Abstract
Hypoxia-inducible factor 1α (HIF-1α) is rapidly degraded by the ubiquitin-proteasome pathway under normoxic conditions. Ubiquitination of HIF-1α is mediated by interaction with von Hippel-Lindau tumor suppressor protein (pVHL). In our previous report, we found that hypoxia-induced active signal transducer and activator of transcription3 (STAT3) accelerated the accumulation of HIF-1α protein and prolonged its half-life in solid tumor cells. However, its specific mechanisms are not fully understood. Thus, we examined the role of STAT3 in the mechanism of pVHL-mediated HIF-1α stability. We found that STAT3 interacts with C-terminal domain of HIF-1α and stabilizes HIF-1α by inhibition of pVHL binding to HIF-1α. The binding between HIF-1α and pVHL, negative regulator of HIF-1α stability, was interfered dose-dependently by overexpressed constitutive active STAT3. Moreover, we found that the enhanced HIF-1α protein levels by active STAT3 are due to decrease of poly-ubiquitination of HIF-1α protein via inhibition of interaction between pVHL and HIF-1α. Taken together, our results suggest that STAT3 decreases the pVHL-mediated ubiquitination of HIF-1α through competition with pVHL for binding to HIF-1α, and then stabilizes HIF-1α protein levels.
Keywords: anoxia; hypoxia-inducible factor1, α subunit; neoplasms; STAT3 transcription factor; ubiquitination; von Hippel-Lindau tumor suppressor protein
Introduction
Hypoxia is a critical condition to initiate many adaptive responses in pathological and physiological processes (Bunn and Poyton, 1996). In many solid tumors, hypoxia is a common feature, and closely related with malignant progression of cancer cells (Hockel and Vaupel, 2001; Joung et al, 2005). Under hypoxic conditions, the diverse genes involved in angiogenesis, erythropoiesis and energy metabolisms are transcriptionally up-regulated by the heterodimeric hypoxia inducible factor-1 (HIF-1) (Semenza, 2001; Wenger, 2002). The α subunit of HIF-1 is the hypoxia-responsive component of the dimer, while HIF-1β is expressed constitutively (Kallio et al., 1999).
Under normoxic conditions, HIF-1α is rapidly degraded by the ubiquitin-proteasome pathway (Ohh et al., 2000). Ubiquitination of HIF-1α is mediated by interaction with von Hippel-Lindau tumor suppressor protein (pVHL) (Kim et al, 2008). HIF-1α is targeted for VHL E3 ligase complex by proline hydroxylation of oxygen-dependent degradation (ODD) domain (Semenza, 2001; Safran and Kaelin, 2003). Hydroxylated HIF-1α is associated with pVHL which has E3 ubiquitin ligase activity and is rapidly degraded by ubiquitin-proteasome pathway (Ivan et al., 2001). Prolyl hydroxylase (PHD) utilize oxygen as a substrate and iron, as a cofactor to hydroxylate the proline residues on HIF-1α (Bruick and McKnight, 2001; Berra et al., 2006). However, under hypoxic conditions, HIF-1α is stabilized due to inhibition of HIF-1α modifying enzymes by low oxygen levels.
Signal transducer and activators of transcription (STAT) proteins play roles in various signaling pathways, including cytokine, growth factor, and hormone signaling (Horvath, 2000). STATs are activated by tyrosine phosphorylation, and phosphorylated STAT forms hetero- or homo-dimer, then translocate to the nucleus (Darnell, 1997). Several constitutively activated STATs have been observed in large wide number of human cancer cell lines and primary tumors (Bowman, 2000; Huang, 2000; Song, 2000). STAT3 proteins play a role in tumorigenesis through up-regulation of genes encoding apoptosis inhibitors, cell-cycle regulators, and inducers of angiogenesis (Sinibaldi et al., 2000; Niu et al., 2002). Aberrant activation of STAT3 is also commonly observed in solid tumors and strongly associated with tumor development and progression (Huang et al., 2000; Song and Grandis, 2000; Schaefer et al., 2002). In our previous report, we found that hypoxia-induced active STAT3 accelerated the accumulation of HIF-1α protein and prolonged its half-life in solid tumor cells (Jung et al., 2005). However, its specific mechanisms are not fully understood. Thus, in this study, we investigated the mechanisms whereby active STAT3 stabilizes and increases HIF-1α protein levels by focusing on pVHL-mediated HIF-1α degradation.
