Abstract
Exposure to light can induce photoreceptor cell death and exacerbate retinal degeneration. In this study, mice with genetic knockout of several genes, including rhodopsin kinase (Rhok-/-), arrestin (Sag-/-), transducin (Gnat1-/-), c-Fos (c-Fos-/-) and arrestin/transducin (Sag-/-/Gnat1-/-), were examined. We measured the expression levels of thousands of genes in order to investigate their roles in phototransduction signaling in light-induced retinal degeneration using DNA microarray technology and then further explored the gene network using pathway analysis tools. Several cascades of gene components were induced or inhibited as a result of corresponding gene knockout under specific light conditions. Transducin deletion blocked the apoptotic signaling induced by exposure to low light conditions, and it did not require c-Fos/AP-1. Deletion of c-Fos blocked the apoptotic signaling induced by exposure to high intensity light. In the present study, we identified many gene transcripts that are essential for the initiation of light-induced rod degeneration and proposed several important networks that are involved in pro- and anti-apoptotic signaling. We also demonstrated the different cascades of gene components that participate in apoptotic signaling under specific light conditions.
Keywords: arrestin, gene expression profiling, oligonucleotide array sequence analysis, proto-oncogene proteins c-fos, retinal degeneration, transducin
Introduction
Apoptosis is the final common pathway in many cases of retinal degeneration or retinal disorders, such as retinitis pigmentosa (RP), and these disorders might be ameliorated by interfering with this process (Chang et al., 1993; Portera-Cailliau et al., 1994). Retinal degeneration and its associated signaling networks have attracted much research interest for many years. Several reports have shown that light induces apoptosis in the retina and that components of AP-1 are involved in this process (Hafezi et al., 1997; Hao et al., 2002). However, the signaling networks that initiate the retinal degeneration cascade are not fully understood. Transducin is a heterotrimeric G protein expressed in the rods and cones of the vertebrate retina, and knockout studies of transducin (Gnat1-/-) showed no alteration in retinal morphology (Makino et al., 2003). However, there was an abrogation of signaling from light-activated rhodopsin (Calvert et al., 2000). Light activates rhodopsin, which is phosphorylated by rhodopsin kinase and the signal is terminated by arrestin, which binds specifically with phosphorylated rhodopsin (Dolph, 2002; Miller and Lefkowitz, 2001).
Rhodopsin signaling is prolonged in transgenic mice with null mutations in the genes encoding rhodopsin kinase (Rhok-/-) (Chen et al., 1999a), arrestin (Sag-/-) (Chen et al., 1999b), or rhodopsin kinase/arrestin (Rhok-/-/Sag-/-) (Choi et al., 2001). In addition, c-Fos is known to be a mediator of apoptosis, but its precise role in light-induced apoptosis is unclear. Therefore, we also used c-Fos-/- mice to identify the genes affected. We screened the gene expression pattern in the retina of mice with knockout of key genes involved in phototransduction. Knockout strategies would provide a great deal of information regarding the functions of genes in mammals. Understanding the mechanism of signaling networks in visual systems will be greatly facilitated by the characterization of transcriptional regulation in genetic knockouts.
There are two types of light-induced apoptotic pathways: one is transducin-dependent and the other is transducin-independent (Hao et al., 2002). The types of gene regulatory networks that are involved or predominant in these two types of apoptosis have not been clearly established. Therefore, we aimed to delineate the signaling network in order to determine which network specifically participates in light-induced apoptosis.
Materials and Methods
Animals
All procedures involving animals were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement on the use of animals in ophthalmic and vision research. Rhodopsin kinase (Rhok-/-), arrestin (Sag-/-), transducin (Gna1-/-), and c-Fos (c-Fos-/-) knockout mice were generated as described elsewhere (Chen et al., 1999a, b; Hafezi et al., 1997; Calvert et al., 2000). Sag-/- and Gna1-/- mice were crossed to each other in order to obtain arrestin/ transducin double-deficient mice (Sag-/-/ Gna1-/-). All of the mice, including the wild type (WT) mice, were reared in darkness until the given experiments were performed. Wild type mice were derived from an initial cross of 129Sv and C57BL/6. The mice used in this study ranged from 6 to 8 weeks of age.
