Abstract
We previously reported that trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor, induced DLC-1 mRNA expression and accumulated acetylated histones H3 and H4 associated with the DLC-1 promoter in DLC-1 non-expressing gastric cancer cells. In this study, we demonstrated the molecular mechanisms by which TSA induced the DLC-1 gene expression. Treatment of the gastric cancer cells with TSA activates the DLC-1 promoter activity through Sp1 sites located at -219 and -174 relative to the transcription start site. Electrophoretic mobility-shift assay (EMSA) revealed that Sp1 and Sp3 specifically interact with these Sp1 sites and showed that TSA did not change their binding activities. The ectopic expression of Sp1, but not Sp3, enhances the DLC-1 promoter responsiveness by TSA. Furthermore, the TSA-induced DLC-1 promoter activity was increased by p300 expression and reduced by knockdown of p300. These results demonstrated the requirement of specific Sp1 sites and dependence of Sp1 and p300 for TSA-mediated activation of DLC-1 promoter.
Keywords: DLC1 protein, human; histone deacetylases; p300-CBP transcription factors; promoter regions, genetic; Sp1 transcription factor; trichostation A
Introduction
The human DLC-1 (deleted in liver cancer) has been known as tumor suppressor showing that DLC-1 inhibits cell growth, colony formation, and invasion capacity in hepatocellular, breast, and non-small cell lung carcinoma (Ng et al., 2000; Yuan et al., 2003a; Kim et al., 2007; Healy et al., 2008). DLC-1 and its rat homolog p122 RhoGAP contain three functional domains such as an amino terminal SAM (sterile α-motif), a central RhoGAP, and a carboxy terminal START (steroidogenic acute regulatory related lipid transfer) domain (Durkin et al., 2007). The RhoGAP domain is well known as a catalyst molecule converting active GTP-bound Rho proteins into inactive GDP-bound protein (Mackay and Hall, 1998), on the other hand the precise functions of the SAM and START domains remain answered.
DLC-1 gene has first been identified to be deleted in primary human hepatocellular carcinoma (Yuan et al., 1998), but further studies demonstrated that DLC-1 expression is down-regulated mainly by DNA hypermethylation in several types of cancer including liver, breast, colon, prostate, gastric, lung, nasopharyngeal, esophageal, cervical carcinomas, and lymphoma (Kim et al., 2003; Yuan et al., 2003b, 2004; Seng et al., 2007; Ying et al., 2007)
We have previously reported that TSA, a HDAC inhibitor, induced DLC-1 mRNA expression in association with an accumulation of acetylated histones H3 and H4 with the DLC-1 promoter in human gastric cancer cell lines (Kim et al., 2003), suggesting the possibility that histone deacetylation of the DLC-1 gene is a new epigenetic mechanism to contribute to down regulation of DLC-1 gene expression. The histone modifying enzymes histone acetyltransferase (HAT) and histone deacetylase (HDAC) have been reported to be involved in modifying chromatin structure and regulating transcription (Eberharter et al., 2002). HATs acetylate the ε-amino group of lysine residues at the N-terminal domain of core histone, resulting in gene activation through relaxation of chromatin, whereas HDACs remove the acetyl group of core histone, leading to gene repression through condensation of chromatin.
In this study, we examined the molecular mechanism of the induction of DLC-1 gene by TSA in human gastric cancer cell lines. We show that Sp1 binding sites located at DLC-1 promoter play important roles in the activation of DLC-1 promoter by TSA. In addition, we found that Sp1, rather than Sp3, and p300 HAT are required for the activation of DLC-1 promoter by TSA, leading to the induction of DLC-1 gene expression. These findings demonstrate the molecular mechanism by which TSA induced the DLC-1 gene expression in gastric cancer cells.
Materials and Methods
Cell lines
Gastric cancer cell lines were obtained from the Korean Cell Line Bank (Seoul). Cell lines were maintained in RPMI-1640 supplemented with 10% (vol/vol) FBS and gentamicin (10 µg/ml) at 37℃ in a humidified 5% CO2 atmosphere.
RT-PCR and Western blot analysis
SNU-5 cells were treated with DMSO (control), 330 nM TSA, 1 µM SK-7068, or 5 mM sodium butyrate (NaBu) for 12 h and RT-PCR was performed as described previously (Kim et al., 2003). Western blot analysis was performed with DMSO or 330 nM TSA-treated SNU-5 and SNU-668 cells as described previously (Kim et al., 2007). Primary antibodies that recognize Sp1 and Sp3 (Santa Cruz Biotechnology) were used.