Materials and Methods
Cell culture
COS7 (monkey kidney cell lines) cells and Caki-I (human renal carcinoma) cells were purchased from ATCC and maintained in DMEM supplemented with 10% FBS (Life Technologies, Gaithersburg, MD) and 1% antibiotics. For induction of HIF-1α protein, cells were treated with 20 µM with MG132 (Calbiochem, CA) after transfection with HA-GFP-HIF-1α.
Plasmid constructs and transient transfection
Wild type of HA-GFP-HIF-1α, series of domain deletion of HIF-1α constructs, and HA-ubiquitin were generously provided by Dr. Jong Wan Park (Seoul National University, Korea). The pCR3-VHL-HA was generously provided by Dr. Hong-Duk Youn (Seoul National University, Korea). The constitutive active form of STAT3 (STAT3 CA) was generously provided by Dr. Hyun-Gyu Lee (Seoul National University, Korea). Cells were transfected with LipofectAMINE reagent (Invitrogen, San Diego, CA) according to the manufacturer's directions.
Coimmunoprecipitation, immunobloting
After 24 h of transfection into COS7 cells, 20 µM of MG132 were treated for 3 h and lysed on ice with lysis buffer [20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% NP-40 and 1 mM PMSF]. Whole cell lysates were immunoprecipitated using suitable antibodies and protein G-Sepharose (Pierce) at 4℃ for 2 h. Binding and washing were performed in the same buffer, except the NaCl concentration was diluted four fold. Immunoprecipitates were separated by SDS-PAGE, electrotransferred to nitrocellulose membrane, and immunoblotted with appropriate antibodies. Anti-HIF-1α was provided by Dr. Jong Wan Park (Seoul National University); anti-phospho STAT3, anti-β-actin, anti-ubiquitin, anti-GFP were purchased from Santa Cruz Biotechnology; anti-pVHL, anti-HA were purchased from BD Transduction Laboratory.
Proteasome inhibition and in vivo ubiquitination assay
COS7 cells or Caki-I cells were transiently transfected with mammalian expression vectors for HA-GFP-HIF-1α, HA-pVHL, and HA-Ub. After 24 h of incubation, cells were treated with 20 µM of MG132 for a further 5 h. Cells were lysed with lysis buffer and immunoprecipitated with anti-HIF-1α antibody. Immunoprecipitated cell lysates were subjected to 6% SDS-PAGE, and ubiquitinylated HIF-1α was detected with anti-ubiquitin antibody.
Statistical analysis
Results are presented as means ± S.E.M. Data are analyzed by using Student's t test. Differences were considered statistically significant at a P value of < 0.05.
