Abstract
The structure, stability, solubility, and function of proteins depend on their net charge and on the ionization state of the individual residues. Consequently, biochemists are interested in the pK values of the ionizable groups in proteins and how these pK values depend on their environment. We review what has been learned about pK values of ionizable groups in proteins from experimental studies and discuss the important contributions they make to protein stability and solubility.
Historical Perspective
Sorensen defined pH in 1909 and in 1917 published the first experimental study of the titration of a protein, egg albumin (1). In succeeding years, hydrogen ion titration curves were determined for several proteins, and it was possible to make rough estimates of the pK values of the ionizable groups of proteins (2). In special cases, it was possible to determine the pK values of individual groups, but it was only when NMR became available that the pK values of individual groups could be readily determined, at least for small proteins (3, 4). This led to rapid progress, and >500 pK values have been determined for individual ionizable groups in folded proteins (5) and a more limited number in unfolded proteins (6).
The landmark paper by Debye and Hückel on the theory of electrolyte solutions was published in 1923 (7), and the ideas were extended to proteins by Linderstrom-Lang in 1924 (8). He recognized that net charge on a protein would influence the ionization of individual groups and incorporated this into the first model developed to understand acid/base properties of proteins. This model was extended by Tanford and Kirkwood (9) in an important paper that triggered an interest in factors that determine pK values of the ionizable groups in proteins that continues to the present day (10).
Protein Ionizable Groups and Their Intrinsic pK Values
Seven amino acid side chains contain groups that ionize between pH 1 and 14. For Asp, Glu, Tyr, and Cys, the ionizable groups are uncharged below their pK and negatively charged above their pK. For His, Lys, and Arg, the ionizable groups are positively charged below their pK and uncharged above their pK. It is useful to know what the pK values of these groups would be in a protein if they are completely exposed to solvent, not hydrogen-bonded, and not affected by the presence of any formal charges. These are generally referred to as the intrinsic pK (pKint) values. The pKint values given in Table 1 are the pK values observed for the ionizable side chains when they are present in blocked pentapeptides with the structure Ala-Ala-X-Ala-Ala, where X is the amino acid whose side chain pK was measured (11). The α-carboxyl and α-amino groups of proteins can also ionize, and their pK values were determined in similar pentapeptides and are also given in Table 1. These pKint values reflect the inductive effects of neighboring peptide bonds but will not be influenced by charge-charge interactions and only minimally by hydrogen bonding or burial of the ionizable group. They should serve as good models for the unperturbed pK values of the ionizable groups in proteins.
TABLE 1.
Characteristics of ionizable side chains in proteins
This is a summary of 541 pK values tabulated from the literature (5). The values were reported under various conditions for 78 folded proteins.
| Group | Contenta | Buriedb | pK value in alanine pentapeptides (pKint)c | Average pK value | Low pK value | High pK value | No. of measurements |
|---|---|---|---|---|---|---|---|
| % | % | ||||||
| Asp | 5.2 | 56 | 3.9 | 3.5 ± 1.2 | 0.5 | 9.2 | 139 |
| Glu | 6.5 | 48 | 4.3 | 4.2 ± 0.9 | 2.1 | 8.8 | 153 |
| His | 2.2 | 72 | 6.5 | 6.6 ± 1.0 | 2.4 | 9.2 | 131 |
| Cys | 1.2 | 90 | 8.6 | 6.8 ± 2.7 | 2.5 | 11.1 | 25 |
| Tyr | 3.2 | 67 | 9.8 | 10.3 ± 1.2 | 6.1 | 12.1 | 20 |
| Lys | 5.9 | 34 | 10.4 | 10.5 ± 1.1 | 5.7 | 12.1 | 35 |
| Arg | 5.1 | 56 | 12.3d | ||||
| C terminus | 3.7 | 3.3 ± 0.8 | 2.4 | 5.9 | 22 | ||
| N terminus | 8.0 | 7.7 ± 0.5 | 6.8 | 9.1 | 16 |
This is the current average % amino acid content of proteins from all three domains of life: Bacteria, Archaea, and Eukaryota (12). For comparison, the most abundant amino acid is Leu (10.3%), and the least abundant is Trp (1.1%).
