Abstract
In light of the anti-inflammatory properties of histone deacetylase (HDAC) inhibitors, such as suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA), we examined a new HDAC inhibitor KBH-A42 for its anti-inflammatory activities. KBH-A42 showed noteworthy anti-inflammatory properties in vitro via suppression of the production of TNF-α, a proinflammatory cytokine, and nitric oxide (NO), a proinflammatory effector molecule, in LPS-stimulated RAW264.7 cells and peritoneal macrophages. It also inhibited TNF-α production in vivo as demonstrated in a LPS-induced mouse endotoxemia model. The levels of TNF-α, IL-1β, IL-6 and iNOS mRNAs determined by RT-PCR propose that the inhibition of these pro-inflammatory mediators by KBH-A42 resulted from inhibiting expression of these genes. However, the EMSA study to see the effect of KBH-A42 on the binding of NF-κB, a transcription factor, to a specific DNA sequence showed that the binding of NF-κB to DNA was not changed regardless of increasing the concentration of KBH-A42 in the presence and absence of LPS stimulation. Interestingly, DNA binding of another transcription factor AP-1 dose-dependently increased by KBH-A42. KBH-A42 differentially regulated the phosphorylation of MAP kinases. While the phosphprylation of ERK1/2 and SAPK/JNK was not affected by KBH-A42, the phosphorylation of p38 decreased by KBH-A42. These results showed that KBH-A42 inhibits production of proinflammatory cytokines in macrophages by decreasing their mRNA levels, and p38 kinase is involved in the KBH-A42-mediated inhibition.
Keywords: anti-inflammatory agents, histone deacetylases, NF-κB, nitric oxide, transcription factor AP-1, tumor necrosis factor-α, vorinostat
Introduction
Histone decetylase (HDAC) and histone acetyltransferase (HAT) have important roles in chromatin remodeling and "epigenetic control" of gene expression (Wu et al., 2000). HDAC catalyzes deacetylation of ε-amino group in lysines located near the N-terminal of core histone proteins. Deacetylation associates to transcriptional repression and abnormal recruitment of HDAC is related to carcinogenesis (Archer et al., 1999; Kouzarides et al., 1999). Studies have shown that inhibition of HDAC elicits anticancer effects in several tumor cells by inhibition of cell growth. This has led to the development of HDAC inhibitors for anti-cancer chemotherapy (Johnstone et al., 2002). Moreover, epigenetic control of chromatin involves various major cell functions such as cell growth, differentiation, and apoptosis (Jaenisch et al., 2003). It has been recently revealed that HDAC inhibitors have anti-inflammatory activities via suppression of cytokines and nitric oxide (NO) (Blanchard et al., 2005). Butyrate reduced IL-12 transcription in T-cells (Diakos et al., 2002) and human blood monocytes (Saemann et al., 2000), and inhibited NO production in RAW macrophage cells (Chakravortty et al., 2000). Suberoylanilide hydroxamic acid (SAHA) inhibited secretion of TNF-α, IL-1β, IL-6, and IFN-γ in LPS-induced PBMC cells, and also inhibited their in vivo production as shown in an LPS induced animal model (Leoni et al., 2002). Appropriate control of TNF-α has been considered as a potential approach for the treatment of rheumatoid arthritis (RA) (Newton et al., 1999; Palladino et al., 2003) along with the success of anti-TNF-α biologics (Moreland et al., 1997; Lipsky et al., 2000). Several "the next generation" anti-TNF-α small molecule approaches have been in progress because of the significant advantage of developing orally active molecules in low cost (Palladino et al., 2003). HDAC inhibitor, Trichostatin A (TSA) caused the apoptosis of osteoclast by up regulating p21WAF1 and is a possible therapeutic agent for osteoporosis (Yi et al., 2007). These results suggest that HDAC could be a potential target for many other non-malignant diseases such as rheumatoid arthritis and osteoporosis.
Recently, we have reported preparation of a novel series of HDAC inhibitors and evaluation of their anti-proliferative and anti-inflammatory activities (Kim et al., 2006, 2007). As a part of our ongoing optimization process of our HDAC inhibitors, we were interested in investigating of KBH-A42 for its anti-inflammatory activity since it was screened from a cell based TNF-α inhibition assay. With its unique HDAC inhibitory activity pattern, KBH-A42 was subjected to the cellular cytokine inhibition, molecular mechanism, and in vivo TNF-α inhibition studies.
