3 wk were recruited and divided into two groups (treated for 3–9 and 10–30 wk). LTBI subjects (n = 24) were recruited among health care workers at Stanford University Medical Center. LTBI subjects were U.S. born and had documentation of a positive (≥10 mm) TST using 5 tuberculin units PPD, a negative chest radiograph, negative history of BCG vaccination, and no clinical evidence of TB. TB patients and LTBI subjects known to be HIV infected or on immunosuppressive regimens were excluded. Healthy TST-negative donors (n = 10) were recruited from laboratory workers at Stanford University Medical Center as negative controls.
Table I.
Clinical parameters of TB patients
| Patient | Gendera | Age (years) | TST | Smear Culture | Anti-TB Treatmentb (wk) |
|---|---|---|---|---|---|
| 01 | F | 58 | NT | + | 1.29 |
| 02 | M | 49 | NT | + | 2.90 |
| 03 | M | 40 | + | + | 2.43 |
| 04 | F | 22 | + | + | 2.86 |
| 05 | M | 19 | + | + | 0.71 |
| 06 | M | 37 | + | + | 1.86 |
| 07 | F | 37 | + | + | 2.57 |
| 08 | M | 53 | + | + | 1.57 |
| 09 | M | 40 | + | + | 0.00 |
| 10 | F | 31 | + | + | 2.14 |
| 11 | F | 40 | + | + | 0.71 |
| 12 | M | 36 | + | + | 0.00 |
| 13 | F | 33 | + | + | 1.00 |
| 14 | F | 36 | + | + | 2.43 |
| 15 | F | 73 | NT | + | 1.14 |
| 16 | F | 76 | + | + | 0.00 |
| 17 | F | 31 | + | + | 1.43 |
| 18 | M | 19 | NT | + | 0.00 |
| 19 | M | 37 | + | + | 2.00 |
| 20 | M | 30 | + | + | 0.00 |
| 21 | F | 38 | − | + | 0.86 |
| 22 | F | 51 | − | + | 1.00 |
| 23 | M | 47 | − | + | 0.00 |
| 24 | M | 66 | − | + | 0.00 |
| 25 | F | 27 | NT | − | 0.00 |
| 26 | M | 48 | + | − | 0.86 |
| 27 | F | 23 | + | − | 0.57 |
| 28 | M | 77 | NT | − | 0.43 |
| 29 | F | 61 | + | − | 0.00 |
| 30 | F | 38 | + | − | 0.00 |
Female; M, male, NT, not tested.
Week of treatment when PBMC were obtained.
PBMC activation and real-time PCR assay
PBMC were isolated by centrifugation over Ficoll and cultured in RPMI 1640 medium (Invitrogen Life Technologies) supplemented with l-glutamine (2 µM), penicillin (100 U/ml)/streptomycin (100 µg/ml), nonessential amino acids (100 µM) (Invitrogen Life Technologies), sodium pyruvate (110 µg/ml) (Irvine Scientific), and 10% heat-inactivated pooled human serum (Gemini) in the absence or presence of 10 µg/ml ESAT-6 (Research Materials and Vaccine Testing Center, Colorado State University) for 15 h. Total RNA was isolated from PBMC using RNeasy mini kits (Qiagen) and was transcribed into cDNA (Invitrogen Life Technologies) according to the manufacturer’s instruction. Expression of 45 immune-related genes was analyzed using qPCR with an ABI Prism 7900 Sequence Detection System (Applied Biosystems; details of PCR protocol at www.appliedbiosystems.com). All of the primers were purchased from Applied Biosystems, including FOXP3, IFNA, IFNG, IL-1A, IL-1B, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12A, IL-12B, IL-13, IL-15, IL-16, IL-17, IL-17B, IL-17C, IL-17E, IL-17F, IL-18, IL-23A, IL-24, IL-27, TNFA, TGFB1, CCL1, CCL4, CCL5, CCL8, CCL13, CCL16, CCL17, CCL18, CCL19, CCL21, CCL23, CCL24, CCL28, CKLF, and CX3CL1. The expression level of a gene in a given sample was represented as fold increase: 2−ΔΔCt, where ΔΔCT = [ΔCT(sample, stimulated)] − [CT(sample, unstimulated)] and ΔCT = [CT(sample)] − [CT(GUS)], where GUS is the housekeeping gene β-glucoronidase.