The % buried is based on an analysis of 61 proteins by Lesser and Rose (13). The % buried is for just the ionizable group: the carboxyl groups of Asp and Glu, the imidazole of His, the –SH of Cys, the –OH of Tyr, the amino group of Lys, and the guanidinium of Arg.
A value of 12.48 is given for Arg in Ref. 49. A correction for the negative charge on the carboxyl group present in Arg gives a pK of ≈ 12.3.
Content and Environment of Ionizable Groups
Amino acids with ionizable side chains make up, on average, 29% of the amino acids in proteins (12). The average content for each is given in Table 1. As discussed below, the extent of burial of the ionizable groups in proteins is important in determining their pK values. The average % burial for the ionizable group in each side chain is given in Table 1 (13). The most buried ionizable groups are the –SH of Cys, the imidazole of His, and the –OH of Tyr. These groups are often buried because they are generally uncharged at pH 7. The least buried are the guanidinium of Arg, the carboxylate groups of Asp and Glu, and the amino groups of Lys. These groups will generally be charged at pH 7. It is surprising that Arg is buried to such an extent because of the high pK and the fact that the Arg side chain can donate five hydrogen bonds. However, in water, the guanidinium group is one of the most weakly hydrated cations, probably because of charge delocalization, and this makes the Arg side chain easier to bury (14). Buried Arg side chains are charged, extensively hydrogen-bonded, and frequently interact by stacking with other planar side chain groups in proteins (15). They make many important contributions to the stability and function of proteins.
Measured pK Values in Folded Proteins
Most of the pK values for ionizable groups in folded proteins were determined by measuring the pH dependence of chemical shifts using NMR (3, 4). A smaller number were measured using indirect techniques (16). Recently, 541 pK values from 78 proteins were compiled (5), and the results are summarized in Table 1. Many of the pK values are perturbed far above and below the pKint values. For example, the pK of one sulfhydryl group is lowered by >6 pK units, and the pK of one carboxyl side chain is raised by >5 pK units.
Perturbation of pK Values
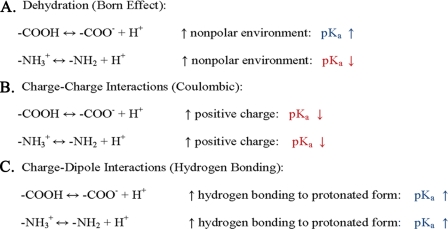
In proteins, the pK values of the ionizable groups may be substantially raised or lowered from the intrinsic pK values by environment effects (see Table 1). The three most important effects are summarized in Fig. 1. Each of these will be discussed in general terms and illustrated with experimental results. Another review discusses the perturbation of the pK values of catalytic groups in enzyme active sites (17).
FIGURE 1.
Factors influencing the pK values of ionizable groups in proteins. A,apK change due to the Born effect results when an ionizable group is buried in the interior of the protein where the dielectric constant is lower than that of water. The lower dielectric constant favors the neutral form of the ionizable group. B, the pK values of all of the ionizable groups in a protein will be decreased by a positively charged environment and increased by a negatively charged environment.C, the pK values of the ionizable groups will be increased when hydrogen bonding is tighter to the protonated form and decreased when hydrogen bonding is tighter to the deprotonated form.
Dehydration (Born Effect)—It is energetically unfavorable to transfer a charged group from water to the interior of a protein where the dielectric constant (εprotein) is lower. Consequently, the neutral state of the ionizable group will be favored, and the pK values of Asp, Glu, Cys, and Tyr will be raised and those of His, Lys, and Arg will be lowered when the groups are buried, partially or completely, in a folded protein. To illustrate this, the pK of acetic acid is increased from 4.8 in water (ε = 78) to 10.1 in ethanol (ε = 24).