Light illumination
The mice reared in the dark were placed in aluminum foil-wrapped polycarbonate cages that were covered with stainless steel wire tops in order to protect them from uncontrolled light exposure. Fluorescent lamps provided light from an opening at the top of the cage. The mice were supplied with food and water through the bottom of the cage. Constant illumination of 2,000 lux without dilation or 6,000 lux on dilated pupils (1% Cyclogyl, Alcon; 5% Phenylephrine, Ciba Vision) was generated by diffused cool white fluorescent lamps and applied for various time periods (24 h for 2,000 lux or 80 min for 6,000 lux). The temperature was kept at 25℃ during irradiation. After light exposure, the mice were either analyzed immediately or after a given period in darkness. Retinas were removed rapidly through a slit in the cornea and frozen in liquid nitrogen until total RNA was extracted using the Trizol method (Invitrogen Life Technologies). Retinas from three to four mice were pooled to make the corresponding sample.
Analysis of retinal morphology
Eyes obtained from anesthetized mice were enucleated and dissected in 1/2 Karnovsky buffer (2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.2). Eyecups were fixed overnight in 1/2 Karnovsky buffer and further fixed for 2 h in 1% osmium tetroxide. Eyecups were embedded in epoxy resin (EPON), and 1-µm thick sections were cut along the vertical meridian at the optic nerve as described previously (Chen et al., 2006). Photomicrographs were acquired at 40× magnification using AxioVision LE Rel. 4.1. software on a Zeiss Axioskop-2 microscope (Zeiss, Göettingen, Germany).
Quantification of apoptosis in the retina
Light-induced apoptosis in the retina was quantified using a cell death detection ELISA assay kit (Roche Molecular Biochemicals) that quantifies the soluble mono- and oligo-nucleosomes released in the cell lysate as a function of apoptosis (Leist et al., 1994; Harada et al., 2000). One retina was homogenized with a 26-½ gauge needle in 200 µl of PBS with 1 mM PMSF. The lysate was centrifuged at 15,000 × g for 10 min at room temperature. A total of 100 µl of supernatant was diluted 10 times with lysis buffer, and 20 µl of diluted supernatant was used in the ELISA measurement according to the manufacturer's instructions.
Microarray analysis
First strand cDNA was synthesized using T7-oligo dT primer and SuperScript II (Invitrogen Life Technologies) with 3 µg of total RNA from retinas. Second strand cDNA was synthesized with second strand buffer (Invitrogen Life Technologies), DNA polymerase I (New England Biolabs, Inc.), DNA ligase (NEB) and RNase H (Invitrogen Life Technologies). cDNA was extracted using phenol:chloroform: isoamyl alcohol, precipitated with ethanol, washed with 80% and 100% cold ethanol, and air dried. The dried pellet was then dissolved in 22 µl of nuclease-free water and stored at -20℃. In vitro transcription was performed using the RNA Transcript Labeling Kit (Enzo Diagnostics) to produce hybridizable biotin-labeled RNA targets. The cDNA was used as a template in the presence of a mixture of unlabeled NTPs and biotinylated CTP and UTP. After in vitro transcription, cRNA was purified using an RNeasy Mini Kit (Qiagen Inc.). The fragmented cRNA, generated by incubation at 94℃ for 35 min, was applied to the Affymetrix GeneChip U74Av2 array (total 12,488 probe sets) and hybridized at 40℃ for 16 h. After hybridization, the array was washed several times and stained with streptavidin-conjugated phycoerythrin in the GeneChip Fluidics Station 400 (Affymetrix, Inc.). The arrays were scanned by the Agilent Scanner (Agilent Technologies) and analyzed using the GeneChip Analysis Suite 5.0 (Affymetrix, Inc.).
