Abstract
Phenylketonuria (PKU; MIM 261600) is an autosomal recessive metabolic disorder caused by a deficiency of phenylalanine hydroxylase (PAH; EC 1.14.16.1). Point mutations in the PAH gene are known to cause PKU in various ethnic groups, and large deletions or duplications account for up to 3% of the PAH mutations in some ethnic groups. However, a previous study could not identify ~14% of the mutant alleles by sequence analysis in Korean patients with PKU, which suggests that large deletions or duplication might be frequent causes of PKU in Koreans. To test this hypothesis, we performed multiplex ligation-dependent probe amplification (MLPA) for the identification of uncharacterized mutant alleles after PAH sequence analysis of 33 unrelated Korean patients with PKU. Bi-directional sequencing of the PAH exons and flanking intronic regions revealed 27 different mutations, including four novel mutations (two missense and two deletion mutations), comprising 57/66 (86%) mutant alleles. MLPA identified a large deletion that encompassed exons 5 and 6 in four patients, another large deletion that extended from exon 4 to exon 7 in one patient, and a duplication of exon 4 in one patient. Chromosomal walking characterized the deletion breakpoint of the most common large deletion that involved exons 5 and 6 (c.456_706+138del). The present study shows that the allelic frequency of exon deletion or duplication is 9% (6/66) in Korean PKU patients, which suggests that these mutations may be frequent causes of PKU in Korean subjects.
Keywords: Asian continental ancestry group, phenylketonurias, phenylalanine hydroxylase, sequence deletion
Introduction
Phenylketonuria (PKU; MIM 261600) is an autosomal recessive metabolic disorder caused by a deficiency of phenylalanine hydroxylase (PAH; EC 1.14.16.1). PAH is a hepatic enzyme that catalyses the hydroxylation of L-Phenylalanine (L-Phe) to L-tyrosine using tetrahydrobiopterin (BH4) as a cofactor.
The human PAH gene, which is located on chromosome 12q, consists of 13 exons spanning 90 kb. To date, more than 520 different mutations in the PAH gene have been characterized in PKU patients and recorded in the PAH Mutation Analysis Consortium Database (http://www.pahdb.mcgill.ca). Although most of these mutations are detectable by sequence analysis, with a detection rate of > 95%, large intragenic deletions or duplications cannot be identified using this method. Rare cases of large genomic deletions involving one or more exons of the PAH gene have been described (Sullivan et al., 1989; Avigad et al., 1990; Guldberg et al., 1993, 1997; Okano et al., 1994; Bosco et al., 1996; Zschocke et al., 1999; Gable et al., 2003).
Recently, multiplex ligation-dependent probe amplification (MLPA) has been used as a sensitive and efficient method for the detection of large deletions and duplications (Schouten et al., 2002). Using MLPA, several studies have identified large deletions and duplications of the PAH gene in PKU patients lacking PAH mutations in one or both alleles, based on sequence analyses (Desviat et al., 2006; Kozak et al., 2006; Birk Moller et al., 2007). However, deletions or duplications of the PAH gene have been detected in fewer than half of the patients tested, and the estimated allelic frequency of exon deletions have been reported as < 3% in a Western population (Desviat et al., 2006; Kozak et al., 2006; Birk Moller et al., 2007).
In a previous study, only 86% of the mutant alleles were identified by sequence analysis of the entire PAH coding region, suggesting that large deletions or duplication are frequent causes of PKU in Koreans (Lee et al., 2004). To elucidate this hypothesis, we performed both sequencing and gene dosage analyses of the PAH gene in 33 unrelated Korean PKU patients. After identifying a recurrent large deletion that encompasses exons 4 and 5, the deletion breakpoint was analyzed by chromosomal walking. In addition, the relationship between genotype and phenotype, e.g., BH4 responsiveness, was also evaluated.