Construction of the DLC-1 promoter linked to luciferase gene with deletions and site mutations
A fragment of the DLC-1 gene promoter region (-577/+117) was generated by PCR using fetal lung fibroblast genomic DNA as a template and the resulting PCR product was inserted into the pGL3-basic luciferase reporter vector (pGL3b) (Promega) as described previously (Kim et al., 2003). Sequence numbering is relative to the transcription start site (+1) (Genbank Accession NM_006094). The serial deletion constructs of the DLC-1 promoterluciferase constructs were generated from the -577/+117-pGL3b by PCR and inserted into the pGL3b vector. The site mutation constructs of the DLC-1 promoter were generated using a Quick-Change site-directed mutagenesis kit (Stratagene), based on the DLC-1 promoter-luciferase construct (-219/+117-pGL3b) that contains Sp1 binding sites. The sequences and orientations of the inserts were determined by direct sequencing.
Transient transfection and luciferase assay
To assess basal DLC-1 promoter activity in gastric cancer cells, the DLC-1 promoter-luciferase construct (-577/+117-pGL3b) was transfected into gastric cancer cells using Lipofectamine 2000 (Invitrogen). To study the effects of TSA on DLC-1 promoter activity, SNU-5 and SNU-668 cells were tranfected with the indicated DLC-1 promoterluciferase construct or cotransfected with the DLC-1 promoter-luciferase constructs and expression vectors for Sp1, Sp3 (pEVR2/Sp1 and pRC/CMV/Sp3 respectively, provided by Dr. G. Suske) (Hagen et al., 1994), or p300, or pSuper vector carrying the shRNA against p300 sequence (pSuper-p300) (Liu et al., 2003) using Lipofectamine 2000. Twenty-four hours post transfection, cells were treated with 330 nM TSA, or DMSO as control, for additional 12 h. Cell lysates were collected for luciferase assays. Luciferase activities of cell lysates were measured using a TR717 Microplate Luminometer (Tropix, Inc., Bedford, MA) and a Bioluminescent Reporter Gene Assay System (Tropix, Inc.) according to the manufacturer's instructions. Luciferase activities were normalized for total protein concentration determined using BCA assay.
Nuclear exreact preparation and electrophoretic mobility shift assay
Nuclear extracts from DMSO- or TSA-treated SNU-5 or SNU-668 cells were prepared as described previously (Hiramoto et al., 1998). For EMSA, oligonucleotide probes from -213 to -190 (5'-GAG GCGGGGCCTGGGCGGGGCGGGGC-3') were annealed and labeled with [γ-32P]ATP and T4-polynucleotide kinase (NEB). Five µg of nuclear extract and 1 µg poly (dI-dC) were preincubated for 20 min at 4℃ in an appropriate binding buffer (20 mM HEPES pH 7.5, 5 mM KCl, 1 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 0.1% NP-40, 10% glycerol). The mixture was then incubated on ice for 30 min with 0.5 ng of the labeled oligonucleotide. For supershift experiments, 2 µg of anti-Sp1 and/or anti-Sp3 antibodies were added during preincubation. For competition experiments, 20-fold molar excess cold oligonucleotide was added to the binding mixture 20 min before addition of the labeled probe. Samples were loaded onto a 4% polyacrylamide gel and run for 2 h at 200 V in 0.5× TBE. Gels were dried, and protein-DNA binding was visualized by autoradiography.