Materials and Methods
Compounds
KBH-A42 (Kim et al., 2006) and suberoylanilide hydroxamic acid (SAHA) (Gediya et al., 2005) were prepared by the reported procedures.
HDAC assay
HDAC fluorescent activity assays using a Fluror de Lys substrate (Biomol, Plymouth Meeting, PA), which contains an acetylated lysine side chain, were performed according to manufacturer's instructions. In brief, HeLa nuclear extracts, which were used as an HDAC enzyme source, were incubated at 25℃ with 250 mM of Fluror de Lys substrate and various concentrations of KBH-A42 and SAHA. Reactions were stopped after 20 min with Fluror de Lys developer and fluorescence was measured using a microplate spectrofluorometer (LS 50B, Perkin Elmer) with excitation at 360 nm and emission at 460 nm.
Immunoblotting of p21WAF1 and acetylated histone H4
HeLa cells were incubated with apicidin (1 µM), KBH-A42 (10 µM) or 0.1% DMSO in culture medium for 24 h. Cell lysates were boiled in Laemmli sample buffer for 3 min, and 30 µg of each total protein were subjected to SDS-PAGE on 15% slab gels for the analysis of p21WAF1/Cip1 and acetylated histone H4. Proteins were transferred to PVDF membranes, and membranes were blocked for 30 min in PBS containing 0.1% Tween 20 (PBS-T) and 5% (w/v) skim milk, and incubated overnight with anti- p21WAF1/Cip1 (Santa Cruz Biotechnologies, Inc) and acetylated histone H4 (Upstate Biotechnology) antisera. The membranes were then washed with PBS-T and incubated for 2 h with an secondary antibody. Bound antibodies were detected with the enhanced amplified alkaline phosphatase immunoblot system (Bio-Rad).
In vitro NO assay and TNF-α inhibition assay
LPS (200 ng/ml)-stimulated RAW264.7 cells were cultured with KBH-A42 or SAHA for 24 h. NO2- accumulation was used as an indicator of NO production in the medium. The isolated supernatants were mixed with an equal volume of Griess reagent (1% sulfanilamide, 0.1% naphthylethyle-nediamine dihydrochloride, and 2% phosphoric acid) and incubated at room temperature for 10 min. Nitrite production was determined by measuring absorbance at 540 nm versus a NaNO2 standard curve. The concentration of TNF-α secreted in the culture supernatant of RAW264.7 cells was determined by ELISA, according to the manufacture's instruction (R&D Systems, Minneapolis, MN).
Determining cell viability by propidium iodide (PI) staining
LPS (200 ng/ml)-stimulated RAW 264.7 cells were cultured in the presence of various concentrations of KBH-A42 for 24 h. Cells were collected, washed twice in PBS containing 0.1% BSA, and resuspended in 500 µl of PBS containing PI 5 µg/ml for 10 min. Cells were analyzed using an FACSCalibur flow cytometer (BD Biosciences, San Jose, CA), and data were analyzed using WinMidi software (Scripps, LaHoya, CA).
In vivo TNF-α inhibition assay
The compounds were administered intraperitoneally or orally to mice 24 h and 2 h before LPS injection. LPS (20 µg) was injected intraperitoneally to boost the concentration of TNF-α in the blood. Plasma was prepared 30, 60, 90 and 120 min after LPS treatment and the TNF-α level was measured by ELISA, according to the manufacture's instruction (R&D Systems, Minneapolis, MN).
Measurement of mRNA expression by RT-PCR
The expression of cytokines was measured by RT-PCR. The cells were treated with KBH-A42 1 h before LPS stimulation and total RNA was isolated 6 h after LPS stimulation (200 ng/ml). The RNA was reverse transcribed using a GeneAmp RNA PCR kit using 100 ng of total cellular RNA (PerkinElmer). PCR was carried out using 2.5 U of AmpliTaq DNA polymerase in a Bio-Rad Cycler (Bio-Rad Lab). PCR products were electrophoresed on a 3% Nusieve 3:1 agarose gel and photographed after staining with ethidium bromide.
