Abstract
A major mechanism in the cellular defense against oxidative or electrophilic stress is activation of the Nrf2-antioxidant response element signaling pathway, which controls the expression of genes whose protein products are involved in the detoxication and elimination of reactive oxidants and electrophilic agents through conjugative reactions and by enhancing cellular antioxidant capacity. At the molecular level, however, the regulatory mechanisms involved in mediating Nrf2 activation are not fully understood. It is well established that Nrf2 activity is controlled, in part, by the cytosolic protein Keap1, but the nature of this pathway and the mechanisms by which Keap1 acts to repress Nrf2 activity remain to be fully characterized and are the topics of discussion in this minireview. In addition, a possible role of the Nrf2-antioxidant response element transcriptional pathway in neuroprotection will also be discussed.
ARE-mediated Pathway
The induction of many cytoprotective enzymes in response to reactive chemical stress is regulated primarily at the transcriptional level. This transcriptional response is mediated by a cis-acting element termed ARE,2 initially found in the promoters of genes encoding the two major detoxication enzymes, GSTA2 (glutathione S-transferase A2) and NQO1 (NADPH: quinone oxidoreductase 1) (Fig. 1) (1–4). The ARE possesses structural and biological features that characterize its unique responsiveness to oxidative stress (5). It is activated not only in response to H2O2 but specifically by chemical compounds with the capacity to either undergo redox cycling or be metabolically transformed to a reactive or electrophilic intermediate (6). Moreover, compounds that have the propensity to react with sulfhydryl groups such as diethyl maleate, the isothiocyanates, and dithiothiones are also potent inducers of ARE activity. Thus, alteration of the cellular redox status due to elevated levels of ROS and electrophilic species and/or a reduced antioxidant capacity (e.g. glutathione) appears to be an important signal for triggering the transcriptional response mediated by this enhancer.
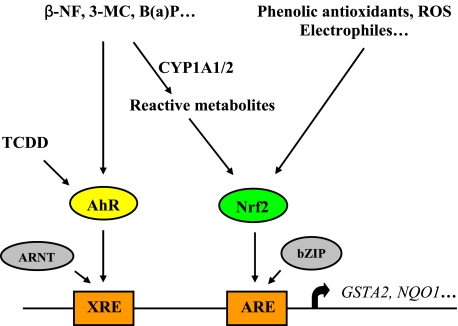
FIGURE 1.
Regulation of rat GSTA2 and NQO1 gene expression. Induction of these two detoxication enzymes is regulated at the transcriptional level mediated by two distinct enhancers, XRE and ARE, controlled by the AhR and Nrf2, respectively. β-Naphthoflavone (β-NF), 3-methylcholanthrene (3-MC), and benzo(a)pyrene (B(a)P) induce expression of these genes through two different pathways, directly by activating the AhR, which, following dimerizing with the aryl hydrocarbon receptor nuclear translocator (ARNT), binds to the XRE to activate transcription, and indirectly via the Nrf2-ARE pathway following their biotransformation to a reactive intermediate. Nrf2 dimerizes with a basic region-leucine zipper (bZIP) protein and binds to the ARE to activate gene transcription. Induction via the AhR-XRE pathway appears to represent an early mechanism of defense against the eventual metabolic transformation of the xenobiotics to their reactive forms. TCDD, 2,3,7,8-tetraclorodibenzo-p-dioxin.
Besides its involvement in inducible gene expression, the ARE is also responsible for the low-level constitutive (or basal) expression of several genes under non-stressed conditions. Because reactive oxygen species and other endogenous reactive molecules are constantly generated from normal aerobic metabolism, the involvement of the ARE in controlling constitutive gene expression implies a critical role of the enhancer in the maintenance of cellular redox homeostasis under both stressed and non-stressed conditions.