Results
Morphological analysis
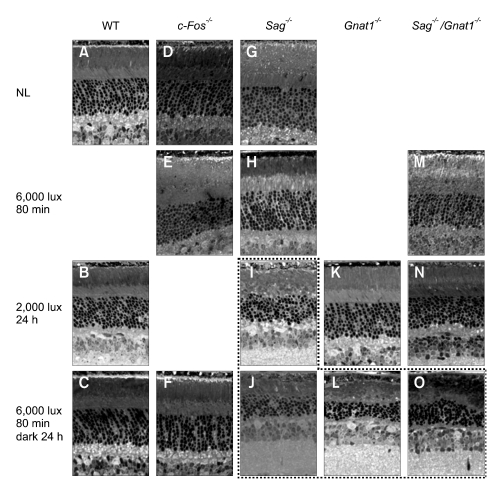
Figure 1 shows retinal sections from mice after exposure to light for various time periods. No damage was detected in the wild type mice under every illumination condition tested: 6,000 lux on dilated pupils for 80 min (data not shown), 2,000 lux without dilation for 24 h (Figure 1B), and 6,000 lux on dilated pupils for 80 min and then dark adaptation for 24 h (Figure 1C). The knockouts that did not show any alteration in morphology on exposure to light are not shown in Figure 1. Little or no damage was discernible in the Sag-/-, Gnat1-/-, Sag-/-/Gnat1-/- or c-Fos-/- mice after exposure to 6,000 lux for 80 min (Figure 1E, H and M) and 2,000 lux for 24 h (Figure 1K and N), except for a significant degeneration in Sag-/- mice exposed to 2,000 lux for 24 h (Figure 1I). Rod outer segments (ROS) and rod inner segments (RIS) were disorganized, the outer nuclear layer (ONL) was thinner, and pyknotic bodies were seen in Sag-/- mice (mainly in inferior region) exposed to 2,000 lux for 24 h. ROS and RIS were disorganized, and the ONL became thin in Sag-/-, Gnat1-/- or Sag-/-/Gnat1-/- mice exposed to 6,000 lux for 80 min and dark adaptation for 24 h (Figure 1J, L and O), indicating that the strong induction of apoptotic signaling by exposure to high intensity light (6,000 lux) started before 80 min, albeit with a delayed kinetics in the knockouts. However, under the same condition (6,000 lux for 80 min and dark adaptation for 24 h), the c-Fos-/- mice showed a completely intact morphology (Figure 1F), while all of the Sag-/-, Gnat1-/- and Sag-/-/Gnat1-/- mice showed severe cell damage (Figure 1J, L and O).
Figure 1.
Retinal sections from mice after exposure to light for various time periods. The dot box indicates severe cell damage. Some pictures taken under certain conditions and at different time points where morphological changes did not occur were not included. NL, no light; WT, wild type; Sag-/-, arrestin knockout; Gnat1-/-, transducin knockout; Sag-/-/Gnat1-/-, arrestin/transducin double knockout; c-Fos-/-, c-Fos knockout.
Apoptosis
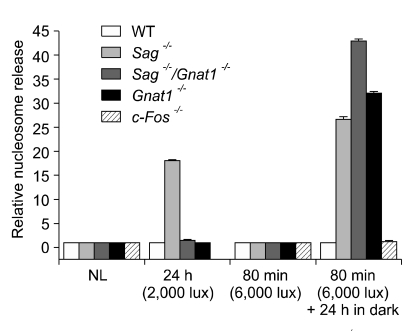
We used the nucleosome release assay to measure apoptotic cell death (Leist et al., 1994; Harada et al., 2000). Whole retinas were obtained from control or irradiated mice under each condition, and damage to DNA derived from the initiation of apoptosis was gauged by measuring the number of nucleosomes released. As shown in Figure 2, nucleosome release was clearly observed in Sag-/- after exposure to 2,000 lux for 24 h and Sag-/-, Gnat1-/- and Sag-/-/Gnat1-/- after exposure to 6,000 lux for 80 min and dark adaptation for 24 h, indicating high levels of apoptosis.
Figure 2.
Nucleosome release was clearly observed in Sag-/- mice exposed to 2,000 lux for 24 h and Sag-/-, Gnat1-/- and Sag-/-/Gnat1-/- exposed to 6,000 lux for 80 min and dark adaptation for 24 h, indicating high levels of apoptosis. NL, no light; WT, wild type.
General gene expression patterns of regulation
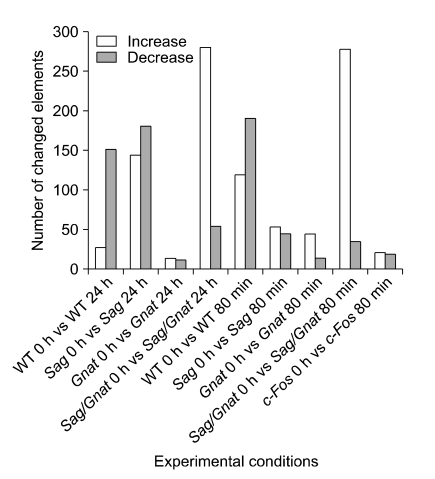
DNA microarray analysis was performed in order to identify the genes regulated by these apoptotic conditions. The numbers of genes with ≥ 2-fold changes in expression level in various knockout mutants were counted under different conditions of light intensity (Figure 3). These mice were raised in a dark room and placed in bright light (2,000 lux) without pupil dilation for 24 h or in very bright light (6,000 lux) with pupil dilatation for 80 min. While many up- or down-regulated genes were detected in wild type, Sag-/- or Sag-/-/Gnat1-/- mice, relatively fewer differentially expressed genes were observed in Gnat1 or c-Fos KO mice as a result of light exposure (2,000 lux), suggesting that both Gnat1 and c-Fos are required in order to mediate light signaling or light-induced apoptotic molecular changes. In addition, it was also clear that a cascade of gene transcripts were differentially expressed as a result of the corresponding gene knockouts, such as in Sag-/-.