Materials and Methods
Subjects
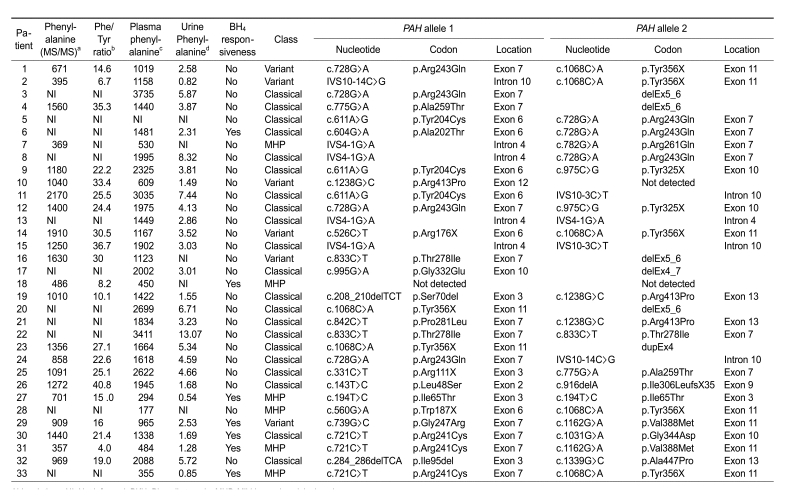
Thirty-three unrelated patients with PAH deficiency were recruited from the Korean PKU family support group. Most of these patients were identified during neonatal screening, and PAH deficiency was diagnosed by conventional biochemical methods. Biochemical diagnosis was based on measurement of the plasma phenylalanine concentration before starting a phenylalanine-restricted diet (Table 1). Disease severity was classified as follows: classical PKU (≥ 1,200 µM); variant PKU (600-1,200 µM), and mild hyperphenylalaninemia (MHP; < 600 µM). Urinary pterin analyses and dihydropteridine reductase (DHPR) assays were performed to exclude 6-pyruvoyltetrahydropterin synthase (PTPS) deficiencies.
Table 1.
Biochemical characteristics and PAH mutations in Korean patients with PKU.
Abbreviations: NI, Not informed; PKU, Phenylketonuria; MHP, Mild hyperphenylalaninemia
aNBS blood spots, reference range: 19-164 µM; bReference range: < 2.0; cPlasma phenylalanine concentration was measured before starting a phenylalanine-restricted diet, Reference range : 54.9-103.1 µM; dReference range : 0.05-0.19 µM
For the BH4 loading test, patients who were not on a phenylalanine-restricted diet were administered BH4 orally at 20 mg/kg (for patients < 36 months of age) or 7.5 mg/kg (> 36 months of age). The blood phenylalanine levels were measured before and at 1, 2, 4, 6, 8, 12, and 24 h after BH4 administration. The BH4 loading test was considered positive when the initial plasma phenylalanine concentration was decreased by at least 40% after 12 h.
Mutation analysis
After obtaining informed consent from the parents, blood samples were collected from the patients and their family members. Genomic DNA was isolated from peripheral blood leukocytes using the Wizard Genomic DNA Purification kit (Promega, Madison, WI)(Lee et al., 2006). All 13 PAH exons and their flanking intronic sequences were amplified by PCR using appropriate primers designed by the authors (sequences available upon request) and a thermal cycler (Model 9700; Applied Biosystems, Foster City, CA). Five microliters of amplification product were treated with 10 U shrimp alkaline phosphatase and 2 U exonuclease I (USB Corp., Cleveland, OH), and direct sequencing was then performed using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) in the ABI Prism 3100 genetic analyzer (Applied Biosystems). Potential novel mutations were defined as being distinct from those in the PAHdb database (http://www.pahdb. mcgill.ca) and the mutations reported previously in PubMed (http://www.ncbi.nlm.nih.gov/PubMed/).