Statistical analysis
Results were expressed as fold increase in ESAT-6-stimulated mRNA levels. The median for each group was determined and differences between groups were tested using the Wilcoxon-Mann-Whitney U test. In all instances, a p < 0.01 was considered significant. Receiver-operating-characteristic (ROC) curves were used to determine the cutoff points yielding the highest combined specificity and sensitivity, and discriminative ability was evaluated by the area under the ROC curve. The 95% confidence interval was estimated by the bootstrap method. Combinations of markers were identified using CART analysis and logistic regression. Using CART, a best tree was chosen by cross-validation and the minimum error rule, i.e., minimum misclassification error as provided by cross-validation after an optimal tree was grown and pruning was performed. Using the CART default options, equal misclassification costs, Gini splitting criterion, equal priors, and 10 as the minimum number of groups per nodes, a single CART prediction model was derived. Genes selected by CART analysis were used to establish a multivariate logistic regression model, and then the ROC analysis was performed for the resultant model. The sensitivity and specificity were derived from the jackknifed probabilities. All analyses were conducted with STATA 8.0 (STATA) and CART 5.0 software (Salford Systems).
Results
qPCR can be used to measure ESAT-6-stimulated expression of IFN-γ mRNA in PBMC of Mtb-infected individuals
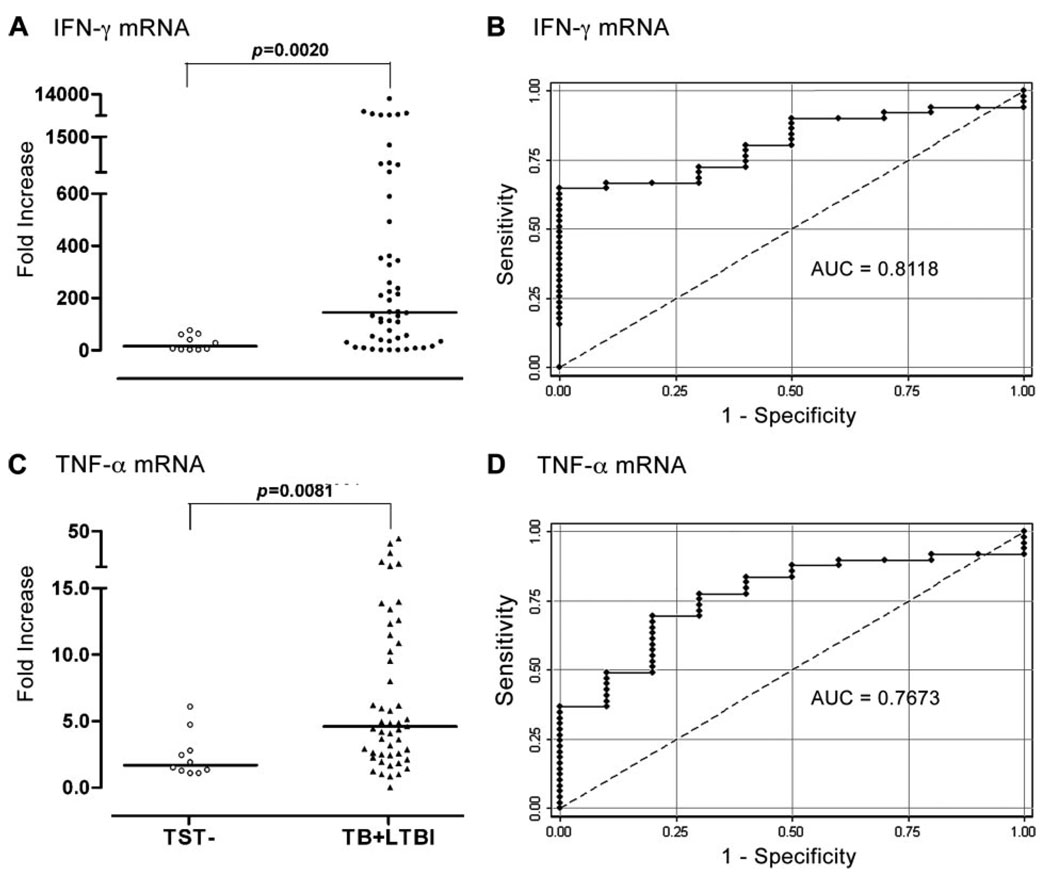
Currently, identification of individuals infected with Mtb relies on the TST or on assays that measure IFN-γ production by ELISA or ELISPOT following stimulation of PBMC with ESAT-6 plus CFP-10 (10). To determine whether qPCR could accurately identify Mtb-infected individuals, PBMC from Mtb-infected subjects (LTBI plus TB) and TST healthy controls were stimulated with ESAT-6, and mRNA for 45 immune-related genes were assayed. The fold increases in mRNA for IFN-γ was significantly higher in PBMC from Mtb-infected subjects as compared with TST healthy controls (Fig. 1A). These results were then transformed to a ROC curve, which shows the fraction of true positive results (sensitivity) and false positive results (1 – specificity) for various fold increase cutoff levels. For the