Studies of staphylococcal nuclease provide an example of this effect in proteins (18). Val-66 is buried in the hydrophobic core of the enzyme. When it is replaced with Asp, the carboxyl group hasapK of 8.9, 5 pK units higher than the pKint. When it is replaced with Lys, the amino group has a pK of 5.5, 4.9 units lower than the pKint. If these changes resulted only from the Born effect, it would require εprotein = 7.2. It was concluded (18) that, “Regardless of how the pKa calculations were performed, they all showed that the shift in the pKa value of Asp-66 is governed by the loss of hydration of the carboxylic group in the buried state that is not offset by interactions with charges or with polar atoms of the protein.”
Charge-Charge Interactions (Coulombic Interactions)—The net charge on a protein is zero at the isoelectric pH (pI). Below the pI, the net charge on a protein is positive, and above the pI, the net charge is negative. At the pK of a given ionizable group, the net charge will be –½ for Asp, Glu, Tyr, and Cys and +½ for His, Lys, and Arg. The energy of interaction of the ionizable group (i) and the other charges (j) on the protein can be calculated with Coulomb's law: ΔGij = ∑qiqj/εrij, where qi is the charge on the ionizable group of interest, qj is the charge on the other groups at the pH = pK of the ionizing group, ε is the dielectric constant, and rij is the distance between the two charges. (When opposite charges are 4.2 Å apart in water, ΔG =–l kcal/mol, and this is reduced to –0.5 kcal/mol at the ionic strength inside a cell.) The distance between charges can be calculated from the structure of the protein, keeping in mind that the distance between the groups may differ in solution and in a crystal and may vary as the protein is titrated. (For RNase A, structures were determined as a function of pH so that the effect of titration on the distances could be observed (19).) Protein interiors are heterogeneous, so the effective value of ε will depend on which two charges are considered. Values of ε ranging from 2 to 80 have been used (20).
A good example of the effect of coulombic interactions on the pK values of ionizable groups in a protein is provided by a study of RNase Sa (21). RNase Sa is an acidic protein with a pI of 3.5 that contains no Lys residues (0K). By replacing Asp and Glu residues on the surface with 5 Lys residues, a basic protein was created with a pI of 10.2 (5K). At pH 7, the net charge on 0K is –7, and the net charge on 5K is +3, a difference of ∼10 units. Crystal structures and NMR studies show that the structures of 0K and 5K are similar (21). Consequently, except for net charge, the ionizable groups will have similar environments in the two proteins, and coulombic interactions will be the main determinant of pK differences. For the 11 common groups, the pK values were always higher in 5K than in 0K, as expected because of the greater positive charge. The differences ranged from 0.03 to 2.19, with an average difference of 0.75. The pK differences (pK0K – pK5K) calculated as described above with Coulomb's law were in good agreement with the measured values. A value of ε = 45 gave the best agreement between calculated and experimental values. It is not surprising that the ε value that gave the best agreement here is considerably higher than the ε value that gave the best results for the Born effect (22). Based on this, it was concluded (21) that, “Taken together, the results are evidence that charge-charge interactions are the chief perturbant of the pK values of ionizable groups on the protein surface, which is where the majority of the ionizable groups are positioned in proteins.”
These conclusions are supported by other experimental and theoretical studies. A global analysis of available data for short-to-long range coulombic interactions in staphylococcus nuclease led to an effective ε of ≈42 (23). A theoretical analysis was successful in predicting the contribution of electrostatic effects to the stability of four proteins (22). When both the Born effect and coulombic interactions were included in the analysis, the best agreement was found with ε = 20–80 for the Born effect and ε = 40 for coulombic interactions. The significance of these ε values and why a large ε is needed for the Born effect were discussed (22).