MLPA analysis
MLPA was carried out for PKU patients who lacked PAH mutations on one or both alleles, based on previous sequencing analyses. The SALSA PO55 PAH MLPA kit (MRC Holland, Amsterdam, The Netherlands) contains 25 sets of probes: 13 PAH-specific sets, and the remainder comprises control standard probes from other human genes. The assay was carried out in 200-µl tubes in a thermal cycler (Model 9700; Applied Biosystems) according to the manufacturer's instructions. Briefly, a total of 200 ng of genomic DNA from each subject was diluted in 5 µl of TE buffer and denatured at 98℃ for 5 min. MLPA buffer and probe mix (1.5 µl of each) were then added, and the probes were annealed to the target genomic DNA by heating at 95℃ for 1 min, followed by incubation at 60℃ for 16 h. Thirty-two microliters of Ligase-65 mix were added to each sample, and the annealed probes were ligated at 54℃ for 15 min, followed by inactivation at 98℃ for 5 min. Ten microliters of the ligation reaction were removed for multiplex amplification using a pair of common primers, of which one was labeled with the fluorescent dye 6-FAM. Taq polymerase was added to the PCR (total volume of 50 µl) at 60℃, followed by 36 cycles of 95℃ for 30 s, 60℃ for 30 s, 72℃ for 1 min, and a final extension step of 72℃ for 20 min. Between 0.5 µl and 0.75 µl of each reaction was mixed with 0.5 µl of TAMRA-labeled internal size standard and 12.5 µl deionized formamide, and used for fragment analysis on the ABI-3100 capillary sequencer (Applied Biosystems). The conditions used for the fragment analysis were: polymer POP-4 in a 36-cm capillary; run temperature, 60℃; injection voltage, 15 kV; injection time, 3-5 s; run voltage, 15 kV; run time, 24 min. The obtained data were analyzed using the Genescan 3.1.2 software. The peak heights were normalized, and exon deletions were adjudged when the sample peak height was less than 65% of the control peak height.
Long-range PCR
Long-range PCR using the AccuPower TLA PCR premix (Bioneer, Daejeon, Korea) was performed for those patients who had a deletion of exons 5 and 6. By chromosomal walking with different pairs of primers, the smallest PCR amplicon (expected size: 12,522 bp) was detected using the primer pair: F23 (5'-tcgaactcgcaagtttgttg-3') and R6 (3'-cagctggagaggattgaagg-5'). The primer sequences were based on the intronic sequences of the PAH gene obtained from the Ensembl Genome Browser (http://www.ensembl.org).
Molecular modeling
To investigate the effects of the new PAH mutations on the structure of PAH, we used the crystal structure designed with the PAHdb system (http://www.pahdb.mcgill.ca). Models of the PAH monomer mutants were generated with the Biopolymer module of the Insight II modeling software (Molecular Simulations, San Diego, CA). The side-chain conformation of the PAH mutant was determined using the Auto-rotamer command in the Biopolymer module of the Insight II package. The three-dimensional model structure of PAH was visualized with PYMOL (http://www.pymol.org).
Results
Mutation analysis
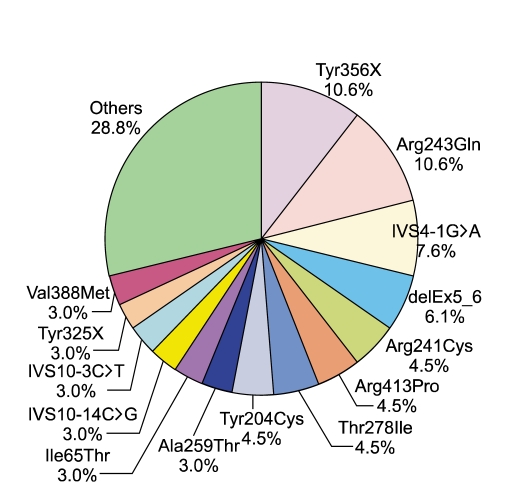
Bi-directional sequencing of the PAH genes of 33 unrelated PKU patients revealed 27 different kinds of mutations with variable frequencies (Table 1, Figure 1). Among the 33 patients, 76% (25/33) carried two mutant alleles, either compound heterozygous (n = 22) or homozygous (n = 3) genotypes. Seven patients carried only one mutant allele, and one patient had no mutations in PAH.