calculated area under the curve (AUC), a value of 0.5 is no better than that expected by chance (the null hypothesis), and a value of 1.0 reflects a perfect indicator. The AUC for using IFN-γ mRNA levels to identify Mtb-infected individuals is 0.81, with 65% sensitivity, 100% specificity, and 70% of cases correctly classified (Fig. 1B). These results are in good agreement with published results from others using ELISPOT or ELISA to assay IFN-γ produced by ESAT-6/CFP-10-stimulated PBMC (10). Thus, measurement of IFN-γ by qPCR can also be used to identify individuals infected with Mtb (10). In addition to IFN-γ, expression of TNF-α mRNA was also significantly increased in Mtb-infected individuals compared with TST– controls (Fig. 1C). ROC curve analysis, however, showed that change in expression of TNF-α was less accurate (Fig. 1D) than that of IFN-γ to predict Mtb infection.
FIGURE 1.
Gene expression and ROC curves for mRNA increases in IFN-γ and TNF-α in ESAT-6-activated PBMC from Mtb-infected individuals (TB + LTBI) and TST– donors. A and C, PBMC from Mtb-infected individuals (TB patients, n = 30; LTBI, n = 24), or TST– controls (n = 10) were cultured with ESAT-6 for 15 h, and mRNAs for IFN-γ and TNF-α were measured by qPCR. Horizontal bar, Median fold increase for each group. B and D, ROC curves for discriminating Mtb infected from TST– individuals. The calculated AUC is 0.81 (95% confidence interval (CI), 0.70–0.92) for IFN-γ and 0.77 (95% CI, 0.62–0.90) for TNF-α.
Identification of differentially expressed genes in response to ESAT-6 in TB and LTBI subjects
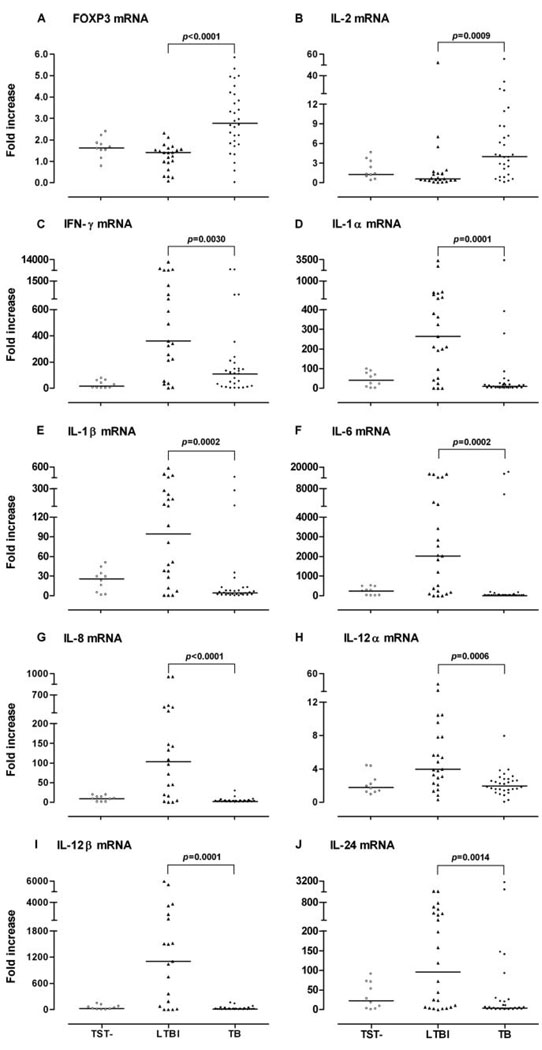
We next divided the Mtb-infected group into those with active TB and those with latent Mtb infection to determine whether qPCR mRNA profiling could distinguish between these two groups. Fold increases in mRNA for FOXP3 and IL-2 were significantly higher in PBMC from TB patients as compared with LTBI individuals (Fig. 2, A and B), while fold increases for IFN-γ, IL-1α, IL-1β, IL-6, IL-8, IL-12α, IL-12β, and IL-24 were significantly higher in LTBI subjects than in TB patients (p < 0.01; Fig. 2, C–J), suggesting that multiple immune mechanisms are involved in the immunopathogenesis of TB. Of note, the fold increase in IFN-γ mRNA levels was significantly lower in TB patients than in LTBI subjects (Fig. 2C).