Charge-Dipole Interactions (Hydrogen Bonds)—Ionizable groups can also interact with the partial charges or dipoles on neighboring polar groups. These interactions will be referred to as hydrogen bonds, but keep in mind that some of these interactions can be important and not meet the definitions generally used for hydrogen bonds. The effect of hydrogen bonding on a pK will depend on whether the interactions are more favorable with the protonated state of the group, in which case the pK will be raised, or with the deprotonated state of the group, in which case the pK will be lowered. Hydrogen bonds generally contribute 1–2 kcal/mol to the stability of a protein (24) and, when the hydrogen bonds are to ionizable groups, they can raise or lower the pK values by several pK units (25). An example is provided by the buried, charged, non-ion-paired carboxyl group of Asp-76 in RNase T1 (16).
The side chain carboxyl of Asp-76 in RNase T1 has a very low pK of 0.6 and forms three intramolecular hydrogen bonds to the side chains of Asn-9, Tyr-11, and Thr-91 (16). To see if these hydrogen bonds were responsible for the low pK, the hydrogen bonds were removed one at a time, and the pK of was Asp-76 measured. When single hydrogen bonds are removed, the average pK increases to 3.3. When two hydrogen bonds are removed, the average pK increases to 5.1. When all three hydrogen bonds are removed, the pK increases to 6.4. Thus, in the absence of the hydrogen bonds, the pK is elevated 2.5 units above the pKint. This increase results from the Born effect because the carboxyl group of Asp-76 is buried and from the negative charge on the protein at pH 6.4. Hydrogen bonding and the positive charge on the protein at low pH combine to lower the pK of this carboxyl group by 5.8 pK units. A comprehensive description of the pK shifts due to hydrogen bonding is available (25). The pK shift per hydrogen bond can be as high as 1.6 for carboxyl groups and higher for sulfhydryl groups.
Contribution of Ionizable Residues to Protein Stability
Major forces favoring folding of globular proteins are the hydrophobic effect and hydrogen bonding, and they are just able to overcome the major force favoring unfolding, conformational entropy, so most globular proteins have a surprisingly low conformational stability of just 2–10 kcal/mol (26). It is now clear that many proteins are unfolded under physiological conditions (27) but fold when it is required for their function. Because of this, minor forces in the 1–3 kcal/mol range become important.
Charge-Charge Interactions—Charged groups in proteins are generally arranged so that coulombic interactions among charges are favorable. However, the arrangement is more favorable on some proteins than on others and is sometimes unfavorable (28). Studies of the pH dependence of protein stability show that coulombic interactions do not make a large contribution to protein stability, probably at most 10 kcal/mol (29). Despite this, coulombic interactions are important to proteins in a number of ways.
There is considerable interest in developing methods to increase protein stability by amino acid substitutions. This is difficult to do by burying hydrophobic surface or adding hydrogen bonds but can be done by improving the charge-charge interactions on the surface of a protein (30). Several groups showed that reversing the charge on a single side chain on the protein surface can improve coulombic interactions and increase stability by >1 kcal/mol (30–34). This approach was used to increase the stability of several proteins, and guidelines are available for doing so with other proteins (30). (Another good approach is to improve the β-turns on the surface of a protein (35).)
Just as attractive charge-charge interactions can stabilize a protein, repulsive charge-charge interactions can destabilize a protein. Recently, it has become clear that many proteins are unfolded or have regions of the polypeptide chain that are disordered under physiological conditions (27). These proteins are referred to as IDPs,2 and the number identified is now >500 (36). This revelation was surprising, but upon reflection, IDPs offer new functions and can improve some known functions of proteins (37). Two factors that are important in determining whether a protein will be folded or unfolded are hydrophobicity and net charge (38). IDPs generally have a low hydrophobicity, a high net charge, or both. The border between folded proteins and unfolded proteins is defined well by the following equation: 〈R〉 = 2.785〈H〉 – 1.151, where 〈R〉 and 〈H〉 are the mean net charge and the mean hydrophobicity of the protein, respectively (38). Recent experimental studies show that favorable and unfavorable coulombic interactions can influence the denatured state ensemble of a protein and influence both protein stability and the mechanism of folding (39).