Figure 1.
Relative frequencies of PAH mutations in 33 independent Korean PKU patients.
Four novel mutations were identified in the present study: two variations that altered the coding sequences (c.604G>A and c.1031G>A, resulting in p.Ala202Thr and p.Gly344Asp, respectively) and two deletions (c.284_286delTCA and c.916delA, resulting in p.Ile95del and p.Ile306LeufsX35, respectively). These four novel mutations were not detected in 200 control chromosomes.
MLPA analysis
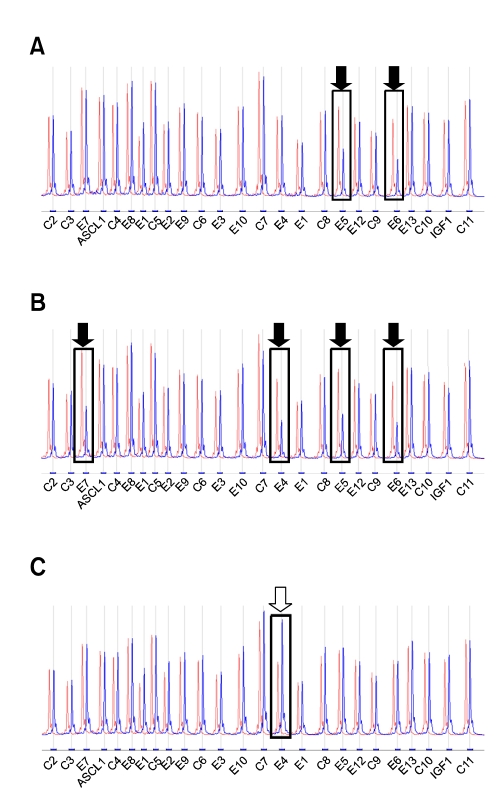
Eight patients who lacked PAH mutations in one or both alleles, based on sequence analyses, were subjected to MLPA analysis. Deletions or duplications were detected in six out of eight patients: five of these patients had exon deletions and one patient had a duplication of exon 4 (Figure 2). Among the five exon deletions, four showed the same pattern in MLPA analysis that encompassed exons 5 and 6, while the remaining deletion involved exons 4 through 7. One patient had a duplication of exon 4. However, gene dosage analysis by MLPA failed to detect any deletions or duplications in two patients. As genomic deletions or duplications were identified in 6/33 unrelated Korean patients with PKU, the allelic frequency of exon deletions or duplications was estimated to be 9% (6/66).
Figure 2.
Multiplex ligation-dependent probe amplification (MLPA) electrophoresis tracings for normal control (red-colored) and PKU subjects (blue-colored) with exon deletions or duplication. The boxes and arrows indicate decreased or increased patient's peaks relative to the control's peaks. A. Decreased MLPA tracing of the PAH gene peaks from a PKU patient with a deletion of exons 5 and 6 (filled arrows). B. Decreased MLPA tracing of the PAH gene peaks from a PKU patient with a deletion extending from exons 4 to 7 (filled arrows). C. Increased MLPA tracing of the PAH gene peaks from a PKU patient with a duplication of exon 4 (open arrow). C, control; E, exon
Characterization of the recurrent large deletion
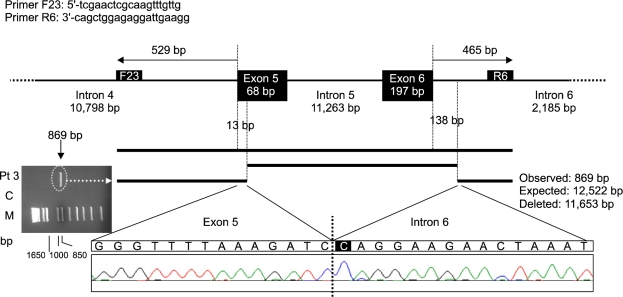
Using different intronic primers that covered the region from intron 4 to intron 6, long-range PCR was used to identify the breakpoint of the recurrent large deletion of exons 5 and 6 (Figure 3). Sequence analysis of the PCR amplicon revealed an 11,653-bp deleted region spanning from nucleotide 14 of exon 5 to nucleotide 138 of intron 6. According to the numbering position of the human PAH reference sequence (http://www.pahdb.mcgill.ca), the systematic name is c.456_706+138del11653 (g.51454_63106del11653).