Results
Histone deacetylase inhibitors activate the DLC-1 promoter and induce DLC-1 gene expression in SNU-5 and SNU-668
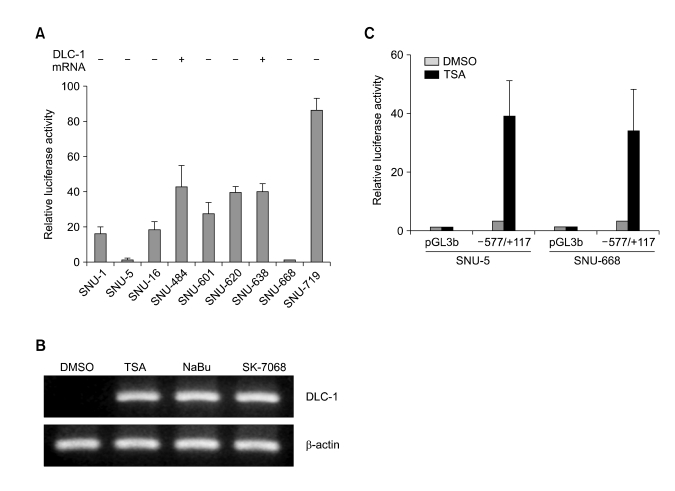
We first investigated basal DLC-1 promoter activity in gastric cancer cells by luciferase assay using DLC-1 promoter-luciferase construct (-577/+117-pGL3b). As shown in Figure 1A, the DLC-1 promoter was active in many gastric cancer cells regardless of their expression status except SNU-5 and SNU-668. The loss of DLC-1 gene expression in the gastric cancer cells whose promoter was active is due to methylation of the DLC-1 promoter region as demonstrated in our previous study (Kim et al., 2003). We have also previously demonstrated that TSA, a specific histone deacetylase (HDAC) inhibitor, induced DLC-1 gene expression in the SNU-5 and SNU-668. Treatment of SNU-5 cells with another HDAC inhibitor, NaBu or SK-7068, also resulted in the induction of DLC-1 gene expression (Figure 1B). To evaluate whether the DLC-1 gene expression by HDAC inhibitors is due to DLC-1 promoter activation, SNU-5 and SNU-668 cells were transiently transfected with empty vector (pGL3b) or -577/+117-pGL3b and treated with DMSO or TSA for 12h before measuring the luciferase acitivity. As shown in Figure 1C, the DLC-1 promoter activity was dramatically increased after TSA treatment in both cells, but no significant induction was observed in cells transfected with empty vector. These results suggest that HDAC inhibitors induce the DLC-1 gene expression through the DLC-1 promoter activation in SNU-5 and SNU-668 cells.
Figure 1.
Induction of DLC-1 gene expression by HDAC inhibitors is via an activation of DLC-1 promoter in human gastric cancer cells. (A) The basal activity of DLC-1 promoter in gastric cancer cells. The DLC-1 promoter luciferase construct (-577/+117-pGL3b) was transiently transfected into gastric cancer cells for 24 h. Luciferase activity was measured and normalized by protein concentration. Data are representative of three independent experiments performed in triplicate. Expression of DLC-1 mRNA of each gastric cancer cells is shown in the top of the graph. (B) HDAC inhibitors induce DLC-1 gene expression. SNU-5 cells were treated with 330 nM TSA, 5 mM NaBu, or 1 µM SK-7068 and cultured for 12 h. DLC-1 and β-actin mRNA expression was determined by RT-PCR as describe in "Materials and Methods" (C). TSA treatment increases the DLC-1 promoter activity. SNU-5 and SNU-668 cells were transiently transfected with empty reporter vector (pGL3b) or DLC-1 promoter luciferase construct (-577/+117-pGL3b) for 24 h followed by treatment with DMSO or 330 nM TSA for 12 h. Luciferase activity was measured and normalized by protein concentration. The data represent the combined results of three independent experiments performed in triplicate.
Activation of the DLC-1 promoter by TSA requires Sp1 sites located between -219 and -174 of the DLC-1 promoter
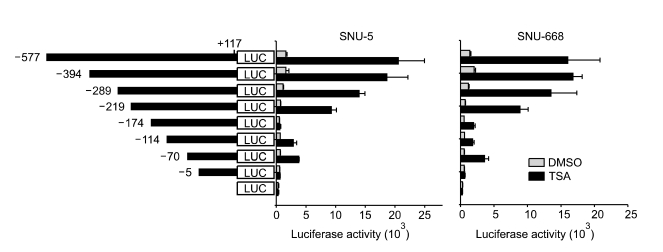
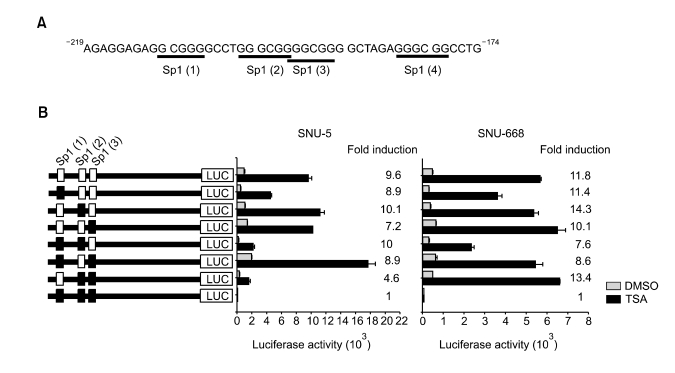
To characterize the region responsible for activation of the DLC-1 promoter by TSA, serial deletion mutants of the DLC-1 promoter were constructed, transiently transfected into SNU-5 and SNU-668, and luciferase activities were measured 12 h after treatment with DMSO or TSA. Deletion of the promoter to position -219 did not significantly affect the TSA-mediated DLC-1 promoter activation, because the fold induction in response to TSA is similar to that of activation by -577/+117-pGL3b. On the other hand, further deletion to position -174 significantly decreased the activation of DLC-1 promoter by TSA in both cells (Figure 2). These results suggest the TSA-responsive element is present within 45bp region between -219 and -174. Identification of potential transcription