Buried Ionizable Residues—As discussed above, many ionizable groups are buried, and these often make important contributions to the function and stability of proteins. An improved version of a finite difference Poisson-Boltzmann method was used to estimate the number of buried ionizable residues (40). For each ionizable group, the dehydration penalty was calculated. If it was great enough to shift the pK value by >5 pK units, the residue was classed as buried. These residues would generally be >80% buried when measured by accessible surface area as in Table 1. Using this criterion, 32% of the Arg residues, 19% of the Asp residues, 13% of the Glu residues, and 6% of the Lys residues are buried. This amounts to 4 buried residues per 100 residues, and the composition is 41% Arg, 28% Asp, 22% Glu, and 9% Lys.
These buried ionizable residues can be used to stabilize or destabilize proteins. Despite being buried, the pK of Asp-76 in RNase T1 is lowered to 0.6 by hydrogen bonding and the positive charge on the protein (16). This buried carboxyl group makes a large contribution to the stability and to the pH dependence of the stability. The D76A mutation of RNase T1 is 3.8 kcal/mol less stable than the wild-type protein, and an analysis suggests that the hydrogen bonding and other interactions of the carboxyl group with the protein contribute ∼8 kcal/mol to the stability. As discussed below, Asp-70 in T4 lysozyme is buried, has a pK of 0.5, and also makes a large contribution to the stability (41).
The two most buried carboxyl groups in RNase Sa are Asp-33, which is 99% buried and has a pK of 2.4, and Asp-79, which is 85% buried and has a pK of 7.4 (16, 42). The D33A mutant is 6 kcal/mol less stable and the D79A mutant is 3.3 kcal/mol more stable than the wild-type protein. The environment of the two carboxyl groups is different: Asp-33 forms three intramolecular hydrogen bonds, and Asp-79 forms none. The net charge is +7 when Asp-33 ionizes, which would lower the pK, and –6 when Asp-79 ionizes, which would raise the pK. These environmental factors combine to give a pK difference of 5 units and markedly different contributions to the stability for these two carboxyl groups. The D79F mutant of RNase Sa has a stability 3.7 kcal/mol greater than the wild-type enzyme, and this raises the Tm (10 °C) (42). This is one of the largest increases in stability observed for a single mutation.
Based on what we have learned about the major forces that contribute to the stability of globular proteins, stability should increase as protein size increases. This is not observed: the conformational stability of globular proteins is independent of size. It has been shown that the number of buried charged groups increases substantially with protein size (40, 43). For example, the number of buried charged groups is 1.9 per 100 residues in proteins with <100 residues and 4.5 per 100 residues in proteins with >300 residues (40). It seems likely that burial of charged side chains that are not hydrogen-bonded or ion-paired is one mechanism that evolution uses to lower protein stability (43).
Ion Pairs—Attractive interactions between nearby (<5Å) oppositely charged groups are called ion pairs or salt bridges. Coulombic interactions in proteins are made favorable by avoiding repulsive charge-charge interactions; on average, there are four attractive ion pairs but only one repulsive ion pair per 100 residues (28). Whether the attractive ion pairs contribute favorably to protein stability is a controversial topic, experimentally (41, 44) and theoretically (45).
An experimental study of a salt bridge between Asp-70 and His-31 in T4 lysozyme showed that a buried salt bridge can make a favorable contribution to protein stability (41). Asp-70 forms a salt bridge with His-31, and this substantially perturbs the pK values: Asp-70 has a pK of 0.5 in the folded protein and 3.5–4 in the unfolded protein, whereas His-31 has a pK of 9.1 in the folded protein and 6.8 in the unfolded protein. This salt bridge contributes ∼4 kcal/mol to the stability of T4 lysozyme.