FIGURE 2.
Gene expression in ESAT-6-activated PBMC from TB patients, LTBI individuals, and TST– donors. A–J, PBMC from TB patients (n = 30), LTBI subjects (n = 24), or TST– controls (n = 10) were cultured with ESAT-6 for 15 h, and mRNA for each cytokine was measured by qPCR. Horizontal bar, Median fold increase.
Identification of biomarkers to differentiate latent from active infection
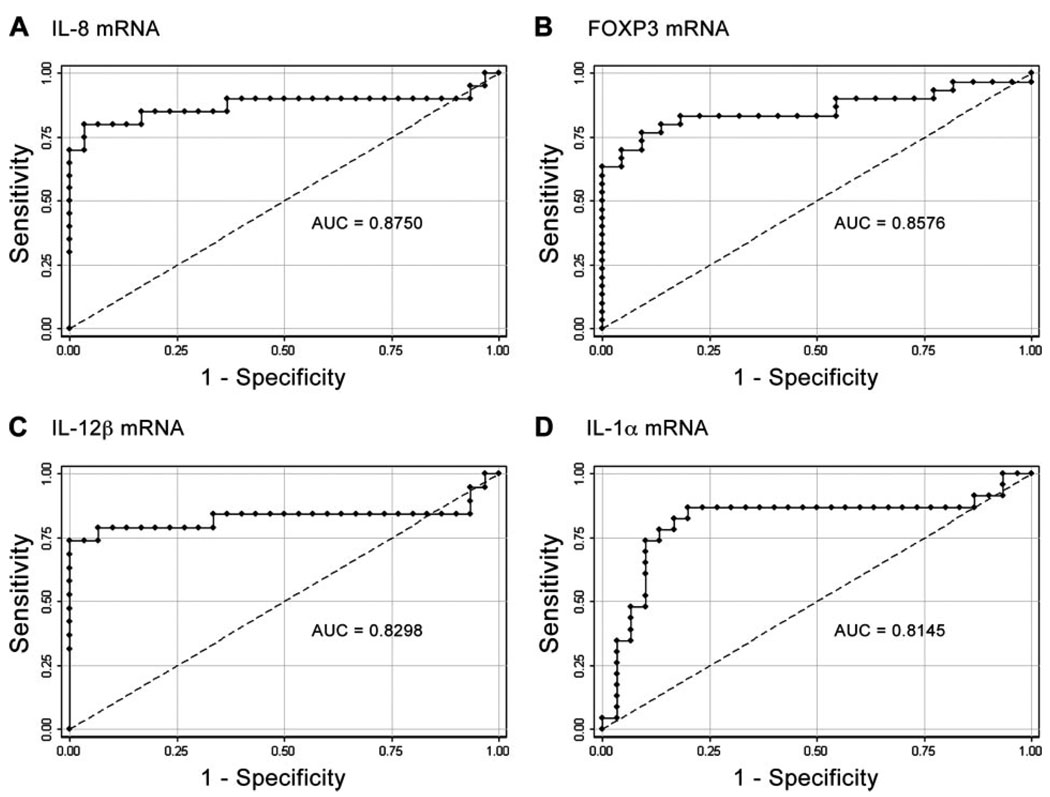
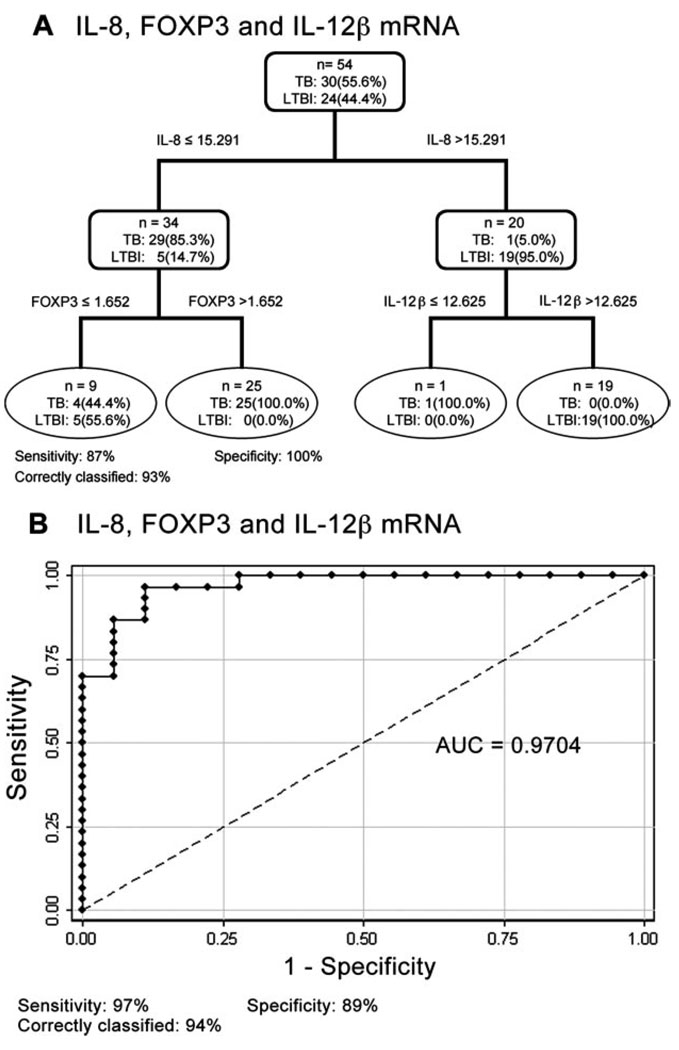
Based on these results, we selected the genes that could best discriminate between TB patients and LTBI individuals ( p ≤ 0.0001). Four genes, IL-8, FOXP3, IL-12β, and IL-1α, were then subjected to ROC analysis (Fig. 3). Individually, ESAT-6-stimulated expression of each gene predicts TB significantly better than chance ( p ≤ 0.0001). We then used CART and logistic regression analyses to determine whether a combination of genes could further improve the predictive outcome. The diagnostic tree generated by CART analysis using 10-fold cross-validation consistently selected IL-8, FOXP3, and IL-12β as the major contributing variables, with 87% sensitivity, 100% specificity, and 93% of cases correctly classified (Fig. 4A). Logistic regression analysis was then used to generate a linear model for IL-8, FOXP3, and IL-12β. Based on the resultant model, further ROC analysis was performed. Using the default cutoff point of 0.5, a linear combination of fold increase in mRNA for IL-8, FOXP3, and IL-12β is the best predictor of active TB (AUC = 0.9704), with 97% sensitivity, 89% specificity, and 94% of cases correctly classified (Fig. 4B). The results of both CART and logistic regression analyses indicate that >93% of Mtb-infected individuals are correctly classified as either active TB or LTBI.
FIGURE 3.
The ability of ESAT-6-stimulated expression of IL-8, FOXP3, IL-12β, and IL-1α mRNA to distinguish TB from latent infection. A–D, The calculated AUC is 0.88 (95% CI, 0.75–1.00) for IL-8, 0.86 (95% CI, 0.75–0.97) for FOXP3, 0.83 (95% CI, 0.67–0.99) for IL-12β, and 0.81 (95% CI, 0.66–0.95) for IL-1α.