Nrf2 Activity and Repression by Keap1
Activation of gene transcription through the ARE is mediated primarily by Nrf2 (nuclear factor E2-related factor 2), first isolated through cloning experiments (7). Following its isolation, Nrf2 was identified as one of the transcription factors acting on the ARE of human NQO1 to activate gene transcription in cell-based transient transfection experiments (8). Similar observations were subsequently made for the AREs of a number of other genes (9). Notably, the involvement of Nrf2 was further corroborated by in vivo studies in which expression of several ARE-dependent genes was found to be severely impaired in nrf2–/– mice (10, 11) and by chromatin immunoprecipitation assays demonstrating direct interaction between endogenous Nrf2 and the ARE in H4IIE cells (12). These studies have also provided evidence that Nrf2 controls both the inducible and constitutive gene expression mediated by the ARE. The significance of Nrf2 having this dual role will be discussed further below, as we attempt to provide a rationale for our understanding of the Nrf2 regulatory pathway.
Nrf2 activity is regulated in part by the actin-associated Keap1 protein, which was initially proposed to act by binding and tethering the transcription factor in the cytoplasm. Activation of Nrf2 in response to stress signals was thought to result from a disruption of this association, releasing Nrf2 for translocation into the nucleus to effect its transcriptional activity (13). Independently, Nrf2 has been found to be a highly unstable protein (t½ ∼ 15 min), subject to proteolytic degradation catalyzed by the 26 S proteasome via the ubiquitin-dependent pathway. In this case, activation of Nrf2 was suggested to be dependent on mechanisms that increase its stability, leading to its accumulation in the cell (14, 15). The unstable nature of the Nrf2 protein and its regulation through this dynamic mechanism suggest that Nrf2 is unlikely to be tethered in a passive complex in the cytoplasm. This was corroborated by a number of studies demonstrating a more active role of Keap1 in its repression of Nrf2 activity. Keap1 appears to promote Nrf2 ubiquitylation in a constitutive manner (12, 16–18) through the cullin-3-dependent pathway (19–22).
That Nrf2 is constantly degraded in non-stressed cells implies that Keap1 is a constitutively active protein and that it promotes Nrf2 ubiquitylation in an unregulated manner. This is supported by the observation that overexpression of Keap1 leads to increased levels of ubiquitin-conjugated forms of Nrf2 in cells (12, 18), indicating that Keap1 is expressed as a functionally active protein. Moreover, the rate of Nrf2 ubiquitylation and its degradation in non-stressed cells appear to be dependent in large part on the abundance of the Keap1 protein in the cell, as suggested by the elevated steady-state levels of Nrf2 observed in keap1–/– animals (23) or following an artificial reduction of the cellular Keap1 protein level by small interfering RNA (24). Notably, these data suggest that upon interaction with Keap1, Nrf2 is targeted directly for ubiquitylation and degradation. Thus, interaction between the two proteins is more likely a transient encounter rather than a sustained association. Given that the steady-state level of Nrf2 in the cell is maintained in part through its constitutive expression, requiring de novo gene transcription and protein synthesis (12, 14), the pathway through which Nrf2 activity is regulated, from synthesis to degradation, requires examination in further detail.
Nrf2-Keap1 Regulatory Pathway
The involvement of Keap1 in promoting Nrf2 degradation led to the recognition that interaction between the two proteins is a dynamic process that must be regulated through a pathway that enables Nrf2 to control both the basal and inducible expression of its genes (12). The fact that Nrf2 controls basal expression of its genes (10, 11) clearly indicates that it is a constitutively and functionally active transcription factor and, notably, implies its presence in the nucleus under homeostatic conditions. That endogenous Nrf2 interacts with the ARE in non-stressed cells not only provides further support to the nuclear nature of Nrf2 but is consistent with its involvement in the basal expression of its genes (12). Thus, the evidence that Nrf2 is localized in the nucleus under constitutive conditions is compelling and does not support the view that Nrf2 co-localizes with Keap1 in the cytoplasm (13, 18, 25–29). The discrepancy of these findings is believed to be due to the nonspecific cross-reactivity of the anti-Nrf2 antibody (C-20, Santa Cruz Biotechnology) used in many, if not all, earlier subcellular localization studies. Compounding this difficulty is the fact that the mobility of the Nrf2 protein on denaturing Tris/glycine-buffered SDS gels does not correspond precisely to its molecular mass (7), making its detection and identification even more tentative. These technical issues therefore call into question of whether localization studies using this antibody provide interpretable and conclusive data, particularly in non-stressed cells, where the Nrf2 protein level is low.