factor binding sites by the MatInspector program, using the TRANSFAC 3.5 matrices, revealed no TATA boxes could be identified. Notably, the DLC-1 promoter region between -219 and -174 has four Sp1 binding sites (Figure 3A), suggesting that Sp1 might be involved in TSA-mediated DLC-1 promoter activation. To evaluate the significance of these putative Sp1 sites, we focused on the first three Sp1 sites and constructed vectors containing various combinations of mutated Sp1 sites. Mutation of all three Sp1 sites abolished both basal and TSA-induced promoter activity in both cells. By contrast, other combinations of mutation showed a 4.6 to 10.1 and a 7.6 to 13.4- fold induction of DLC-1 promoter activity in SNU-5 and SNU-668, respectively (Figure 3B). Taken together, these results suggest that Sp1 sites located between -219 and -174 are essential for basal DLC-1 promoter activity and DLC-1 promoter activation by TSA.
Figure 2.
Activation of the deletion constructs of DLC-1 promoter in SNU-5 and SNU-668 cell. Serial deletion mutants of DLC-1 promoter were constructed as shown in the left panel. Theses constructs were transiently transfected into SNU-5 and SNU-668 cells followed by treatment with DMSO or 330 nM TSA for 12 h. Luciferase activity was measured and normalized to protein concentration in all luciferase reporter experiments. The data represent the combined results of three independent experiments performed in triplicate.
Figure 3.
Identification of the Sp1 binding sites responsible for the activation of DLC-1 promoter by TSA. (A) Sequences of the DLC-1 promoter containing the region -219/-174 bp. The consensus sequences for Sp1 binding sites (GGCGGG or GGGCGG) are underlined. (B) Base substitution mutants using -219/+117 as a template were constructed as shown in the left panel. Theses constructs were transiently transfected into SNU-5 and SNU-668 cells followed by treatment with DMSO or 330 nM TSA for 12 h. Luciferase activity was measured and normalized to protein concentration in all luciferase reporter experiments. The data represent the combined results of three independent experiments performed in triplicate.
Sp1 and Sp3 bind to the -219 to -174 region of the DLC-1 promoter and Sp1 is the main transactivators of the DLC-1 promoter in response to TSA
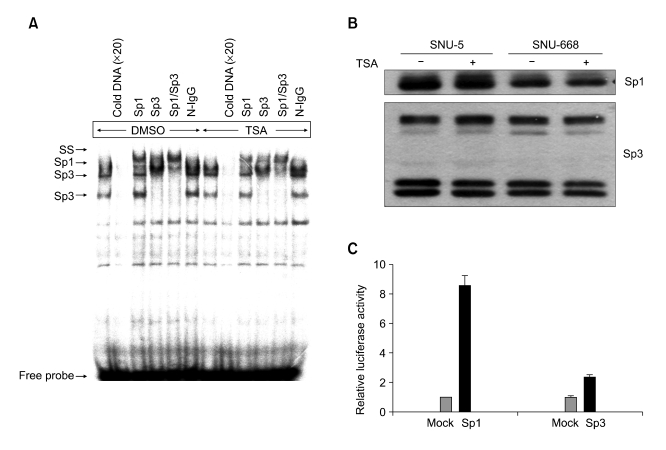
To verify whether transcriptional factors of the Sp family (Sp1/Sp3) bind the Sp1 binding sites and the DLC-1 promoter activation by TSA could be attributed to a change of Sp family protein binding activities to this site, an EMSA was performed with nuclear extracts prepared from DMSO- or TSA-treated SNU-5 cells. Incubation of the 26mer oligonucleotide probe of the region representing the three Sp1 sites with nuclear extracts from SNU-5 cells resulted in the formation of three specific DNA protein complexes (Figure 4A). The addition of an excess of unlabeled oligonucleotide probe completely abolished the formation of all three complexes. To determine whether Sp1 and Sp3 were involved in the formation of the three complexes, antibody supershift assays were performed. The Sp1 and Sp3 antibodies were found to selectively supershift the complexes, whereas the normal rabbit antibody did not show any shifted bands. No apparent changes in complex formation were observed in DMSO- or TSA-treated SNU-5 cells. Similar results were obtained in DMSO- or TSA-treated SNU-668 cells (data not shown). In addition, TSA treatment did not show any significant effect on the expression of Sp1 and Sp3 protein in both cells (Figure 4B). Consequently, these results indicate that Sp1 and Sp3 proteins bind to the Sp1 sites of the DLC-1 promoter, and their expression level and binding affinity to the DLC-1 promoter are not affected by TSA treatment.