Electrophoretic mobility shift assay (EMSA)
The protein content of the nuclear extracts of RAW 264.7 cells was determined using a Bio-Rad protein assay kit according to the manufacturer's instruction (Bio-Rad, Hercules, CA). The oligonucleotide sequence for NF-κB/Rel and AP-1 was 5'-GATCTCAGAGGGGACTTTCCGAGAGA-3' and 5'-GATCTGCATGAGTCAGACACA-3'. Double-stranded oligonucleotides were end-labeled with [γ-32P]ATP. Nuclear extracts (5 µg) were incubated with 2 µg of poly (dI-dC) and a 32P-labeled DNA probe in binding buffer (100 mM KCl, 30 mM HEPES, 1.5 mM MgCl2, 0.3 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM PMSF, 1 µg/ml of aprotinin and 1 µg/ml of leupeptin) for 10 min. DNA binding activity was separated from the free probe using a 4.8% polyacrylamide gel in 0.5 X TBE buffer (44.5 mM Tris, 44.5 mM boric acid and 1 mM EDTA). Following electrophoresis, the gel was dried and subjected to autoradiography.
Western immunoblot analysis
Whole cell lysates (20 µg, for phospho-ERK1/2, phospho-SAPK/JNK, phospho p38, and p38) were separated by 10% SDS-PAGE, then electro-transferred to nitrocellulose membranes (Amersham International, Buckinghamshire, UK). The membranes were preincubated for 1 h at room temperature in Tris-buffered saline (TBS), pH 7.6, containing 0.05% Tween 20 and 3% BSA. The nitrocellulose membranes were incubated with phosphorylated ERK1/2, phosphorylated SAPK/JNK, phosphorylated p38 or p38-specific antibodies. Immunoreactive bands were then detected by incubation with conjugates of anti-rabbit IgG with HRP and enhanced chemiluminescence reagents (Amersham).
Molecular modeling
The program Insight II was used to create a docking model for HDAC-1 based on the crystal structure of HDLP. Both proteins are class 1 HDAC's and share 32% identity. Key residues which are conserved include Asp-168, His-170, and Asp-258, which chelate the Zn2+, and His-131, His-132, and Tyr-297, which interact with the hydroxamic acid (HDLP numbering). His -131 and -132 form charge relays with Asp-166 and Asp-173, respectively, and these residues are also conserved. A narrow channel in the binding pocket is formed by Phe-141 and Phe-198, residues which are also conserved in HDAC-1. At the top of the binding pocket, Glu-92 in HDLP is mutated to Asp in HDAC-1 and Tyr-91 is mutated to Glu. Docking calculations for KBH-A42 were carried out with the program Discover (McMartin et al., 1997). The hydroxamic acid moiety was constrained to the conformation seen in the crystal structure, while the rest of the inhibitor was free to move. Mutated residues were allowed to relax during the minimization steps.
Results
In vitro assays
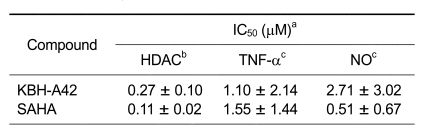
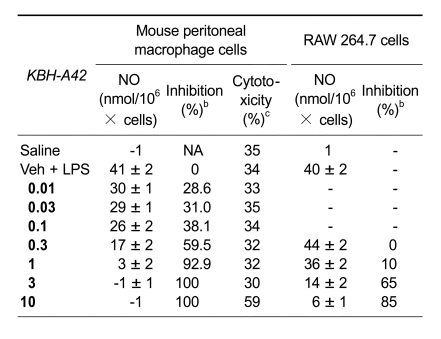
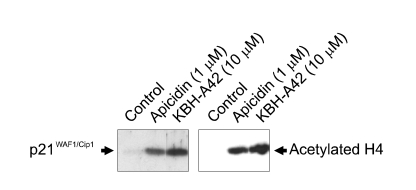
The preliminary evaluation of a HDAC inhibitor KBH-A42 on inhibition of TNF-α and NO in RAW 264.7 cells was in our previous communication (Kim et al., 2007). SAHA, a known HDAC inhibitor, was used as a reference compound. KBH-A42 showed a good inhibitory activity to the partially purified HDAC enzyme, obtained from HeLa cell lysates, and its IC50 value was 0.27 µM. Inhibition of TNF-α and NO production were performed in the LPS induced murine macrophage RAW 264.7 cells. IC50 values of KBH-A42 against TNF-α and NO production were 1.10 and 2.71 µM, respectively, which were comparable to those of SAHA (Table 1). Nitric oxide inhibition assay in peritoneal macrophage cells showed that KBH-A42 inhibited 38% of NO production at 0.1 µM (Table 2). Further reduction was observed; 60% and 93% at 0.3 and 1 µM, respectively. KBH-A42 also showed typical characteristics of HDAC inhibitors, such as increasing the hyperacetylation level of histone 4 (H4) and the expression of cyclin-dependent kinase inhibitor p21WAF1 (Figure 2) (Archer et al., 1998).