We have reported raising an antibody that reacts with high affinity and specificity to Nrf2 and with minimal cross-reactivity in HepG2 cells (14). Using this antibody, endogenous Nrf2 was observed to localize predominantly in the nucleus of HepG2 and H4IIEC3 cells in the absence of any stress inducers (12). This nuclear localization is not unique to these two cell lines, as similar observations were also made for various other cell types, including primary cells.3 An example of the nuclear localization of Nrf2 in human umbilical vein endothelial cells is shown in Fig. 2. These findings, combined with those obtained from studies using nrf2–/– mice and from chromatin immunoprecipitation experiments discussed above, suggest that Nrf2 is a nuclear protein and is transcriptionally active under both constitutive and stress conditions. Translocation of Nrf2 into the nucleus, following its biosynthesis on ribosomes, therefore does not appear to be a regulated process. As such, activation and accumulation of Nrf2 in the nucleus in response to stress signals are most likely a result of its stabilization, mediated by mechanisms that decrease the rate of its degradation.
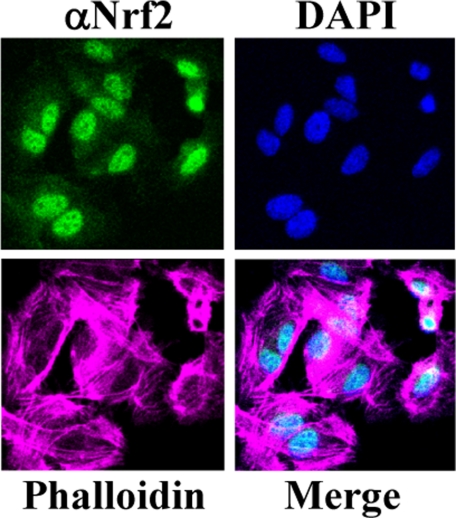
FIGURE 2.
Nuclear localization of Nrf2 in human umbilical vein endothelial cells. Localization of Nrf2 was performed by immunocytochemistry and confocal microscopy. Nrf2 was stained with an anti-Nrf2 antibody (14) and visualized with a secondary antibody conjugated with Alexa Fluor 488. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI), and the F-actin filaments were stained with Alexa Fluor 680-conjugated phalloidin.
An important question is how Keap1 targets Nrf2 for ubiquitylation, given their localization in distinct cellular compartments. It appears that Keap1 is capable of engaging in nucleocytoplasmic shuttling activity, as shown by several studies (12, 30, 31) and similarly observed in a more recent report (32). This shuttling activity by Keap1 was not observed, however, in another study (33), and the reason for this discrepancy is not understood. Work from our laboratory suggests that Keap1 transiently enters the nucleus and targets Nrf2 for ubiquitylation in this compartment. This was based on the observation that inhibition of proteasome activity by MG132 caused accumulation of both intact and ubiquitylated forms of Nrf2 in the nucleus, indicating that both the ubiquitylation and degradation processes are likely initiated and end in this compartment (12). In contrast, it has been suggested that, in addition to its role in promoting cytoplasmic Nrf2 degradation under non-stressed conditions, Keap1 also enters the nucleus and escorts Nrf2 out to the cytoplasm for degradation under stressed conditions. This conclusion was based on the observation that Nrf2 was localized primarily in the cytoplasm in cells cotransfected with wild-type Keap1 but was localized mainly in the nucleus in cells cotransfected with a nuclear export signal mutant analog of Keap1, which was trapped in the nucleus. However, in subcellular fractionation analyses from the same study, Nrf2 was found to be distributed largely in the nuclear fraction compared with the cytoplasmic fraction, whether from cells cotransfected with wild-type Keap1 or with the nuclear export signal mutant form of Keap1. The reason for these contradictory results is not clear (32).