Figure 4.
Sp1 and Sp3 bind to the DLC-1 promoter and Sp1 is mainly involved in activation of the DLC-1 promoter by TSA. (A) Sp1/Sp3 binds to the Sp1 sites located within DLC1 promoter. EMSA was carried out using [γ-32P] ATP -labeled oligonucleotide spanning Sp1 binding sites of DLC1 promoter and nuclear extracts prepared from SNU-5 cells treated with DMSO or 330 nM TSA for 12 h. The reaction was carried out in the absence (lanes1 and 7) or presence of 20-fold molar excess of unlabeled probe (lanes2 and 8) or in the presence of specific antibodies to Sp1 (lanes3 and 9), Sp3 (lanes4 and 10), both (lanes5 and 11), or normal rabbit IgG (lanes6 and 12). (B) TSA has no effect on the expression of Sp1 and Sp3 protein. Protein extracts from DMSO-or 330 nM TSA-treated SNU-5 and SNU-668 cells were analyzed by Western blotting using antibodies directed to Sp1 or Sp3. (C) Sp1 is the main transactivators of the DLC-1 promoter in response to TSA. Empty vector (mock), Sp1, or Sp3 expression vectors were cotransfected with the DLC-1 promoter luciferase construct (-219/+117-pGL3b) into SNU-668 cells and treated cells with 330 nM TSA for 12 h before measuring luciferase activity. Data are representative of two independent experiments performed in triplicate.
To determine whether both Sp1 and Sp3 are involved in the activation of DLC-1 promoter in response to TSA, we cotransfected Sp1 or Sp3 expression vectors with the DLC-1 promoter luciferase construct into SNU-668. Sp1 expression obviously increased the responsiveness of the DLC-1 promoter to TSA, but Sp3 expression had little effect on the induction of DLC-1 promoter activity by TSA (Figure 4C). This result suggests that Sp1 is the main transactivator of the DLC-1 promoter in response to TSA.
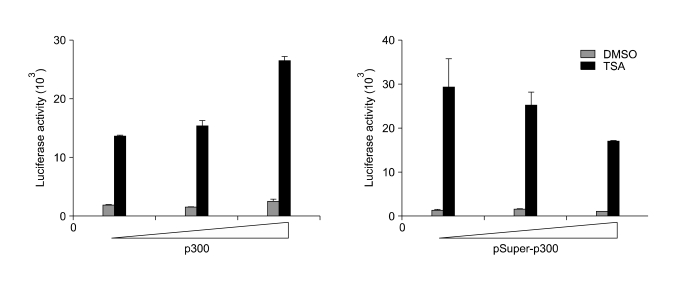
p300, a histone acetyltransferase, participates in the DLC-1 promoter activation by TSA
Because no changes in the expression level and binding affinity of Sp1 to DLC-1 promoter after TSA treatment were found, we considered that the activation of DLC-1 promoter by TSA results from interaction of Sp1 with other proteins. To examine whether p300 HAT is involved in the activation of DLC-1 promoter by TSA, a p300 expression vector or shRNA for p300 were cotransfected with the DLC-1 promoter luciferase construct into SNU-668. As shown in Figure 5, the expression of p300 increased stimulation of the DLC-1 promoter by TSA, whereas knock down of p300 inhibited DLC-1 promoter responsiveness to TSA in a dose-dependent manner. These results suggest that p300 acts as a co-activator and/or possibly the histone actyltrasferase activity of p300 is involved in the acetylation of histone protein around the DLC-1 promoter and the concomitant activation of DLC-1 promoter.