Proteins from thermophiles are stabilized in different ways (46), but the strategy used most often is improving charge-charge interactions and adding ion pairs (46, 47). Thermophilic proteins generally contain more charged groups and salt bridges than proteins from mesophiles, and several lines of evidence suggest that the contribution of salt bridges to protein stability will increase at higher temperatures (47).
The most stable protein known is the CutA1 protein from the hyperthermophile Pyrococcus horikoshii, with a Tm of ∼150 °C at pH 7 (48). This protein is found in bacteria, plants, and animals, including humans. All of the CutA1 proteins studied are remarkably stable, and this is thought to be due in part to a common trimeric structure. The most striking difference between P. horikoshii CutA1 and the same protein from Escherichia coli is the number of ion pairs in the monomer: 30 are found in the protein from the hyperthermophile, but only one is found in the protein from the mesophile. This suggests that ion pairs are of crucial importance to the stability of the most stable proteins.
Contribution of Ionizable Residues to Protein Solubility
Protein solubility is a concern to biochemists in experimental studies, and the recognition of its role in protein folding diseases makes it even more important. Because many proteins are now used as drugs, solubility is a concern in the biopharmaceutical industry. Key principles of protein solubility were summarized by Cohn in 1943 (49): “A given protein is least soluble in the neighborhood of its isoelectric point in the presence, as well as the absence, of neutral salts... The solubility of proteins in the uncombined, salt-free state varies widely. This is true among those that separate in the crystalline state and in the amorphous state... The forces between molecules in the solid state as well as those between solvent and solute molecules determine solubility.”
The approach generally used to increase the solubility of a protein is to replace the most hydrophobic residue on the surface with a charged or polar residue. Recent studies provide insights into protein solubility (50). Thr-76 has the most exposed side chain in RNase Sa. This Thr was replaced with the other 19 amino acids, and the solubility was measured. The most soluble variant was T76D (43 mg/ml), and least soluble was T76W (3.6 mg/ml), a 12-fold difference! One surprising finding was that His, Asn, Thr, and Gln make unfavorable contributions to the solubility relative to Ala near the pI of RNase Sa. Similarly, Arg and Lys make unfavorable contributions to the solubility relative to Ala when solubility is measured at pH 7, where the protein has a charge of approximately –5. In contrast, Asp and Ser always make favorable contributions to the solubility. These results suggest that the best approach for increasing solubility is to replace the most hydrophobic residue on the protein surface with Asp or Ser (50).
Supplementary Material
This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviation used is: IDPs, intrinsically disordered proteins.
References
- 1.Sorensen, S. P. L., Hoyrup, M., Hempel, J., and Palitzsch, S. (1917) C. R. Trav. Lab. Carlsberg 12 68–163 [Google Scholar]
- 2.Tanford, C. (1962) Adv. Protein Chem. 17 69–165 [Google Scholar]
- 3.Markley, J. L. (1975) Acc. Chem. Res. 8 70–80 [Google Scholar]