Figure 3.
Breakpoint analysis and schematic figure of a deletion of exons 5 and 6 of the PAH gene (Ex5_6del11653). Long-range PCR encompassing exons 5 and 6 using intronic primers results in an amplicon of 12.5 kb, which is not amplified from a control sample (C), while a shorter 869-bp product is detected in patient 3, who has a heterozygous deletion of exons 5 and 6, as determined by MLPA. Sequence analysis shows a deletion of 11,653 bp that spans from the nucleotide 14 of exon 5 to nucleotide 138 of intron 6, designated as c.456_706+138del11653 (g.51454_63106del11653).
Molecular modeling
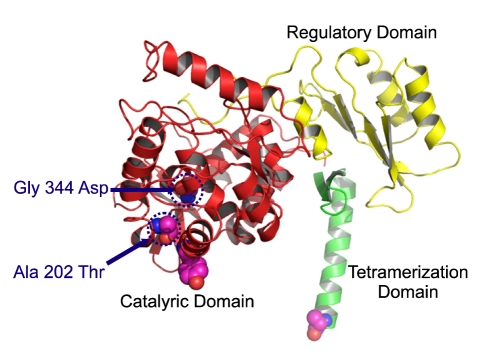
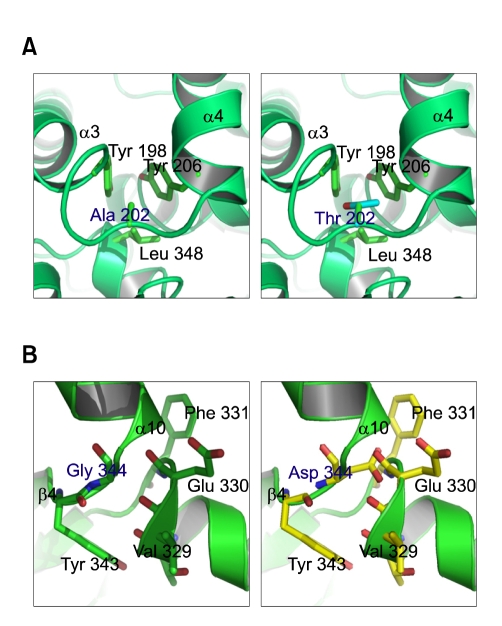
The locations of the two novel missense variants (p.Ala202Thr and p.Gly344Asp) identified in the present study in the 3-D structure of the PAH monomer are depicted in Figure 4. These variations are located in the catalytic domain. Ala202 belongs to the loop between the α3 and α4 helical domains. Changing the alanine at amino acid position 202 to threonine is expected to weaken the hydrophobic interactions among Ala202, Tyr198, Tyr206, and Leu348, and the activity of PAH may be decreased as a result of alterations to the side-chain and polarity that contribute to structural conformation (Figure 5A). Gly344 is located between the β4 strand and the α10 helical domain. Changing the glycine at amino acid position 344 to aspartic acid is expected to distort the backbone interactions among Gly344, Leu347, Glu330, Phe331, Val329, Thr328, and Gly346 over the β4 strand and the α10 helical domain by inducing strong repulsive interactions (Figure 5B).
Figure 4.
Ribbon diagram of a monomer of human PAH depicting the locations of the amino acid replacements newly identified in Korean PAH patients. Substituted amino acid residues are denoted with red-colored balls.
Figure 5.