Figure 5.
p300 is involved in the TSA-mediated DLC-1 promoter activation. p300 expression vector or pSuper vector carrying the shRNA against p300 (pSuper-p300) were cotransfected with the DLC-1 promoter luciferase construct (-577/+117-pGL3b) into SNU-668 cells and treated cells with DMSO or 330 nM TSA for 12 h before measuring luciferase activity. Data are representative of two independent experiments performed in triplicate.
Discussion
Several studies have shown that transcriptional silencing of DLC-1 gene is one of several mechanisms for tumor progression. Many human cancers express significantly reduced level of DLC-1 by genetic and epigenetic mechanism (Dunkin et al., 2007). Reexpression of DLC-1 gene in DLC-1 null cells inhibits tumor cell growth and metastasis (Goodison et al., 2005; Kim et al., 2007). Inhibition of DLC-1 expression using RNA interference promoted cell proliferation and migration of in the colon cancer cell line (Jin et al., 2008) and hepatocellular carcinoma in mice (Xue et al., 2008). These results imply that DLC-1 acts as tumor suppressor gene and may be a candidate therapeutic target for cancer.
We have previously found that TSA induced DLC-1 gene expression in DLC-1 non-expressing gastric cancer cell lines, SNU-5 and SNU-668, which have the unmethylated alleles (Kim et al.,2003). Since our report, several studies have demonstrated that DLC-1 gene expression was induced by treatment with either TSA alone or in combination with 5-aza-dC, a demethylating agent, in several cancers such as prostate cancer, multiple myeloma, and non-Hodgkin's lymphoma (Guan et al., 2006; Song et al., 2006; Shi et al., 2007), suggesting that histone deacetylation of DLC-1 gene in not restricted to gastric cancer cells and is a new epigenetic mechanism for silencing of DLC-1 gene in human cancers. However, details of molecular mechanism underlying TSA-induced DLC-1 gene expression have not been elucidated. In this study, we have investigated the molecular mechanism of the induction of DLC-1 gene expression by TSA in SNU-5 and SNU-668 cells. We found that the DLC-1 induction by TSA requires Sp1 sites located between -219 and -174 of DLC-1 promoter and revealed that Sp1, rather than Sp3, and p300 HAT play an important role in the activation of DLC-1 promoter by TSA, resulting in DLC-1 gene expression.
Recently, the treatment of mammalian cells with HDAC inhibitors resulted in transcriptional activation of a variety of genes, particalarly the genes which contain the Sp1 binding site such as p21 (WAF1), p19 (INK4D), TGF-β RII, and KLF4 (Sowa et al., 1997; Zhao et al., 2003; Chen et al., 2004; Yokota et al., 2004; Yi et al., 2007). It has been also shown that p300 collaborates with Sp1 in the activation of genes by TSA such as p21 (WAF1) (Xiao et al., 2000) and pyruvate dehydrogenase kinase 4 (PDK4) (Kwon et al., 2006). Based on the observations that HDACs and p300 can directly interacts with Sp1 (Owen et al., 1998; de Ruijter et al., 2003), we here suggest a possible mechanism for induction of DLC-1 gene expression by TSA. HDACs bound to DLC-1 promoter region via Sp1 renders histone hypoacetylation around DLC-1 promoter, leading to represses DLC-1 gene expression due to the compact chromatin structure. After TSA treatment, HDACs are released and p300 is recruited into the Sp1 to permit histone acetylation associated with the DLC-1 promoter region, resulting in the gene expression.
This is the first study showing the mechanism of induction of DLC-1 gene expression by a histone deacetylase inhibitor in human gastric cancer cell lines. Our present study suggests that the restoration of DLC-1 by HDAC inhibitors could be a major mechanism for suppressing tumor growth.
Acknowledgements
We thank Dr. Rudy Juliano for critical review of the manuscript and helpful discussions. This work was supported in part by grants from the Ministry of Science and Technology of Korea through the National Research Laboratory Program for Cancer Epigenetics and by BK21 Project for Medicine, Dentistry, and Pharmacy.