Figure 3.
Number of up-regulated or down-regulated genes in Sag-/-, Gnat1-/-, Sag-/-/Gnat1-/- or c-Fos-/- conditions.
Expression levels of target genes in wild type and knockout mice
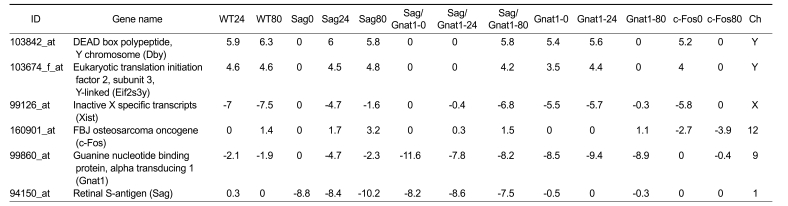
We confirmed the expression of target genes in order to determine whether they were effectively knocked out in the corresponding knockout (Table 1). The Sag gene was effectively knocked out by -log2 8.8 at 0 h, -log2 8.4 at 24 h and -log2 10 in the 80-min sample of Sag-/- retinas. Significant downregulation of Gnat1 was indicated by -log2 9 at 0 h, -log2 9.4 at 24 h, -log2 9 in the 80 min sample.
Table 1.
Confirmation of target gene knockout in the mouse retina. Values are given as log2 ratio in comparison with non-treated wild type mice. WT24, exposure to 2,000 lux light for 24 h in wild type; WT80, exposure to 6,000 lux light for 80 min in wild type; Sag0, non-treated in Sag-/- knockout; Sag24, exposure to 2,000 lux light for 24 h in Sag-/- knockout; Sag80, exposure to 6,000 lux light for 80 min in Sag-/- knockout; Gnat1-0, non-treated in Gnat1-/- knockout; Gnat1-24, exposure to 2,000 lux light for 24 h in Gnat1-/- knockout; Gnat1-80, exposure to 6,000 lux light for 80 min in Gnat-/- knockout; c-Fos0, non-treated in c-Fos-/- knockout; c-Fos80, exposure to 6,000 lux light for 80 min in c-Fos-/- knockout. Ch, chromosome.
Crystallin genes were induced only by bright light
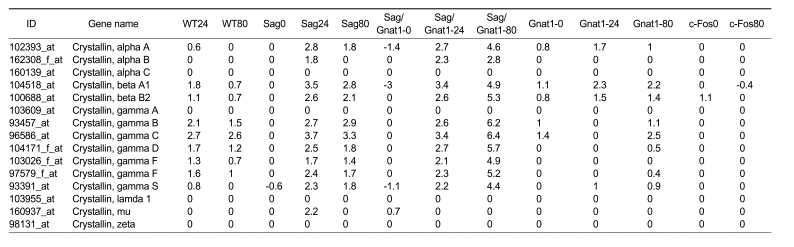
Intense light exposure was reported to increase crystallin content in the rat retina (Sakaguchi et al., 2003). The Coomassie blue staining intensity of crystallin 2D gel components was 2 to 3 times greater in the light-exposed retinas than in the control retinas. Neither wild type nor Rhok-/-/Sag-/- mice exposed to low light (450 lux) showed any significant induction of crystalline gene species (alpha B & C, beta B2, gamma A, B, C, D, & F, lamda 1, and mu) (Krishnan et al., 2008). However, when we surveyed the expression levels at 24 h after continuous illumination of 2,000 lux without dilation or 80 min after illumination of 6,000 lux with dilation, the expression levels of crystallin alpha A, alpha B, beta A1, beta B2, gamma B, gamma C, gamma D, gamma F, and gamma S were remarkably induced in both wild type and knockout mice, including the Sag-/-, Sag-/-/Gnat1-/-, and Gnat1-/- mice (Table 2). This finding supports the fact that only bright light can induce crystallin genes.Recent studies have shown that an increase in the chaperone ability of alpha-crystallin at higher temperatures can protect target proteins from aggregating and precipitating (Ecroyd et al., 2007; Rekas et al., 2007). There are two main crystallin gene families: the alpha-crystallins and beta/gamma-crystallins. The beta/gamma-crystallin family has been suggested to affect lens development (Andley, 2007). The increased levels of beta/gamma-crystallins in bright light conditions may indicate a new function of beta/gamma-crystallins that is related to stress.