Table 1.
In vitro inhibition of HDAC enzyme and the inhibition of TNF-α and NO by KBH-A42 and SAHA.
aValues are the means of a minimum of three experiments. bHDAC enzyme obtained from HeLa cell lysate. cRAW264.7 cells were used.
Table 2.
In vitro inhibition of NO by KBH-A42 in mouse peritoneal macrophage cells and RAW 264.7 cells.
aValues are the means of five experiments. bMouse peritoneal macrophage cells were used. cCytotoxicity was measured by PI staining method.
Figure 2.
Induction of p21WAF1 expression and histone H4 hyperacetylation by apicidin and KBH-A42. Lysates (30 µg) of the HeLa cells exposed to 1 µM apicidin, or 10 µM KBH-A42 (Lanes 2-3 respectively) or 0.1% DMSO (Lane 1) were examined by 15% SDS-PAGE and analyzed with immunoblotting using antibodies for p21WAF1/Cip1 and acetylated histone H4.
KBH-A42 inhibits production of TNF-α in in vivo model
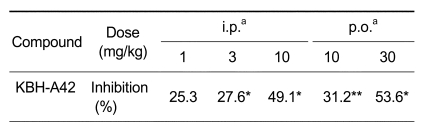
TNF-α inhibitory effect of KBH-A42 was then examined in vivo (Table 3) with an insight of promising results of TNF-α and NO inhibition. In this model, KBH-A42 was administered (i.p or p.o) to mice 24 h and 2 h before intraperitoneal injection of LPS for the purpose of inducing TNF-α production in mice. KBH-A42 inhibited TNF-α production in a dose dependent manner and showed 50% inhibition at the tested dose of 10 mg/kg (i.p.), compared to the TNF-α level in mice receiving LPS stimulation alone. In the case of p.o. administration, the production of TNF-α was also inhibited dose-dependently and displayed more than 50% inhibition at the tested dose of 30 mg/kg. However, no further increase in inhibition was observed at the higher dose level of 100 mg/kg.
Table 3.
In vivo inhibition of LPS induced TNF-α by KBH-A42.
aBALB/c mice (four per group) were treated. KBH-A42 was pretreated twice (i.p. or p.o.) at 24 h and 2 h before LPS (1.5 mg/kg, i.p.) was injected. Serum TNF-α was measured after 60 min. *P < 0.05; **P < 0.01.
Mechanism study
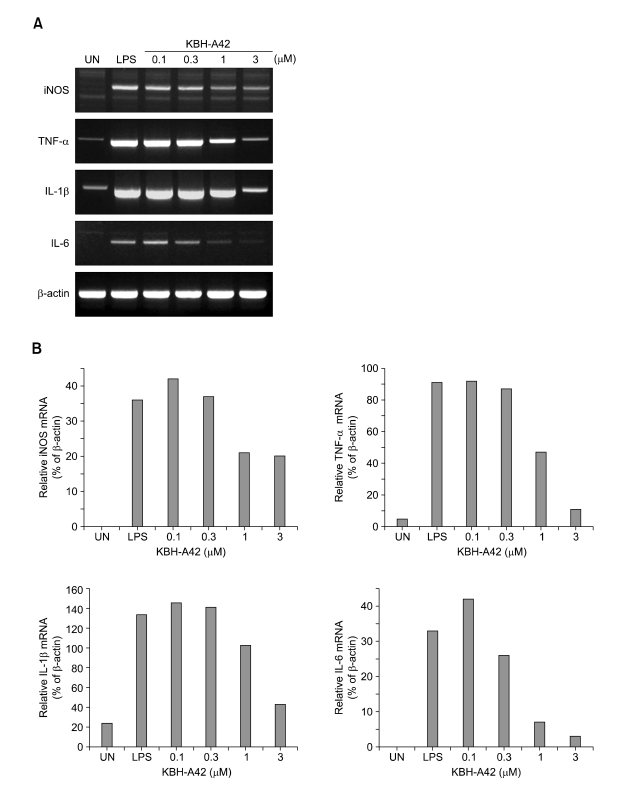
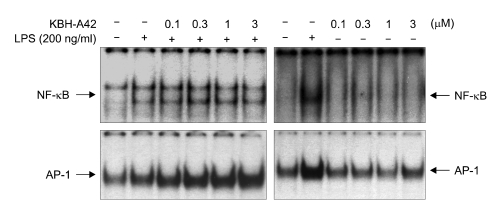
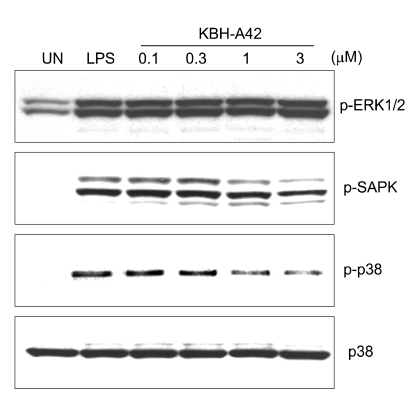