- 4.Matthew, J. B., Gurd, F. R., Garcia-Moreno, B., Flanagan, M. A., March, K. L., and Shire, S. J. (1985) Crit. Rev. Biochem. Mol. Biol. 18 91–197 [DOI] [PubMed] [Google Scholar]
- 5.Grimsley, G. R., Scholtz, J. M., and Pace, C. N. (2009) Protein Sci. 18 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tollinger, M., Crowhurst, K. A., Kay, L. E., and Forman-Kay, J. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4545–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debye, P., and Hückel, E. (1923) Phys. Z. 24 185–206 [Google Scholar]
- 8.Linderstrom-Lang, K. U. (1924) C. R. Trav. Lab. Carlsberg 15 1–29 [PubMed] [Google Scholar]
- 9.Tanford, C., and Kirkwood, J. G. (1957) J. Am. Chem. Soc. 79 5333–5339 [Google Scholar]
- 10.Spassov, V. Z., and Yan, L. (2008) Protein Sci. 17 1955–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurlkill, R. L., Grimsley, G. R., Scholtz, J. M., and Pace, C. N. (2006) Protein Sci. 15 1214–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan, I. K., Kondrashov, F. A., Adzhubei, I. A., Wolf, Y. I., Koonin, E. V., Kondrashov, A. S., and Sunyaev, S. (2005) Nature 433 633–638 [DOI] [PubMed] [Google Scholar]
- 13.Lesser, G. J., and Rose, G. D. (1990) Proteins 8 6–13 [DOI] [PubMed] [Google Scholar]
- 14.Mason, P. E., Neilson, G. W., Dempsey, C. E., Barnes, A. C., and Cruickshank, J. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4557–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti, P., and Bhattacharyya, R. (2007) Prog. Biophys. Mol. Biol. 95 83–137 [DOI] [PubMed] [Google Scholar]
- 16.Thurlkill, R. L., Grimsley, G. R., Scholtz, J. M., and Pace, C. N. (2006) J. Mol. Biol. 362 594–604 [DOI] [PubMed] [Google Scholar]
- 17.Harris, T. K., and Turner, G. J. (2002) IUBMB Life 53 85–98 [DOI] [PubMed] [Google Scholar]
- 18.Karp, D. A., Gittis, A. G., Stahley, M. R., Fitch, C. A., Stites, W. E., and Garcia-Moreno, E. B. (2007) Biophys. J. 92 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berisio, R., Lamzin, V. S., Sica, F., Wilson, K. S., Zagari, A., and Mazzarella, L. (1999) J. Mol. Biol. 292 845–854 [DOI] [PubMed] [Google Scholar]
- 20.Warshel, A., Sharma, P. K., Kato, M., and Parson, W. W. (2006) Biochim. Biophys. Acta 1764 1647–1676 [DOI] [PubMed] [Google Scholar]
- 21.Laurents, D. V., Huyghues-Despointes, B. M., Bruix, M., Thurlkill, R. L., Schell, D., Newsom, S., Grimsley, G. R., Shaw, K. L., Trevino, S., Rico, M., Briggs, J. M., Antosiewicz, J. M., Scholtz, J. M., and Pace, C. N. (2003) J. Mol. Biol. 325 1077–1092 [DOI] [PubMed] [Google Scholar]
- 22.Roca, M., Messer, B., and Warshel, A. (2007) FEBS Lett. 581 2065–2071 [DOI] [PubMed] [Google Scholar]
- 23.Baran, K. L., Chimenti, M. S., Schlessman, J. L., Fitch, C. A., Herbst, K. J., and Garcia-Moreno, B. E. (2008) J. Mol. Biol. 379 1045–1062 [DOI] [PubMed] [Google Scholar]
- 24.Takano, K., Scholtz, J. M., Sacchettini, J. C., and Pace, C. N. (2003) J. Biol. Chem. 278 31790–31795 [DOI] [PubMed] [Google Scholar]
- 25.Li, H., Robertson, A. D., and Jensen, J. H. (2005) Proteins 61 704–721 [DOI] [PubMed] [Google Scholar]