Table 2.
Regulation of crystallin genes by light exposure. Values are given as log2 ratio in comparison with non-treated wild type mice. Abbreviations are the same as in Table 1.
Photoreceptor genes were affected by different mechanisms
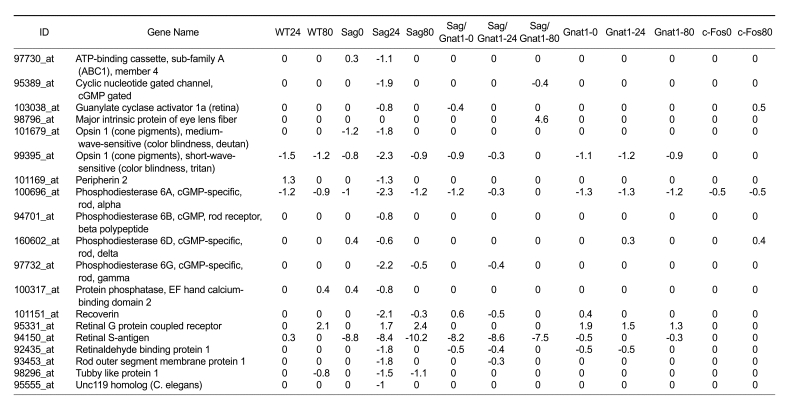
As shown in Table 3, the expression of photoreceptor genes was significantly down-regulated in Sag-/- mice (2,000 lux without pupil dilation for 24 h), indicating that the progression of apoptosis (Figure 2) induced remarkable structural changes (Figure 1I). The surprising observation was that this down-regulation of photoreceptor signaling genes, presumably due to photoreceptor cell damage, did not occur in Sag-/-/Gnat1-/- mice under the same condition (2,000 lux without dilation for 24 h) (Figure 1N and 2). Bright light (5,000 lux for 20 min or 1,700 lux for 7 days) has been reported to trigger apoptosis of photoreceptor cells through the induction of AP-1. On the other hand, photoreceptor cell apoptosis in Rhok-/- or Sag-/- mice exposed to low intensity light is dependent on transducin and does not require AP-1 (Hao et al., 2002). When exposed to 2,000 lux for 24 h, the molecular responses of Sag-/-/Gnat1-/- mice to photoreceptor genes were quite different from those of Sag-/- mice but similar to those of wild type or Gnat1-/- mice (Figure 1 and 2), indicating that an additional knockout of the Gnat1 gene causes a delayed response and/or ramification of signaling pathways in Sag-/-/Gnat1-/- mice. Therefore, it is more likely that apoptotic signaling requires the involvement of transducin under this condition.
Table 3.
Expression of the photoreceptor genes in wild type and knockout mice. Values are given as log2 ratio in comparison with non-treated wild type mice. Abbreviations are the same as those in Table 1.
Signal transduction networks
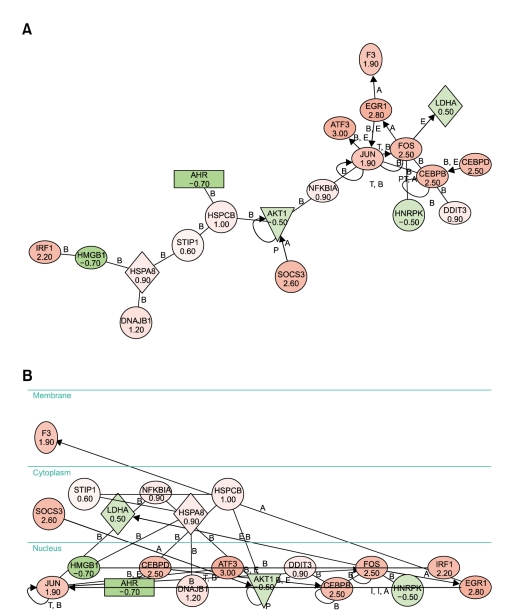
In light-induced retinal degeneration, as well as in inherited retinal degeneration, apoptosis is the "common pathway" of photoreceptor cell death. Therefore, we studied the signaling of stress-induced photoreceptor apoptosis mechanisms by utilizing the pathway analysis program (Ingenuity). We used a subset of regulated genes (> 0.5 or < -0.5 in log2 ratio) under specific conditions in order to obtain insights into the mechanisms of cell death and signaling. In 1 h illumination of 2,000 lux in Rhok-/- mice, we observed 2 major networks (scored equal to or above 7). The 1st network constitutes EGR1, Fos, and NR4A1, which seem to be the major proteins of apoptosis. It also includes BDNF, CEBP delta, CTSB (Cathepsin B), and DUSP1 (dual specificity phosphatase 1), which function in the developmental process. As the amount of dark adaptation time increases, the number of major networks increases. Six major networks (scored equal to or above 7) were observed after 3 h of dark adaptation followed by an hour of light illumination. After 3 h of dark adaptation the first network, which consisted of AHR, AKT1, ATF3, CEBPbeta, CEBPdelta, DDIT3, DNAJB1, EGR1, F3, Fos, HMGB1, HNRPK, HSPA8, HSPCB, IRF1, Jun, LDHA, NFκBIA, SOCS3, and STIP1, obtained the highest score of 35 (Figure 4). In addition, all 20 members of this particular network were being regulated in that condition. Therefore, it is most likely that this network plays a very critical role in the mutant, and its role may be in apoptosis.