Result
STAT3 interacts with HIF-1α and increases HIF-1α protein levels
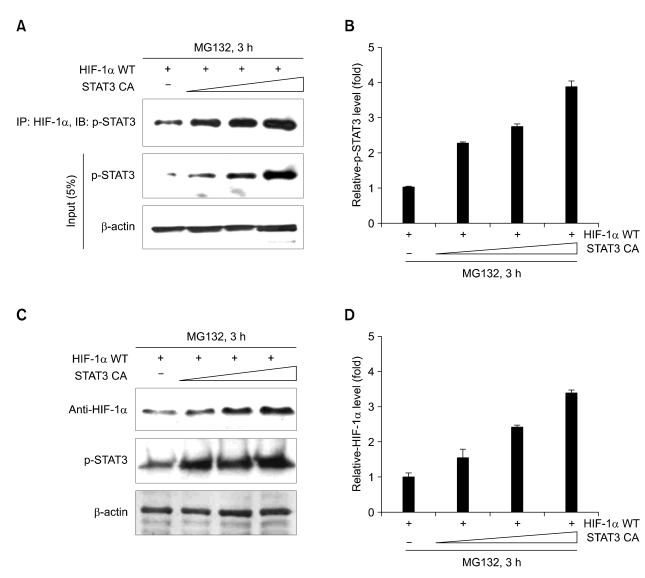
To confirm the involvement of hypoxia-induced active STAT3 in HIF-1α stabilization, we first examined the physical interaction between constitutive active STAT3 and HA-GFP-HIF-1α. The interaction between HIF-1α and STAT3 in vivo was confirmed by coimmunoprecipitation assays. When the overexpressed HA-GFP-HIF-1α in COS7 cells or Caki-I cells (data not shown) was stabilized by MG132 under normoxic conditions, the overexpressed constitutive active STAT3 was immunoprecipitated (Figure 1A). Also, to examine whether active STAT3 increases the HIF-1α protein stability, COS7 cells were transiently transfected with constitutive active STAT3 in a dose dependent manner. As shown in Figure 1C, along with increasing amount of constitutive active STAT3, the protein levels of HA-GFP-HIF-1α increased gradually. This result indicates that STAT3 is involved in HIF-1α stabilization.
Figure 1.
STAT3 interacts with HIF-1α and increases HIF-1α protein levels. (A) COS7 cells were transiently transfected with HA-GFP-HIF-1α and various amounts of constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with 20 µM of MG132 for 3 h. Cell lysates were immunoprecipitated with anti-HIF-1α antibody, and immunoblotted with anti-p-STAT3 antibody. 5% of input cell lysates were SDS-PAGED and immunoblotted with anti-p-STAT3 antibody and anti-β-actin antibody. (B) Relative band intensities of Figure 1A, first panel. mean ± S.E.M. of at least three independent experiments. (C) COS7 cells were transiently transfected with HA-GFP-HIF-1α and various amounts of constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with 20 µM of MG132 for 3 h. Cell lysates were immunoblotted with anti-HIF-1α, anti-phospho-STAT3 and anti-β-actin antibody. (D) Relative band intensities of Figure 1C. mean ± S.E.M. of at least three independent experiments.
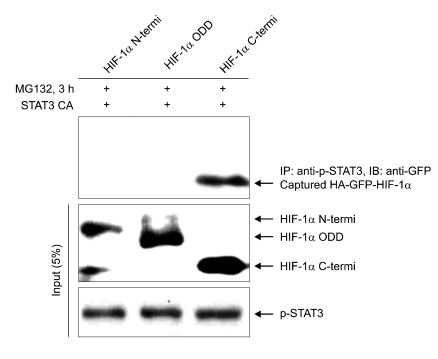
Next, to investigate which domain of HIF-1α interacts with STAT3, we performed the coimmunoprecipitation assay using each series of deletion constructs of HIF-1α. When the overexpressed each series of deletion constructs of HIF-1α in COS7 cells was stabilized by MG132 under normoxic conditions, the overexpressed C-terminal domain of HIF-1α was immunoprecipitated by the overexpressed constitutive active STAT3 (Figure 2). It was not found that the overexpressed other deletion domains of HIF-1α, N-termini and ODD were not precipitated by active STAT3.
Figure 2.
STAT3 interacts with C-termini of HIF-1α in vivo. COS7 cells were transiently transfected with each series of deletion domain of HA-GFP-HIF-1α and constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with 20 µM of MG132 for 3 h. Cell lysates were immunoprecipitated with anti-p-STAT3 antibody, and immunoblotted with anti-GFP antibody. 5% of input cell lysates were analyzed by SDS-PAGE and immunoblotting with anti-GFP antibody and anti-p-STAT3 antibody.