- 26.Pace, C. N., Trevino, S., Prabhakaran, E., and Scholtz, J. M. (2004) Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radivojac, P., Iakoucheva, L. M., Oldfield, C. J., Obradovic, Z., Uversky, V. N., and Dunker, A. K. (2007) Biophys. J. 92 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spassov, V. Z., Karshikoff, A. D., and Ladenstein, R. (1994) Protein Sci. 3 1556–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pace, C. N., Laurents, D. V., and Thomson, J. A. (1990) Biochemistry 29 2564–2572 [DOI] [PubMed] [Google Scholar]
- 30.Schweiker, K. L., Zarrine-Afsar, A., Davidson, A. R., and Makhatadze, G. I. (2007) Protein Sci. 16 2694–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi, Y., Cho, J. H., Kim, E. Y., Shan, B., Schindelin, H., and Raleigh, D. P. (2007) Biochemistry 46 7497–7505 [DOI] [PubMed] [Google Scholar]
- 32.Strickler, S. S., Gribenko, A. V., Keiffer, T. R., Tomlinson, J., Reihle, T., Loladze, V. V., and Makhatadze, G. I. (2006) Biochemistry 45 2761–2766 [DOI] [PubMed] [Google Scholar]
- 33.Schwehm, J. M., Fitch, C. A., Dang, B. N., Garcia-Moreno, E. B., and Stites, W. E. (2003) Biochemistry 42 1118–1128 [DOI] [PubMed] [Google Scholar]
- 34.Grimsley, G. R., Shaw, K. L., Fee, L. R., Alston, R. W., Huyghues-Despointes, B. M., Thurlkill, R. L., Scholtz, J. M., and Pace, C. N. (1999) Protein Sci. 8 1843–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trevino, S. R., Schaefer, S., Scholtz, J. M., and Pace, C. N. (2007) J. Mol. Biol. 373 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sickmeier, M., Hamilton, J. A., LeGall, T., Vacic, V., Cortese, M. S., Tantos, A., Szabo, B., Tompa, P., Chen, J., Uversky, V. N., Obradovic, Z., and Dunker, A. K. (2007) Nucleic Acids Res. 35 D786–D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell Biol. 6 197–208 [DOI] [PubMed] [Google Scholar]
- 38.Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000) Proteins 41 415–427 [DOI] [PubMed] [Google Scholar]
- 39.Cho, J. H., Sato, S., Horng, J. C., Anil, B., and Raleigh, D. P. (2008) Arch. Biochem. Biophys. 469 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, J., Mao, J., and Gunner, M. R. (2005) J. Mol. Biol. 348 1283–1298 [DOI] [PubMed] [Google Scholar]
- 41.Anderson, D. E., Becktel, W. J., and Dahlquist, F. W. (1990) Biochemistry 29 2403–2408 [DOI] [PubMed] [Google Scholar]
- 42.Trevino, S. R., Gokulan, K., Newsom, S., Thurlkill, R. L., Shaw, K. L., Mitkevich, V. A., Makarov, A. A., Sacchettini, J. C., Scholtz, J. M., and Pace, C. N. (2005) J. Mol. Biol. 354 967–978 [DOI] [PubMed] [Google Scholar]
- 43.Kajander, T., Kahn, P. C., Passila, S. H., Cohen, D. C., Lehtio, L., Adolfsen, W., Warwicker, J., Schell, U., and Goldman, A. (2000) Struct. Folding Des. 8 1203–1214 [DOI] [PubMed] [Google Scholar]
- 44.Horovitz, A., Serrano, L., Avron, B., Bycroft, M., and Fersht, A. R. (1990) J. Mol. Biol. 216 1031–1044 [DOI] [PubMed] [Google Scholar]
- 45.Kumar, S., and Nussinov, R. (1999) J. Mol. Biol. 293 1241–1255 [DOI] [PubMed] [Google Scholar]
- 46.Petsko, G. A. (2001) Methods Enzymol. 334 469–478 [DOI] [PubMed] [Google Scholar]
- 47.Pace, C. N. (2000) Nat. Struct. Biol. 7 345–346 [DOI] [PubMed] [Google Scholar]
- 48.Sawano, M., Yamamoto, H., Ogasahara, K., Kidokoro, S., Katoh, S., Ohnuma, T., Katoh, E., Yokoyama, S., and Yutani, K. (2008) Biochemistry 47 721–730 [DOI] [PubMed] [Google Scholar]
- 49.Cohn, E. J., and Edsall, J. T. (1943) Proteins, Amino Acids and Peptides, Hafner Publishing Co., New York
- 50.Trevino, S. R., Scholtz, J. M., and Pace, C. N. (2007) J. Mol. Biol. 366 449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.