Predicted structural changes in human PAH induced by novel mutations. (A) Changing p.Ala202 to threonine is expected to weaken the hydrophobic interactions among Ala202, Tyr198, Tyr206 and p.Leu348. (B) Changing p.Gly344Asp to aspartic acid is expected to distort the backbone interactions among Gly344, Leu347, Glu330, Phe331, Val329, Thr328, and Gly346 over the β4 strand and the α10 helical domain by inducing strong repulsive interactions.
BH4 responsiveness
As shown in Table 1, all three patients with the p.Arg241Cys mutation showed BH4 responsiveness: patient 30 with p.Arg241Cys and p.Gly344Asp, patient 31 with p.Arg241Cys and p.Val388Met, and patient 33 with p.Arg241Cys and p.Tyr356X showed 42%, 72%, and 40% reductions in plasma phenylalanine levels, respectively. An additional three patients with p.Gly247Arg and p.Val388Met, p.Ala202Thr and p.Arg243Gln, and homozygous p.Ile65Thr mutation also showed BH4 responsiveness (Table 1).
Discussion
In the present study, we identified 27 different mutations in 33 unrelated Korean PKU patients, including four novel mutations. p.Arg243Gln, p.Tyr356X, IVS4-1G>A, and p.Tyr204Cys were the most prevalent mutations which accounted for 35% (22/63) of the total mutant alleles. This finding was consistent with the previous report of Lee et al. (2004). In Europe, there are several prevalent founder alleles that represent the expansion, migration, and genetic drift of European population (Zschocke, 2003). Particularly, the p.Arg408Trp mutation has been reported to be one of the most frequent mutation in PKU patients in Eastern Europe and Germany, with a frequency of 20-84%. However, this mutation was not found in the present study. Instead, p.Arg243Gln was found to be a recurring mutation in Korean PKU patients (8/63 mutant alleles).
In the present study, we also identified four novel mutations in 33 unrelated PKU Korean patients. Two deletion mutations led to an in-frame deletion or frameshifting, while two missense mutations were predicted to affect enzymatic function and/or stability. However, eight patients (24%) were negative for PAH mutations in one or both alleles. Three of the 66 alleles (4.5%) were not identified by either sequencing or MLPA analysis. Studies using exon analysis of different populations worldwide have shown PAH mutation detection rates of 90~99% (Guldberg et al., 1996; Zschocke, 2003; Song et al., 2005). It remains to be seen whether uncharacterized alleles correspond to molecular defects that are embedded in the intronic regions and cause splicing aberrations or to impairments of the 5'-end promoter or regulatory regions and 3'-end polyadenylation regions. Large deletions or duplications of the PAH gene may also be sources of the defective alleles that remain to be specified.
In the present study, large deletions or duplications were detected in 75% (6/8) of the patients who were subjected to MLPA, which is comparable to the rates detected for other ethnic groups (Spain, 3/22; Czech Republic, 31/59; Denmark and Germany, 7/34; Japan, 2/8) (Okano et al., 1998; Desviat et al., 2006; Kozak et al., 2006; Birk Moller et al., 2007). In particular, larger gene rearrangements that consist mainly of deletions of exons 5 and 6 have been found in 4/66 (6.1%) PKU alleles. Although deletions of exon 5 and 6 have been detected in 2/8 Japanese PKU patients with uncharacterized alleles by Southern hybridization, the breakpoint junction fragment (c.442_706 + 152del) was different to that in Koreans (c.456_706 + 138del) (Okano et al., 1998). The breakpoint junction of the deletion mutation identified in the present study was also different from those of European populations, i.e., EX5del4232ins268 and EX3del4765 in Czech PKU patients, and EX3del6604ins8 and EX5del1881 in Spanish PKU patients (Desviat et al., 2006; Kozak et al., 2006). Considering the relatively high number of deletions of exons 5 and 6 in our current population, we postulate a founder effect of this mutation in the Korean population.