Figure 4.
Graphic view of biological network 1, which was generated using the Ingenuity Pathway Analysis program. Genes with a log2 ratio of 0.5 or greater and -0.5 or lower were used to generate biological networks in Rhok-/- mice exposed to 1 h of 2,000 lux and 3 h of dark adaptation. Pink and red denote the up-regulated genes while dark and light green correspond to the downregulated genes. (A) Spatial layout. (B) Subcellular layout.
Lessons from knockout experiments
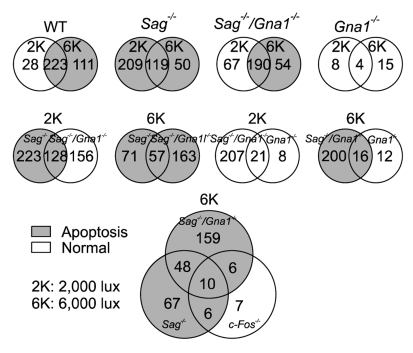
The Venn diagrams (Figure 5) show the numbers of regulated genes (more than 2 fold) between conditions. Several conclusions can be derived from these Venn diagrams. First, there were 362 gene elements in wild type retinas regulated by light exposure itself. Second, a different set of genes was regulated by exposure to 2,000 lux or 6,000 lux of light in all conditions tested. Third, the transducin knockout seems to block phototransduction signals triggered by exposure to both 2,000 lux and 6,000 lux of light. Fourth, transducin appears to play an important role in apoptotic signaling in arrestin knockout (Sag-/-) mice exposed to 2,000 lux of light but not in those exposed to 6,000 lux of light. Fifth, the c-Fos knockout appears to block photo signal transduction under 6,000 lux of light.
Figure 5.
Venn diagrams of regulated (≥ 2 fold) genes in Sag-/-, Gnat1-/-, Sag-/-/Gnat1-/- or c-Fos-/- mice. Shaded regions represent light-induced apoptotic conditions while white regions correspond to normal conditions.
Discussion
Retinal degeneration has long been a subject of intense investigation due to its important clinical applications. In the present study, we examined the effects of constant light-induced phototoxicity and signaling networks. We performed gene expression analyses to facilitate the identification of regulated genes in the retinas of different knockout mice under different conditions. When the number of regulated genes was measured in each knockout or under each light condition, significant molecular changes, which eventually induced retinal cell degeneration, were observed in Sag-/-, Gnat1-/- or Sag-/-/Gnat1-/- knockouts (Figure 1I, J, L, O and 2). These results suggest that such knockouts may be at greater risk of light-induced damage.
Intense light exposure was reported to change the crystallin content in the retina (Andley, 2007; Ecroyd et al., 2007; Rekas et al., 2007). Alpha-crystallin has antioxidant as well as antiapoptotic functions, and it can protect enzymes and other crystallins against both chemically- and thermally-induced inactivation or aggregation, which may play an important role in maintaining the transparency of the lens (Manzanares et al., 2001; Xi et al., 2003; Liu et al., 2004; Mao et al., 2004). When we surveyed the expression levels 24 h after continuous illumination of 2,000 lux without dilation or 80 min after illumination of 6,000 lux with dilation, the expression levels of crystalline alpha A, alpha B, beta A1, beta B2, gamma B, gamma C, gamma D, gamma F and gamma S were remarkably increased. As a chaperone and heat shock protein, alpha-crystallins can serve as a regulator of protein conformation and as stress sensors. The ability of crystallins to interact with free radicals and remove hypochlorous acid could potentially contribute to the maintenance of the lens functionally. Many chaperones play a role in regulating cell proliferation and apoptosis (Mosser and Morimoto, 2004).