Along with in vitro and in vivo results, KBH-A42 showed inhibitory activities against TNF-α and NO production in RAW 264.7 cells. Thus, mRNA levels of iNOS, TNF-α, IL-1β, and IL-6 in LPS induced RAW 264.7 cells were evaluated by RT-PCR (Figure 3). KBH-A42 was treated 1 h before LPS and total RNAs of RAW 264.7 cells were isolated 6 h after LPS. All mRNA levels of iNOS, TNF-α, IL-1β, and IL-6 decreased in a dose dependent manner by KBH-A42 compared to those with LPS stimulated cells. These results indicate that KBH-A42 exerts its inhibitory activities on inflammatory cytokine production at mRNA expression level. To see if the inhibition of the mRNAs by KBH-A42 was due to inhibition of the activity of transcription factors, the possibility that KBH-A42 affects transcription factors, such as NF-κB, was investigated. NF-κB has a very important role in expression of pro-inflammatory mediators such as iNOS, interleukins, and TNF-α. Many anti-inflammatory drugs are known to target to inhibit an activation of this transcription factor. (D'Acuisto et al., 2002; Kyung et al., 2008) However, the effects on activity of transcription factors by KBH-A42 are quite mixed-up in RAW 264.7 cells (Figure 4). The effect of KBH-A42 on transcription factors was determined by measuring the extent of DNA binding of transcription factors. KBH-A42 was treated in the presence and absence of LPS. When cells were treated with LPS, KBH-A42 was treated 1 h before LPS stimulation. The extent of DNA binding of NF-κB and AP-1 was examined 1 h after LPS stimulation using the EMSA method. The DNA binding activity of NF-κB was not changed with increasing the concentration of KBH-A42 (0.1-3 µM). KBH-A42 alone did not have any effect on the binding of NF-κB to DNA. In contrast, the DNA binding of another transcription factor AP-1 dose-dependently increased by KBH-A42 in the LPS stimulated cells. In case of the cells without LPS stimulation, the increase in DNA binding of AP-1 was observed at 3 µM of KBH-A42. Next, the effect of KBH-A42 on the phosphorylation of MAP kinases (ERK1/2, SAPK/JNK, p38) was examined in the RAW 264.7 cell line using Western blotting method (Figure 5). KBH-A42 was pre-treated 1 h before LPS stimulation. The phosphorylation of p38, ERK1/2, and SAPK/JNK was examined at 10 min after LPS stimulation. The expression of p38 was not affected by the treatment of KBH-A42, but the phosphorylation of p38 was decreased with the increase of concentration of KBH-A42. The phosphorylation of SAPK/JNK slightly decreased at 3 µM of KBH-A42. The phosphorylation of ERK1/2 MAP kinase, however, showed no change with the increasing concentration of KBH-A42.
Figure 3.