FIGURE 4.
The combination of IL-8, FOXP3, and IL-12β provides the best discrimination between TB and latent infection. A, Classification tree: rectangles = internal nodes, ovals = terminal nodes. Percentages in each node represent the proportion of specified observations in each group. B, The calculated AUC is 0.97 (95% CI, 0.93–1.00). The values under each panel indicate the sensitivity, specificity, and percent correctly classified.
To further test the ability of ESAT-6-stimulated expression of IL-8/FOXP3/IL-12β to differentiate TB patients from LTBI subjects, a predictive analysis based on the logistic regression model, was expanded to patients treated with anti-TB therapy for longer than 3 wk (data not shown). These patients were subdivided into those treated for an intermediate period (3–9 wk) or for a longer period (10–30 wk). The combination of IL-8/FOXP3/IL-12β expression correctly classified 88% of the patients treated for 3–9 wk and 95% of the patients treated for 10–30 wk. To determine whether the duration of treatment changes the immune profile, expression of ESAT-6-stimulated IL-8, FOXP3, and IL-12β was individually examined in patients treated for <3, 3–9, or 10–30 wk. No difference was observed (data not shown), suggesting that ESAT-6-stimulated expression of all three genes during the first 30 wk after diagnosis/treatment in TB patients is independent of anti-TB therapy.
Discussion
Currently, confirmation of a diagnosis of pulmonary TB requires either isolation of Mtb or identification of specific Mtb DNA from respiratory specimens. Both tests have important drawbacks: culture requires 2–3 wk, and nucleic acid amplification tests, although faster, are not as sensitive as culture. Commonly, treatment for active TB is begun before there is bacteriologic confirmation of the diagnosis, and it is not unusual for clinical criteria to be met without bacteriologic proof, leading to empiric treatment and its attendant risks if the person does not actually have active TB. Two diagnostic tests, the TST and the expression of IFN-γ by PBMC in response to ESAT-6 and CFP-10, are widely used to identify Mtb-infected individuals (10), but neither differentiates individuals with LTBI from those with active TB. In the present study, we found that the measurement of the combination of ESAT-6-stimulated IL-8, FOXP3, and IL-12β mRNA in PBMC distinguishes individuals with active TB from latent Mtb infection.
Although the results presented in this study may lead to potentially useful new diagnostic tests, much additional work is needed. For this initial survey, we excluded TB patients with HIV/AIDS, reasoning that various degrees of immunosuppression might complicate data interpretation. However, patients with HIV/AIDS contribute most of the cases with culture-negative TB for whom a new diagnostic test is urgently needed (16). Studies on newly diagnosed TB patients before treatment are also planned. Lastly, a less expensive, technically demanding test must be developed, such as a multiplex or dipstick format in which the cytokines themselves are measured in response to stimulation.
Our findings provide new insights into the immune responses to Mtb. The patterns of cytokine expression imply that the transition from latent infection to active disease is not merely a “switch” from a Th1 to Th2 response. Expression of FOXP3, a transcription factor specific for Tregs, correlates with the suppressive activity of these cells, and activation of human CD4+CD25− T cells leads to expression of FOXP3 and acquisition of regulatory function (17, 18). There have been reports that Tregs are increased in patients with active TB or H. pylori infection (6, 7), suggesting that Tregs may underlie the suppressed cellular immunity described in these chronic infections. The mechanism by which Tregs suppress other T cell responses is controversial, but requires either cell-cell contact in vitro or the release of soluble factors such as IL-10 and TGF-β (19). In the patients with active TB in our cohort, FOXP3+ cells were induced in an Ag-specific manner because there was no difference in FOXP3 expression in unstimulated PBMC between the TB and LTBI groups (data not shown), but there was a significant increase in FOXP3 expression in ESAT-6-stimulated PBMC from TB patients.