STAT3 competes with pVHL for binding to HIF-1α
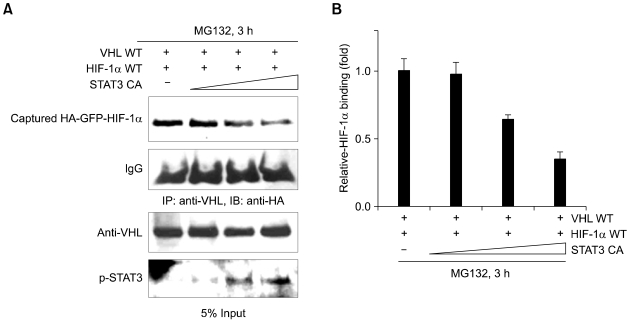
It is well known that pVHL plays an important role in hypoxia-induced HIF-1α stability via direct interaction. HIF-1α protein levels are mainly regulated by oxygen-dependent pVHL-mediated ubiquitination-proteasome degradation pathway (Ivan et al, 2001; Safran and Kaelin, 2003). Therefore, we investigated whether STAT3 increase HIF-1α protein levels by inhibiting pVHL-mediated degradation. As shown in Figure 3A, constitutive active STAT3 was co-expressed in a dose dependent manner with the wild type of pVHL and HIF-1α in COS7 cells. Along with increasing amount of overexpressed constitutive active STAT3, the bindings of overexpressed pVHL to overexpressed HIF-1α were decreased gradually (Figure 3A).
Figure 3.
STAT3 competes with pVHL for binding to HIF-1α. (A) COS7 cells were transiently transfected with HA-VHL, HA-GFP-HIF-1α and various amounts of constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with with 20 µM MG132 for 3 h. Cell lysates were immunoprecipitated with anti-VHL antibody, and immunoblotted with anti-HA antibody. 5% of input cell lysates were analyzed by SDS-PAGE and immuno-blotting with anti-pVHL and anti-phospho-STAT3 antibody. (B) Relative band intensities of Figure 3A, first panel, means ± S.E.M. of at least three independent experiments.
STAT3 decrease poly-ubiquitination of HIF-1α via inhibition of VHL binding
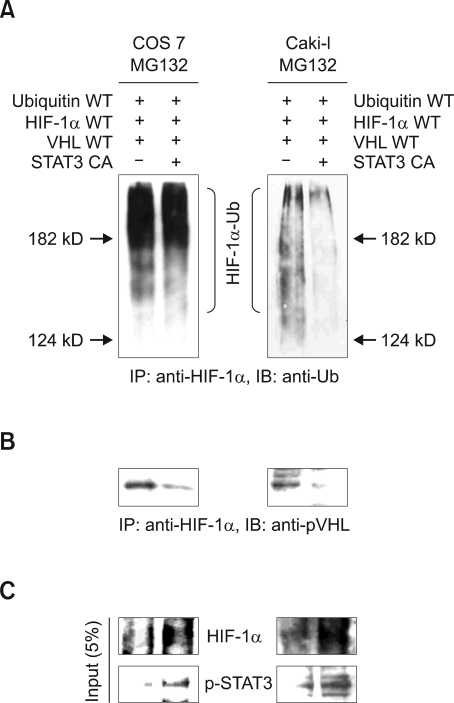
The overexpression of constitutive active STAT3 abolished the interaction between HIF-1α and pVHL in a dose dependent manner (Figure 3A). Thus, we next examined whether STAT3 is involved in the pVHL-mediated ubiquitination of HIF-1α. HA-ubiquitin, HA-pVHL and HA-GFP-HIF-1α were co-expressed with or without overexpression of constitutive active STAT3 in COS7 cells (Figure 4A, left panel) or Caki-I cells (Figure 4A, right panel). After 24 h of transient transfection, cells were treated with 20 µM of MG132 for 5 h. Cells lysates were immunoprecipitated with anti-HIF-1α antibody and immunoblotted with anti-ubiquitin antibody (Figure 4A). The poly-ubiquitination of HA-GFP-HIF-1α was significantly reduced in cells overexpressed with constitutive active STAT3 (Figure 4A, lane 2) and the overexpressed HA-GFP-HIF-1α protein levels were more enhanced in cells with overexpressed constitutive active STAT3 (Figure 4C). This result indicates that STAT3 blocks pVHL-mediated ubiquitination of HA-GFP-HIF-1α, because the binding of HA-pVHL to the HA-GFP-HIF-1α was significantly reduced in cells overexpressed with constitutive active STAT3 (Figure 4B). Taken our results, we suggest that STAT3 enhances HIF-1α stability through inhibition of the pVHL-mediated ubiquitination of HIF-1α.