In a previous study, PAH with Arg241Cys substitution was shown to have 28% residual activity in the COS cell expression system (Kim et al., 2006). In addition, Korean PKU patients with p.Arg241Cys showed BH4 responsiveness (Lee et al., 2004). Arg241 is located near the cofactor binding region but does not interact directly with the cofactor, so the p.Arg241Cys mutation may lead to relatively mild structural deformities. The patient with p.Ala202Thr/p.Arg243Gln showed BH4 responsiveness. Even though p.Arg243Gln was associated with classical PKU in a previous study, this patient showed variant PKU, which suggests that p.Ala202Thr has sufficient residual enzyme activity to ensure responsiveness. Although there was only one p.Ile65Thr homozygote in the present study, this patient showed MHP and BH4 responsiveness, which suggests that p.Ile65Thr is associated with near-normal levels of enzymatic activity and BH4 responsiveness. Of the 27 different mutations described herein, the p.Ile65Thr and p.Ala202Thr mutations were responsive to BH4. These defects are located at opposite sides of the BH4-binding pocket. Specifically, the composite structural model of the PAH tetramer reveals that these mutations are in close proximity to the dimer interface and occur along the interface region of the regulatory domain. This observation suggests that, upon mutation of p.Ile65Thr, p.Ala202Thr, and p.Arg241Cys, dimer stability is reduced. The increased BH4 levels rescue dimer stability and, consequently, enzyme activity recovers to the normal level.
In conclusion, exon deletions or duplication may be more frequent causes of PKU in Korean patients than in other ethnic groups. Therefore, a gene dosage analysis should be considered as a complementary diagnostic tool for Korean PKU patients without point mutations in one or two chromosomes, and the long-range PCR method developed in the present study may be useful for screening Koreans for deletions of exons 5 and 6.
Acknowledgements
This work was supported by a grant from the IN-SUNG foundation for Medical Research and by the Samsung Biomedical Research Institute grant, #SBRI C-A8-205-1.
Abbreviations
- BH4
tetrahydrobiopterin
- DHPR
dihydropteridine reductase
- L-Phe
L-phenylalanine
- MLPA
multiplex ligation-dependent probe amplification
- PAH
phenylalanine hydroxylase
- PKU
phenylketonuria
References
- 1.Avigad S, Cohen BE, Bauer S, Schwartz G, Frydman M, Woo SL, Niny Y, Shiloh Y. A single origin of phenylketonuria in Yemenite Jews. Nature. 1990;344:168–170. doi: 10.1038/344168a0. [DOI] [PubMed] [Google Scholar]
- 2.Birk Moller L, Nygren AO, Scott P, Hougaard P, Bieber Nielsen J, Hartmann C, Guttler F, Tyfield L, Zschocke J. Low proportion of whole exon deletions causing phenylketonuria in Denmark and Germany. Hum Mutat. 2007;28:207. doi: 10.1002/humu.9481. [DOI] [PubMed] [Google Scholar]
- 3.Bosco P, Ceratto N, Cali F, Goltsov AA, Eisensmith RC, Novelli G, Dalla Piccola B, Romano V. RFLP discordance in a PKU family due to a deletion in the PAH gene. Turk J Pediatr. 1996;38:497–504. [PubMed] [Google Scholar]
- 4.Desviat LR, Perez B, Ugarte M. Identification of exonic deletions in the PAH gene causing phenylketonuria by MLPA analysis. Clin Chim Acta. 2006;373:164–167. doi: 10.1016/j.cca.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Gable M, Williams M, Stephenson A, Okano Y, Ring S, Hurtubise M, Tyfield L. Comparative multiplex dosage analysis detects whole exon deletions at the phenylalanine hydroxylase locus. Hum Mutat. 2003;21:379–386. doi: 10.1002/humu.10199. [DOI] [PubMed] [Google Scholar]