When we further explored the fate of the photoreceptor genes, we found that their expression levels were significantly down-regulated in Sag-/- mice, representing remarkable structural changes due to the progression of apoptosis (Figure 1I). A surprising observation was that this down-regulation of photoreceptor signaling genes, presumably due to photoreceptor cell damage, did not occur in Sag-/-/Gnat1-/- mice under the same condition (Figure 1N). The deletion of transducin seems to block the light-related apoptotic signal. This result supports that photoreceptor cell apoptosis in Rhok-/- or Sag-/- mice exposed to low light is dependent on transducin and does not require c-Fos (Hao et al., 2002).
Relatively fewer genes were differentially expressed in Gnat1-/- and c-Fos-/- mice exposed to light (Figure 3). c-Fos is a component of the dimeric transcription factor AP-1, and it must combine with Jun proteins to form a functional unit (Angel and Karin, 1991). Ablation of c-Fos protects photoreceptors from light-induced damage (Hafezi et al., 1997). In most of the conditions tested, including the wild type, c-Fos was significantly up-regulated by exposure to bright light (except in c-Fos knockouts) (Table 1), confirming the important role of AP-1 in light-induced stress.
In the present study, we found out that different cascades of gene components were induced or inhibited as a result of corresponding gene knockouts under specific light conditions. For example, the expression of the crystalline gene was increased in the retina by exposure to bright light. The deletion of transducin seems to block light-related apoptotic signaling in Sag-/- mice. c-Fos has been significantly up-regulated as a light stress signal, while knockout of the c-Fos gene appears to block bright light-induced retinal degeneration. These results, therefore, provide an impetus to construct retinal photoreceptor signaling pathways.
Acknowledgement
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2006-311-C00482), and it was also partly supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MOST) (R01-2007-000-20533-0).
Abbreviations
- GPCR
G protein coupled receptor
- Rhok
Rhodopsin kinase
- Sag
S antigen (Arrestin)
References
- 1.Andley UP. Crystallins in the eye: Function and pathology. Prog Retin Eye Res. 2007;26:78–98. doi: 10.1016/j.preteyeres.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Calvert PD, Krasnoperova NV, Lyubarsky AL, Isayama T, Nicolo M, Kosaras B, Wong G, Gannon KS, Margolskee RF, Sidman RL, Pugh EN, Jr, Makino CL, Lem J. Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha -subunit. Proc Natl Acad Sci USA. 2000;97:13913–13918. doi: 10.1073/pnas.250478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang GQ, Hao Y, Wong F. Apoptosis: final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- 5.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci USA. 1999a;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness) Invest Ophthalmol Vis Sci. 1999b;40:2978–2982. [PubMed] [Google Scholar]
- 7.Chen J, Shi G, Concepcion FA, Xie G, Oprian D, Chen J. Stable rhodopsin/arrestin complex leads to retinal degeneration in a transgenic mouse model of autosomal dominant retinitis pigmentosa. J Neurosci. 2006;26:11929–11937. doi: 10.1523/JNEUROSCI.3212-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi S, Hao W, Chen CK, Simon MI. Gene expression profiles of light-induced apoptosis in arrestin/rhodopsin kinase-deficient mouse retinas. Proc Natl Acad Sci USA. 2001;98:13096–13101. doi: 10.1073/pnas.201417498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolph PJ. Arrestin: roles in the life and death of retinal neurons. Neuroscientist. 2002;8:347–355. doi: 10.1177/107385840200800410. [DOI] [PubMed] [Google Scholar]