Figure 4.
STAT3 decreases poly-ubiquitination of HIF-1α via inhibition of pVHL binding. (A) COS7 cells (left panel) or Caki-I cells (right panel) were transiently transfected with HA-ubiqutin, HA-pVHL, HA-GFP-HIF-1α and various amounts of constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with with 20 µM MG132 for 5 h. Cell lysates were immunoprecipitated with anti-HIF-1α antibody, and immunoblotted with anti-ubiquitin antibody. (B) COS7 cells (left panel) or Caki-I cells (right panel) cells were transiently transfected with HA-ubiqutin, HA-pVHL, HA-GFP-HIF-1α and various amounts of constitutive active STAT3 constructs. After incubation for 24 h, cells were treated with 20 µM of MG132 for 5 h. Cell lysates were immunoprecipitated with anti-HIF-1α antibody, and immunoblotted with anti-pVHL antibody. (C) 5% of input cell lysates were analyzed by immuno-blotting with anti-HIF-1α antibody and anti-p-STAT3 antibody (COS 7 cells, left panel; Caki-I cells, right panel).
Discussion
STAT3 is considered to be an oncogene since it cause the activation of cyclin D1, c-myc, and bcl-xL expression, and also is involved in promotion of cell-cycle progression, cellular transformation, and the prevention of apoptosis (Darnell, 1997; Bowman et al., 2000). In our previous report, we have demonstrated that STAT3 is a pivotal regulator of HIF-1-mediated VEGF expression in hypoxic solid tumor cells (Jung et al., 2005). Thus, STAT3 is considered to be an oncogenic inducer in solid tumor as well as HIF-1. Also, we found that hypoxia-induced active STAT3 accelerated the accumulation of HIF-1α protein and prolonged its half-life in solid tumor cells (Jung et al., 2005). However, its specific mechanisms are not fully understood.
In this study, we investigated the role of STAT3 in the mechanism of pVHL-mediated HIF-1α stability. We found that STAT3 interacts with the C-terminal domain of HIF-1α and stabilizes HIF-1α by competition with pVHL for binding to HIF-1α. The interaction between HIF-1α and negative regulator of HIF-1α stability, pVHL was interfered by dose-dependently overexpressed constitutive active STAT3. Moreover, we found that the enhanced HIF-1α protein levels by active STAT3 are due to decrease of poly-ubiquitination of HIF-1α protein via inhibition of interaction between pVHL and HIF-1α. Taken together, our results suggest that STAT3 decreases the pVHL-mediated ubiquitination of HIF-1α through competition with pVHL for binding to HIF-1α, and then stabilizes HIF-1α protein levels.
Under normoxic conditions, HIF-1α is rapidly degraded by the ubiquitin-proteasome pathway (Ohh et al., 2000). Ubiquitination of HIF-1α is mediated by interaction with pVHL. HIF-1α is targeted for VHL E3 ligase complex by proline hydroxylation of oxygen-dependent degradation (ODD) domain (Semenza, 2001; Safran and Kaelin, 2003). Hydroxylated HIF-1α is associated with pVHL which has E3 ubiquitin ligase activity and is rapidly degraded by the ubiquitin-proteasome pathway (Ivan et al., 2001). The ODD domain of HIF-1α is mainly critical domain for regulation of HIF-1α stabilization, because it is reported that ODD domain is interacted with pVHL in HIF-1α degradation by ubiquitin-proteasome pathway. Thus, it is conceivable that the binding of STAT3 to HIF-1α occurs in the ODD domain of HIF-1α and consequently, the inhibition of pVHL binding to HIF-1α by competent binding of STAT3 occurs in the ODD domain. However, interestingly, our result showed that STAT3 interacts with HIF-1α in the C-terminal region, the very next region of ODD domain of HIF-1α (Figure 2). Since STAT3 protein has a high molecular weight and forms hetero or homo-dimer structure in its active state, there is the possibility that the heavy and complicated structured STAT3 protein can interfere with the binding of pVHL to HIF-1α. It still remains to be solved by in vitro assays whether the interaction between STAT3 and HIF-1α is direct or indirect.