(A) mRNA levels of iNOS, TNF-α, IL-1β and IL-6 by RT-PCR (RAW 264.7 macrophage cells). HDAC inhibitor, KBH-A42 was treated 1h before LPS stimulation. Total RNA's of RAW 264.7 cells were isolated 6 h after LPS stimulation (200 ng/ml) and RT-PCR was performed on iNOS, TNF-α, IL-1β, IL-6 and β-actin mRNA's. (B) Densitometric analysis of autoradiographic signals corresponding to iNOS, THF-α, IL-1β, IL-6 mRNAs corrected on the basis of β-actin mRNA signals in the same lane.
Figure 4.
EMSA (RAW 264.7 cells, 5 × 105 cell/ml) for NF-κB and AP-1 at 1 h after LPS stimulation (200 ng/ml). HDAC inhibitor, KBH-A42 was treated 1 h before LPS stimulation.
Figure 5.
Western blotting for MAP kinases (phosphorylated ERK1/2, phosphorylated SAPK phosphorylated p38 and p38) at 10 min after LPS stimulation (200 ng/ml). HDAC inhibitor, KBH-A42 was treated 1 h before LPS stimulation.
Docking study
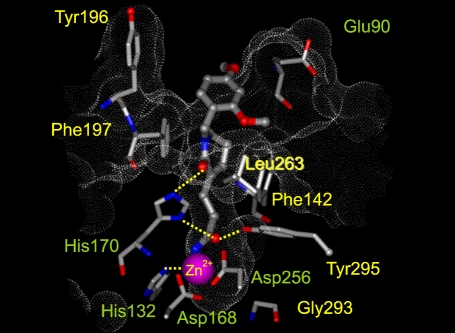
Docking study with a model of human HDAC-1 based on the crystal structure of a bacterial HDAC homologue (HDLP, PDB code 1C3R) showed that KBH-A42 docked within the active site (Figure 6). The aliphatic chain between hydroxamate and δ-lactam ring was bounded in the tubular hydrophobic pocket, and the hydroxamic acid moiety chelates to the catalytic Zn2+ ion bound in the active site. Although HDLP has a large deletion at entrance to the active site, the docking study suggests that KBH-A42 binds in the active site of HDAC1 by interacting the pharmacophoric groups with the corresponding amino acids in binding pocket.
Figure 6.
The docked orientations for KBH-A42 bound to the HDAC1 catalytic core.
Discussion
In the present study, we have demonstrated that the new HDAC inhibitor, KBH-A42 had a promising anti-inflammatory activity by showing its abilities to reduce cellular TNF-α and NO production in RAW 264.7 cells and to inhibit TNF-α production in an in vivo mouse endotoxemia model. KBH-A42 also inhibited NO production in LPS-induced peritoneal macrophages and did not affect cell viability in effective concentrations. The RT-PCR study on mRNA levels of TNF-α, IL-1β, IL-6, and iNOS proposed that the inhibition of these cytokines and NO might be resulted from the inhibition of mRNA expression. Thus, it appears that this inhibitor displayed in vitro and in vivo anti-inflammatory properties via suppression of a potent pro-inflammatory cytokine, TNF-α, and a pro-inflammatory key mediator, NO. The EMSA study examined the effect of KBH-A42 on binding to specific DNA sequences of NF-κB and AP-1. The binding of NF-κB to the DNA was not changed with increasing concentration of KBH-A42 in the presence and absence of LPS stimulation, but DNA binding of another transcription factor AP-1, interestingly, dose-dependently increased with LPS stimulation. KBH-A42 differentially regulated the phosphorylation of MAP kinases. While the phosphorylation of ERK1/2 and SAPK/JNK was not affected by KBH-A42, the phosphorylation of p38 decreased with increasing the concentration of KBH-A42.