IL-8 (CXCL8) induces chemotaxis of inflammatory cells and directs immune responses. Multiple stimuli induce the secretion of IL-8, including TNF-α and IL-1 (20). Granulocytes from patients with TB produce IL-8 in response to lipoarabinomannan isolated from Mtb (21). Elevated levels of IL-8 in bronchoalveolar lavage fluid have been described in patients with TB compared with uninfected controls (22, 23). We show in this study that the fold increase in IL-8 mRNA in ESAT-6-stimulated PBMC is significantly elevated in LTBI compared with active TB (Fig. 2G), and a similar increase was also observed for IL-1α and IL-1β (Fig. 2, D and E). Of note, neutralization of IL-1 blocks IL-8 production in Mtb infection (24), suggesting that the enhanced IL-8 expression in LTBI subjects may be controlled by IL-1.
IL-12, a heterodimer composed of two subunits encoded by IL-12α and IL-12β, is a major inducer of IFN-γ. IL-12p40-deficient children suffer from mycobacterial infection primarily because their IFN-γ-mediated immunity is impaired (25). In the present study, expression of IL-12β was significantly decreased in patients with TB compared with the LTBI group, indicating that low expression of IFN-γ in TB patients might be due to the reduction of IL-12β. Indeed, addition of exogenous IL-12 enhanced the IFN-γ response of ESAT-6 stimulated PBMC from TB patients (data not shown), underscoring the importance of IL-12β in the induction of IFN-γ.
In summary, our study suggests that measuring the ESAT-6-stimulated expression of mRNA of immune related genes in PBMC can be used to identify Mtb-infected individuals. Expression of IL-8/FOXP3/IL-12β can distinguish latent infection from active TB regardless of anti-TB treatment time. In newly diagnosed or short-term treated TB patients, decreased IL-8 and IL-12β expression may reflect impairment of immune cell recruitment and Th1 responses, while increased levels of FOXP3 suggest involvement of regulatory T lymphocyte-mediated immunosuppression. Our findings imply that the cellular immune response changes as latent infection transitions to active disease and that monitoring expression of the genes identified here may allow identification of individuals needing anti-TB treatment, particularly in developing countries.
Acknowledgments
We thank Colorado State University and the National Institutes of Health (National Institute of Allergy and Infectious Diseases Contract NO1 AI-75320) for providing recombinant proteins; I. Rudoy and C. Merrifield of the TB Clinic at the San Francisco Department of Public Health for recruitment of TB patients, blood drawing, and data collection; S. Lyu for technical assistance; R. A. Olshen for statistical advice; and G. Schoolnik and P. Small for helpful discussions.
Footnotes
This work was supported by grants from the National Institutes of Health (RO1 AI051356 to C.C. and RO1 AI034238 to P.C.H.).
Abbreviations used in this paper: Mtb, Mycobacterium tuberculosis; TB, tuberculosis; LTBI, latent TB infected; Treg, T regulatory cell; TST, tuberculin skin test; PPD, purified protein derivative; BCG, bacille Calmette-Guérin; ESAT-6, early secretory antigenic target 6; CPF-10, culture filtrate protein 10; qPCR, quantitative real-time PCR; CART, classification and regression tree; ROC, receiver-operating-characteristic; AUC, area under the curve; Ct, cycle threshold; CI, confidence interval.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Flynn JL, Chan J. Immunology of tuberculosis. Annu. Rev. Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 2.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann SH. Is the development of a new tuberculosis vaccine possible? Nat. Med. 2000;6:955–960. doi: 10.1038/79631. [DOI] [PubMed] [Google Scholar]
- 4.Bacchetta R, Gregori S, Roncarolo MG. CD4+regulatory T cells: mechanisms of induction and effector function. Autoimmun. Rev. 2005;4:491–496. doi: 10.1016/j.autrev.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat. Immunol. 2005;6:331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 6.Lundgren A, Stromberg E, Sjoling A, Lindholm C, Enarsson K, Edebo A, Johnsson E, Suri-Payer E, Larsson P, Rudin A, et al. Mucosal FOXP3-expressing CD4+D25high regulatory T cells in Helicobacter pylori-infected patients. Infect. Immun. 2005;73:523–531. doi: 10.1128/IAI.73.1.523-531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guyot-Revol V, Innes JA, Hackforth S, Hinks T, Lalvani A. Regulatory T cells are expanded in blood and disease sites in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 2006;173:803–810. doi: 10.1164/rccm.200508-1294OC. [DOI] [PubMed] [Google Scholar]