- 10.Ecroyd H, Meehan S, Horwitz J, Aquilina JA, Benesch JL, Robinson CV, Macphee CE, Carver JA. Mimicking phosphorylation of alphaB-crystallin affects its chaperone activity. Biochem J. 2007;401:129–141. doi: 10.1042/BJ20060981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafezi F, Steinbach JP, Marti A, Munz K, Wang ZQ, Wagner EF, Aguzzi A, Reme CE. The absence of c-fos prevents light-induced apoptotic cell death of photoreceptors in retinal degeneration in vivo. Nat Med. 1997;3:346–349. doi: 10.1038/nm0397-346. [DOI] [PubMed] [Google Scholar]
- 12.Hao W, Wenzel A, Obin MS, Chen CK, Brill E, Krasnoperova NV, Eversole-Cire P, Kleyner Y, Taylor A, Simon MI, Grimm C, Reme CE, Lem J. Evidence for two apoptotic pathways in light-induced retinal degeneration. Nat Genet. 2002;32:254–260. doi: 10.1038/ng984. [DOI] [PubMed] [Google Scholar]
- 13.Harada T, Harada C, Nakayama N, Okuyama S, Yoshida K, Kohsaka S, Matsuda H, Wada K. Modification of glial-neuronal cell interactions prevents photoreceptor apoptosis during light-induced retinal degeneration. Neuron. 2000;26:533–541. doi: 10.1016/s0896-6273(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan J, Lee G, Han SU, Choi S. Characterization of phototransduction gene knockouts revealed important signaling networks in the light-induced retinal degeneration. J Biomed Biotechnol. 2008;2008:327468. doi: 10.1155/2008/327468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leist M, Gantner F, Bohlinger I, Germann PG, Tiegs G, Wendel A. Murine hepatocyte apoptosis induced in vitro and in vivo by TNF-alpha requires transcriptional arrest. J Immunol. 1994;153:1778–1788. [PubMed] [Google Scholar]
- 16.Liu JP, Schlosser R, Ma WY, Dong Z, Feng H, Lui L, Huang XQ, Liu Y, Li DW. Human alphaA- and alphaB-crystallins prevent UVA-induced apoptosis through regulation of PKCalpha, RAF/MEK/ERK and AKT signaling pathways. Exp Eye Res. 2004;79:393–403. [PubMed] [Google Scholar]
- 17.Makino CL, Wen XH, Lem J. Piecing together the timetable for visual transduction with transgenic animals. Curr Opin Neurobiol. 2003;13:404–412. doi: 10.1016/s0959-4388(03)00091-6. [DOI] [PubMed] [Google Scholar]
- 18.Manzanares D, Bauby C, de la Pena R, Garcia JC, Sanchez R, Martinez S, Romay CH, Lopez-Reconde JL, Pino E, Lissi EA. Antioxidant properties of alpha-crystallin. J Protein Chem. 2001;20:181–189. doi: 10.1023/a:1010996528884. [DOI] [PubMed] [Google Scholar]
- 19.Mao YW, Liu JP, Xiang H, Li DW. Human alphaA- and alphaB-crystallins bind to Bax and Bcl-X(S) to sequester their translocation during staurosporine-induced apoptosis. Cell Death Differ. 2004;11:512–526. doi: 10.1038/sj.cdd.4401384. [DOI] [PubMed] [Google Scholar]
- 20.Miller WE, Lefkowitz RJ. Arrestins as signaling molecules involved in apoptotic pathways: a real eye opener. Sci STKE. 2001;2001:PE1. doi: 10.1126/stke.2001.69.pe1. [DOI] [PubMed] [Google Scholar]
- 21.Mosser DD, Morimoto RI. Molecular chaperones and the stress of oncogenesis. Oncogene. 2004;23:2907–2918. doi: 10.1038/sj.onc.1207529. [DOI] [PubMed] [Google Scholar]
- 22.Portera-Cailliau C, Sung CH, Nathans J, Adler R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc Natl Acad Sci USA. 1994;91:974–978. doi: 10.1073/pnas.91.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rekas A, Jankova L, Thorn DC, Cappai R, Carver JA. Monitoring the prevention of amyloid fibril formation by alpha-crystallin. Temperature dependence and the nature of the aggregating species. FEBS J. 2007;274:6290–6294. doi: 10.1111/j.1742-4658.2007.06144.x. [DOI] [PubMed] [Google Scholar]
- 24.Sakaguchi H, Miyagi M, Darrow RM, Crabb JS, Hollyfield JG, Organisciak DT, Crabb JW. Intense light exposure changes the crystallin content in retina. Exp Eye Res. 2003;76:131–133. doi: 10.1016/s0014-4835(02)00249-x. [DOI] [PubMed] [Google Scholar]
- 25.Xi J, Farjo R, Yoshida S, Kern TS, Swaroop A, Andley UP. A comprehensive analysis of the expression of crystallins in mouse retina. Mol Vis. 2003;9:410–419. [PubMed] [Google Scholar]