HDAC inhibitors, SAHA and butyrate showed in vitro and in vivo anti-inflammatory activity profiles comparable to those of KBH-A42. SAHA reduced serum levels of TNF-α, IL-1β, IL-6, and IFN-γ in an in vivo LPS-induced endotoxemia animal model and also inhibited these cytokines in the LPS-stimulated PBMC cell line (Leoni et al., 2002). Butyrate also inhibited the level of IL-12 in human monocytes (Saemann et al., 2000) and NO production in RAW 264.7 macrophage cells (Chakravortty et al., 2000). Although anti-inflammatory effects by these HDAC inhibitors are quite similar, the mechanisms involved in the regulation of NF-κB by these inhibitors are different in various types of cells. In LPS-stimulated monocytes and macrophages, butyrate inhibited NO production by stabilizing IκB, a natural NF-κB inhibitor, and inhibiting iNOS expression (Chakravortty et al., 2000). TSA and butyrate inhibited the proteosomal degradation of IκB, NF-κB translocation and DNA binding in a colon cancer cell line. Meanwhile, TSA and SAHA enhanced the production of TNF-α, IL-6 and NO by LPS stimulation in primary microglial cells (Yin et al., 2001). Taken together, the anti-inflammatory effects and the mode of action of the various HDAC inhibitors vary with cell types and the classes of HDAC inhibitors. This variability may be caused by the presence of many kinds of tissue specific HDAC isoforms. Docking results of KBH-A42 with HDAC-1 homologous HDLP may suggest a possible molecular design to isoform selective HDAC inhibitors upon elucidation of the crystal structures of HDAC isoforms.
KBH-A42 is a new class HDAC inhibitor. More understanding of the role of each HDAC isoform and mechanistic aspects of new classes of HDAC inhibitors could give more precise answers on these questions. Further study with new classes of HDAC inhibitors could be very valuable to understand the role of each HDAC isoforms.
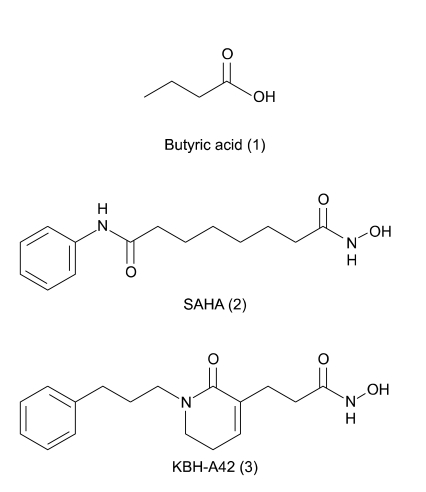
Figure 1.
Structures of HDAC inhibitors.
Acknowledgments
This work was supported by a grant from KRIBB Research Initiative Program, a grant (0405-NS01-0704-0001) of the Korean Health 21 R&D Project, Ministry of Health and Welfare, and the Brain Korea 21 project, the Republic of Korea.
Abbreviations
- HDAC
histone deacetylase
- HDLP
histone deacetylase like protein
- SAHA
suberoylanilide hydroxamic acid
- TSA
trichostatin A
References
- 1.Archer SY, Meng S, Shei A, Hodin RA. p21(WAF1) is required for butyrate-mediated growth inhibition of colon cancer cells. Proc Natl Acad Sci USA. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer SY, Hodin RA. Histone acetylation and cancer. Curr Opin Genet Dev. 1999;9:171–174. doi: 10.1016/s0959-437x(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard F, Chipoy C. Histone deacetylase inhibitors: new drug for the treatment of inflammatory diseases? Drug Discovery Today. 2005;3:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 4.Chakravortty D, Koide N, Kato Y, Sugiyama T, Mu MM, Yoshida T, Yokochi T. The inhibitory action of butyrate on lipopolysccharide induced nitric oxide production in RAW 264.7 murine macrophage cells. J Endotoxin Res. 2000;6:243–247. [PubMed] [Google Scholar]
- 5.D'Acuisto F, May MJ, Ghosh S. Inhibition of nuclear factor kappa (NF-κB): an emerging theme in anti-inflammatory therapies. Mol Interv. 2002;2:22–35. doi: 10.1124/mi.2.1.22. [DOI] [PubMed] [Google Scholar]
- 6.Diakos C, Prieschl EE, Saemann M, Novotny V, Bohmig G, Csonga R, Baumruker T, Zlabinger GJ. Novel mode of interference with nuclear factor of activated T-cells regulation in T-cell by bacterial metabolite n-butyrate. J Biol Chem. 2002;277:24243–24251. doi: 10.1074/jbc.M200191200. [DOI] [PubMed] [Google Scholar]