- 8.Brodie D, Schluger NW. The diagnosis of tuberculosis. Clin. Chest Med. 2005;26:247–271. doi: 10.1016/j.ccm.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Diel R, Nienhaus A, Lange C, Meywald-Walter K, Forssbohm M, Schaberg T. Tuberculosis contact investigation with a new, specific blood test in a low-incidence population containing a high proportion of BCG-vaccinated persons. Respir. Res. 2006;7:77. doi: 10.1186/1465-9921-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai M, Riley LW, Colford JM., Jr Interferon-γ assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect. Dis. 2004;4:761–776. doi: 10.1016/S1473-3099(04)01206-X. [DOI] [PubMed] [Google Scholar]
- 11.Cole ST. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology. 2002;148:2919–2928. doi: 10.1099/00221287-148-10-2919. [DOI] [PubMed] [Google Scholar]
- 12.Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- 13.Demissie A, Abebe M, Aseffa A, Rook G, Fletcher H, Zumla A, Weldingh K, Brock I, Andersen P, Doherty TM. Healthy individuals that control a latent infection with Mycobacterium tuberculosis express high levels of Th1 cytokines and the IL-4 antagonist IL-4δ2. J. Immunol. 2004;172:6938–6943. doi: 10.4049/jimmunol.172.11.6938. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher HA, Owiafe P, Jeffries D, Hill P, Rook GA, Zumla A, Doherty TM, Brookes RH. Increased expression of mRNA encoding interleukin (IL)-4 and its splice variant IL-4δ2 in cells from contacts of Mycobacterium tuberculosis, in the absence of in vitro stimulation. Immunology. 2004;112:669–673. doi: 10.1111/j.1365-2567.2004.01922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Thoracic Society and the Centers for Disease Control and Prevention. Diagnostic standards and classification of tuberculosis in adults and children: this official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999; this statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am. J. Respir. Crit. Care Med. 2000;161:1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 16.Yusuph H, Garbati MA, Gashau W. Mantoux reaction in patients with HIV-related pulmonary tuberculosis in Maiduguri, Nigeria. Afr. J. Med. Med. Sci. 2005;34:125–128. [PubMed] [Google Scholar]
- 17.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 18.Walker MR, Kasprowicz DJ, Gersuk VH, Benard A, Van Landeghen M, Buckner JH, Ziegler SF. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25− T cells. J. Clin. Invest. 2003;112:1437–1443. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bach JF. Regulatory T cells under scrutiny. Nat. Rev. Immunol. 2003;3:189–198. doi: 10.1038/nri1026. [DOI] [PubMed] [Google Scholar]
- 20.Standiford TJ, Kunkel SL, Basha MA, Chensue SW, Lynch JP, III, Toews GB, Westwick J, Strieter RM. Interleukin-8 gene expression by a pulmonary epithelial cell line: a model for cytokine networks in the lung. J. Clin. Invest. 1990;86:1945–1953. doi: 10.1172/JCI114928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedel DD, Kaufmann SH. Chemokine secretion by human polymorphonuclear granulocytes after stimulation with Mycobacterium tuberculosis and lipoarabinomannan. Infect. Immun. 1997;65:4620–4623. doi: 10.1128/iai.65.11.4620-4623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am. J. Respir. Cell Mol. Biol. 1998;19:513–521. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 23.Kurashima K, Mukaida N, Fujimura M, Yasui M, Nakazumi Y, Matsuda T, Matsushima K. Elevated chemokine levels in bronchoalveolar lavage fluid of tuberculosis patients. Am. J. Respir. Crit. Care Med. 1997;155:1474–1477. doi: 10.1164/ajrccm.155.4.9105097. [DOI] [PubMed] [Google Scholar]
- 24.Rom WN, Schluger N, Law K, Condos R, Zhang Y, Weiden M, Harkin T, Tchou-Wong KM. Human host response to Mycobacterium tuberculosis. Schweiz Med. Wochenschr. 1995;125:2178–2185. [PubMed] [Google Scholar]
- 25.Frucht DM, Holland SM. Defective monocyte costimulation for IFN-γ production in familial disseminated Mycobacterium avium complex infection: abnormal IL-12 regulation. J. Immunol. 1996;157:411–416. [PubMed] [Google Scholar]