- 6.Guldberg P, Romano V, Ceratto N, Bosco P, Ciuna M, Indelicato A, Mollica F, Meli C, Giovannini M, Riva E, et al. Mutational spectrum of phenylalanine hydroxylase deficiency in Sicily: implications for diagnosis of hyperphenylalaninemia in southern Europe. Hum Mol Genet. 1993;2:1703–1707. doi: 10.1093/hmg/2.10.1703. [DOI] [PubMed] [Google Scholar]
- 7.Guldberg P, Levy HL, Hanley WB, Koch R, Matalon R, Rouse BM, Trefz F, de la Cruz F, Henriksen KF, Guttler F. Phenylalanine hydroxylase gene mutations in the United States: report from the Maternal PKU Collaborative Study. Am J Hum Genet. 1996;59:84–94. [PMC free article] [PubMed] [Google Scholar]
- 8.Guldberg P, Henriksen KF, Mammen KC, Levy HL, Guttler F. Large deletions in the phenylalanine hydroxylase gene as a cause of phenylketonuria in India. J Inherit Metab Dis. 1997;20:845–846. doi: 10.1023/a:1005352725283. [DOI] [PubMed] [Google Scholar]
- 9.Kim SW, Jung J, Oh HJ, Kim J, Lee KS, Lee DH, Park C, Kimm K, Koo SK, Jung SC. Structural and functional analyses of mutations of the human phenylalanine hydroxylase gene. Clin Chim Acta. 2006;365:279–287. doi: 10.1016/j.cca.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Kozak L, Hrabincova E, Kintr J, Horky O, Zapletalova P, Blahakova I, Mejstrik P, Prochazkova D. Identification and characterization of large deletions in the phenylalanine hydroxylase (PAH) gene by MLPA: evidence for both homologous and non-homologous mechanisms of rearrangement. Mol Genet Metab. 2006;89:300–309. doi: 10.1016/j.ymgme.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Lee DH, Koo SK, Lee KS, Yeon YJ, Oh HJ, Kim SW, Lee SJ, Kim SS, Lee JE, Jo I, Jung SC. The molecular basis of phenylketonuria in Koreans. J Hum Genet. 2004;49:617–621. doi: 10.1007/s10038-004-0197-5. [DOI] [PubMed] [Google Scholar]
- 12.Lee KA, Kim JW. Heterozygosities of 735 microsatellite markers, background linkage disequilibrium in the Korean population. Exp Mol Med. 2006;38:662–667. doi: 10.1038/emm.2006.78. [DOI] [PubMed] [Google Scholar]
- 13.Okano Y, Asada M, Kang Y, Nishi Y, Hase Y, Oura T, Isshiki G. Molecular characterization of phenylketonuria in Japanese patients. Hum Genet. 1998;103:613–618. doi: 10.1007/s004390050877. [DOI] [PubMed] [Google Scholar]
- 14.Okano Y, Hase Y, Shintaku H, Araki K, Furuyama J, Oura T, Isshiki G. Molecular characterization of phenylketonuric mutations in Japanese by analysis of phenylalanine hydroxylase mRNA from lymphoblasts. Hum Mol Genet. 1994;3:659. doi: 10.1093/hmg/3.4.659. [DOI] [PubMed] [Google Scholar]
- 15.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song F, Qu YJ, Zhang T, Jin YW, Wang H, Zheng XY. Phenylketonuria mutations in Northern China. Mol Genet Metab. 2005;86(Suppl 1):S107–S118. doi: 10.1016/j.ymgme.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan SE, Moore SD, Connor JM, King M, Cockburn F, Steinmann B, Gitzelmann R, Daiger SP, Woo SL. Haplotype distribution of the human phenylalanine hydroxylase locus in Scotland and Switzerland. Am J Hum Genet. 1989;44:652–659. [PMC free article] [PubMed] [Google Scholar]
- 18.Zschocke J, Quak E, Knauer A, Fritz B, Aslan M, Hoffmann GF. Large heterozygous deletion masquerading as homozygous missense mutation: a pitfall in diagnostic mutation analysis. J Inherit Metab Dis. 1999;22:687–692. doi: 10.1023/a:1005527731397. [DOI] [PubMed] [Google Scholar]
- 19.Zschocke J. Phenylketonuria mutations in Europe. Hum Mutat. 2003;21:345–356. doi: 10.1002/humu.10192. [DOI] [PubMed] [Google Scholar]