In conclusion, the results of this study demonstrate that active STAT3 enhances HIF-1α protein stability through inhibition of pVHL binding to the HIF-1α and pVHL-mediated ubiquitinantion of HIF-1α. In addition, it is suggested that STAT3 is a superior target for the development of anticancer drugs which has focused on targeting HIF-1α.
Acknowledgements
This work was supported by a grant from the 2007 National R&D Program for Cancer Control, Ministry of Health and Welfare (Project No.: 800-20070230), and Korean Science and Engineering Foundation (R01-2006-000-10977-0).
Abbreviations
- HIF-1α
hypoxia inducible factor-1α
- STAT3
signal transducer and activator of transcription 3
- VHL
von Hippel-Lindau
References
- 1.Bunn HF. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 2.Bowman T, Garcia R, Turkson J, Jove R. Stats in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 3.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 4.Berra E, Ginouves A, Pouyssegur J. The hypoxia-inducible-factor hydroxylases bring fresh air into hypoxia signaling. EMBO Rep. 2006;7:41–45. doi: 10.1038/sj.embor.7400598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 6.Huang M, Page C, Reynold RK, Lin J. Constitutive activation of STAT3 oncogene product in human ovarian carcinoma cells. Gynec Oncol. 2000;79:67–73. doi: 10.1006/gyno.2000.5931. [DOI] [PubMed] [Google Scholar]
- 7.Hockel M, Vaupel P. Tumor hypoxia; definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 8.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.Joung YH, Lim EJ, Lee MY, Park JH, Ye SK, Park EU, Kim SY, Zhang Z, Lee KJ, Park DK, Park TK, Moon WK, Yang YM. Hypoxia activates the cyclin D1 promoter via the Jak2/STAT5b pathway in breast cancer cells. Exp Mol Med. 2005;37:353–364. doi: 10.1038/emm.2005.45. [DOI] [PubMed] [Google Scholar]
- 11.Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, Lee JW, Choi S, Park JW, Ye SK, Chung MH. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19:1296–1298. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 12.Kallio PJ, Wilson WJ, O Brien S, Makino Y, Poellinger L. Regulation of the hypoxia inducible transcription factor 1 alpha by the ubiquitin proteasome pathway. J Biol Chem. 1999;274:6519–6525. doi: 10.1074/jbc.274.10.6519. [DOI] [PubMed] [Google Scholar]
- 13.Kim BY, Kim H, Cho EJ, Youn HD. Nur77 upregulates HIF-α by inhibiting pVHL-mediated degradation. Exp Mol Med. 2008;40:71–83. doi: 10.3858/emm.2008.40.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, Heller R, Ellis LM, Karras J, Bromberg J, Pardoll D, Jove R, Yu H. Constitutive stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 15.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 16.Safran M, Kaelin WG., Jr HIF hydroxylation and the mammalian oxygen-sensing pathway. J Clin Invest. 2003;111:779–783. doi: 10.1172/JCI18181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer LK, Ren Z, Fuller GN, Schaefer TS. Constitutive stat3 alpha in brain tumors; Localization to tumor endothelial cells and activation by endothelial tyrosine kinase receptor (VEGFR-2) Oncogene. 2002;21:2058–2065. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 19.Sinibaldi D, Wharton W, Turkson J, Boeman T, Peters G, Pledger WJ, Jove R. Induction of p21 WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblast; Role of activated STAT3 signaling. Oncogene. 2000;19:5419–5427. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 20.Song JI, Grandis JR. Stat signaling in head and neck cancer. Oncogene. 2000;19:2489–2495. doi: 10.1038/sj.onc.1203483. [DOI] [PubMed] [Google Scholar]
- 21.Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]