Abbreviations
- DLC-1
deleted in liver cancer-1
- EMSA
electrophoretic mobility-shift assay
- GAP
GTPase-activating protein
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- NaBu
sodium butyrate
- TSA
trichostatin A
References
- 1.Chen ZY, Rex S, Tseng CC. Krüppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–798. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 2.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, Popescu NC. DLC-1:a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 7.Hagen G., Müller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Healy KD, Hodgson L, Kim TY, Shutes A, Maddileti S, Juliano RL, Hahn KM, Harden TK, Bang YJ, Der CJ. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog. 2008;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramoto M, Shimizu N, Sugimoto K, Tang J, Kawakami Y, Ito M, Aizawa S, Tanaka H, Makino I, Handa H. Nuclear targeted suppression of NF-kappa B activity by the novel quinone derivative E3330. J Immunol. 1998;160:810–819. [PubMed] [Google Scholar]
- 10.Jin Y, Tian X, Shang Y, Huang P. Inhibition of DLC-1 gene expression by RNA interference in the colon cancer LoVo cell line. Oncol Rep. 2008;19:669–674. [PubMed] [Google Scholar]
- 11.Kim TY, Jong HS, Song SH, Dimtchev A, Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M, Bang YJ. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22:3943–3951. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 12.Kim TY, Lee JW, Kim HP, Jong HS, Kim TY, Jung M, Bang YJ. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;355:72–77. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- 13.Kwon HS, Huang B, Jeoung NH, Wu P, Steussy CN, Harris RA. Retinoic acids and trichostatin A (TSA), a histone deacetylase inhibitor, induce human pyruvate dehydrogenase kinase 4 (PDK4) gene expression. Biochim Biophys Acta. 2006;1759:141–151. doi: 10.1016/j.bbaexp.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Liu G., Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 15.Mackay DJ, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 16.Ng IO, Liang ZD, Cao L, Lee TK. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. [PubMed] [Google Scholar]
- 17.Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21 (WAF1) cyclindependent kinase inhibitor gene through Sp1 and CBP/p300. J Biol Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 18.Pang JC, Chang Q, Chung YF, Teo JG, Poon WS, Zhou LF, Kong X, Ng HK. Epigenetic inactivation of DLC-1 in supratentorial primitive neuroectodermal tumor. Hum Pathol. 2005;36:36–43. doi: 10.1016/j.humpath.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, Hsieh WS, Ambinder RF, Lin X, Chan AT, Tao Q. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andreski M, Wooldridge JE, Caldwell CW. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 21.Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, Xie Y. High-frequency promoter hypermethylation of the deleted in liver cancer-1 gene in multiple myeloma. J Clin Pathol. 2006;59:947–951. doi: 10.1136/jcp.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 23.Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21 (waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J Biol Chem. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
- 24.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers S, Zender L, Lowe SW. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi T, Baek JH, Kim HJ, Choi MH, Seo SB, Ryoo HM, Kim GS, Woo KM. Trichostatin A-mediated upregulation of p21 (WAF1) contributes to osteoclast apoptosis. Exp Mol Med. 2007;39:213–221. doi: 10.1038/emm.2007.24. [DOI] [PubMed] [Google Scholar]
- 26.Ying J, Li H, Murray P, Gao Z, Chen YW, Wang Y, Lee KY, Chan AT, Ambinder RF, Srivastava G, Tao Q. Tumor-specific methylation of the 8p22 tumor suppressor gene DLC1 is an epigenetic biomarker for Hodgkin, nasal NK/T-cell and other types of lymphomas. Epigenetics. 2007;2:15–21. doi: 10.4161/epi.2.1.3883. [DOI] [PubMed] [Google Scholar]
- 27.Yokota T, Matsuzaki Y, Miyazawa K, Zindy F, Roussel MF, Sakai T. Histone deacetylase inhibitors activate INK4d gene through Sp1 site in its promoter. Oncogene. 2004;23:5340–5349. doi: 10.1038/sj.onc.1207689. [DOI] [PubMed] [Google Scholar]
- 28.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- 29.Yuan BZ, Zhou X, Durkin ME, Zimonjic DB, Gumundsdottir K, Eyfjord JE, Thorgeirsson SS, Popescu NC. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003a;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- 30.Yuan BZ, Durkin ME, Popescu NC. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet. 2003b;140:113–117. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- 31.Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 32.Zhao S, Venkatasubbarao K, Li S, Freeman JW. Requirement of a specific Sp1 site for histone deacetylase-mediated repression of transforming growth factor beta Type II receptor expression in human pancreatic cancer cells. Cancer Res. 2003;63:2624–2630. [PubMed] [Google Scholar]