- 7.Gediya LK, Chopra P, Purushottamachar P, Maheshwari N, Najar VCO. A New Simple and High-Yield Synthesis of Suberoylanilide Hydroxamic Acid and Its Inhibitory Effect Alone or Combination with Retinoids on Proliferation of Human Prostate Cancer Cells. J Med Chem. 2005;48:5047–5051. doi: 10.1021/jm058214k. [DOI] [PubMed] [Google Scholar]
- 8.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 9.Johnstone RW. Histone decetylase inhibitors: Novel drugs for the treatment of cancer. Nature Rev Drug Discovery. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 10.Kim HM, Lee K, Park BW, Ryu DK, Kim K, Lee CW, Park S-K, Han JW, Lee HY, Han G. Synthesis, Enzymatic Inhibition and Cancer Cell Growth Inhibition of Novel δ-Lactam-Based Histone Deacetylase (HDAC) Inhibitors. Bioorg Med Chem Lett. 2006;16:4068–4070. doi: 10.1016/j.bmcl.2006.04.091. [DOI] [PubMed] [Google Scholar]
- 11.Kim HM, Ryu DK, Choi Y, Park BW, Lee K, Han SB, Lee CW, Kang MR, Kang JS, Boovanahalli SK, Park SK, Han JW, Chun TG, Lee HY, Nam KY, Choi EH, Han G. Structure-Activity Relationship Studies of a Series of Novel δ-Lactam based Histone Deacetylase Inhibitors. J Med Chem. 2007;50:2737–2741. doi: 10.1021/jm0613828. [DOI] [PubMed] [Google Scholar]
- 12.Kouzarides T. Histone acetylase and deacetylase in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 13.Kyung TW, Lee JE, Shin HH, Choi HS. Rutin inhibits osteoclast formation by decreasing reactive oxygen species and TNF-α by inhibiting activation of NF-κB. Exp Mol Med. 2008;40:52–58. doi: 10.3858/emm.2008.40.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Dona G, Fossati G, Sozzani S, Azam T, Bufler P, Fantuzzi G, Goncharov I, Kim S-H, Pomerantz BJ, Reznikov LL, Siegmund B, Dinarello CA, Mascagni P. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsky PE, van der Heijde DM, St Clair W, Furst DE, Breedveld FC, Kalden JR, Smolen JS, Weisman M, Emery P, Feldmann M, Harriman GR, Maini RN. Infliximab and methotrexate in the treatment of rheumatoid arthritis. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N Engl J Med. 2000;343:1594–1560. doi: 10.1056/NEJM200011303432202. [DOI] [PubMed] [Google Scholar]
- 16.McMartin C, Bohacek RS. QXP: Powerful, rapid computer algorithms for structure-based drug design. J Comput Aided Mol Des. 1997;11:333–344. doi: 10.1023/a:1007907728892. [DOI] [PubMed] [Google Scholar]
- 17.Moreland LW, Baumgartner SW, Schiff MH, Tindall EA, Fleischmann RM, Weaver AL, Ettlinger RE, Cohen S, Koopman WJ, Mohler K, Widmer MB, Blosch CM. Treatment of rheumatoid arthritis with a recombinant human tumor necrosis factor receptor (p75)-Fc fusion protein. N Engl J Med. 1997;337:141–147. doi: 10.1056/NEJM199707173370301. [DOI] [PubMed] [Google Scholar]
- 18.Newton RC, Decicco CP. Therapeutic potential and strategies for inhibiting tumor necrosis factor-α. J Med Chem. 1999;42:2295–2314. doi: 10.1021/jm980541n. [DOI] [PubMed] [Google Scholar]
- 19.Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. Anti-TNF-α therapies: the next generation. Nature Rev Drug Discovery. 2003;2:736–746. doi: 10.1038/nrd1175. [DOI] [PubMed] [Google Scholar]
- 20.Saemann MD, Bohmig GA, Osterreicher CH, Burtscher H, Parolini O, Diakos C, Stockl J, Walter H, Horl WH, Zlabinger GJ. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14:2380–2382. doi: 10.1096/fj.00-0359fje. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 22.Yi T, Baek J-H, Kim H-J, Choi M-H, Seo S-B, Ryoo H-M, Kim G-S, Woo KM. Trichostatin A-mediated upregulation of p21WAF1 contributes to osteoclast apoptosis. Exp Mol Med. 2007;39:213–221. doi: 10.1038/emm.2007.24. [DOI] [PubMed] [Google Scholar]
- 23.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-κB activation and cellular proteosome activation. J Biol Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]