Besides the experimental evidence discussed above, there are several other observations that are difficult to reconcile with the proposal that Keap1 targets Nrf2 for degradation in the cytoplasm and upstream of its transactivation function (i.e. without Nrf2 being able to drive transcription of its genes at the basal level) and that inhibition of Keap1 activity by stress signals promotes Nrf2 translocation into the nucleus (13, 18, 25, 26, 34–36). For instance, because Nrf2 is constitutively expressed in the cell, such a regulatory pathway would imply that Nrf2 is targeted for degradation by Keap1 directly following its de novo synthesis, therefore representing a rather inefficient mechanism of gene regulation with respect to cellular energy usage. In addition, despite its high rate of turnover, the fact that Nrf2 is expressed at a steady-state level in the cell indicates that there is a time-elapsed interval (i.e. t½ ∼ 15 min) following its biosynthesis and prior to its degradation. Such an interval might represent a time window during which Nrf2 transactivates its genes, fulfilling its role in driving their constitutive expression. Nuclear shuttling by Keap1 would provide an efficient mechanism by facilitating the rapid degradation of Nrf2 following transcriptional activation of its genes. The precise mechanism by which Nrf2 is targeted for degradation by Keap1 is not well understood. However, it is noteworthy that the stability of many transcription factors has been linked to the potency of their transactivation domain, which is frequently found to encompass or overlap with their degron (37). Because Nrf2 is highly unstable and possesses a potent transactivation domain adjacent to the Keap1-interacting Neh2 domain (38, 39), it is possible that Nrf2 is targeted for degradation by mechanisms linked to its transcriptional activity. On the basis of experimental and biological evidence discussed here, the regulatory pathway controlling Nrf2 stability by Keap1 is summarized in Fig. 3.
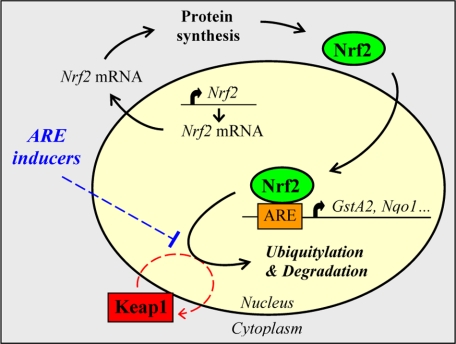
FIGURE 3.
Proposed Nrf2-ARE signaling pathway. Nrf2 is expressed constitutively in the cell and translocates directly to the nucleus following its synthesis. Following transactivation of its genes, Nrf2 is targeted for degradation by Keap1 in the nucleus, a process that requires the transient shuttling of Keap1 into this compartment. In cells under stress, stabilization of Nrf2 is thought to be dependent on mechanisms that either prevent or reduce access of Keap1 to Nrf2. This figure was reproduced from Ref. 12.
Molecular Mechanisms Controlling Nrf2 Stabilization
The stabilization of Nrf2 in response to reactive chemical stress is most likely regulated by mechanisms leading to a decrease in the rate of its degradation. In earlier studies examining Nrf2 stability using the translation inhibitor cycloheximide (14, 15), the half-life of Nrf2 was found to extend from ∼15 min in untreated cells to ∼30 min in cells exposed to a stress-inducing agent, indicating that Nrf2 is still subject to a high, albeit slower, rate of degradation under stress conditions. Thus, besides a regulatory mechanism promoting Nrf2 stabilization in response to stress, there appears to be a constitutive mechanism promoting its degradation in cells under stress, presumably to prevent its accumulation in an uncontrolled manner. Whether these two mechanisms operate independently or in a coordinate fashion is not currently understood and remains to be investigated.
A well established mechanism that controls Nrf2 stability is that mediated by Keap1. Because of the large number of Cys residues within its primary structure, Keap1 presents an attractive target for potential regulation by thiol-reactive chemical species, and hence, inhibitory modulation of its activity was suggested to be an important mechanism for Nrf2 activation (13, 18, 25, 26). Indeed, a number of reactive Cys residues (at positions 257, 273, 288, and 297) in Keap1 have been identified by in vitro labeling studies as putative sites of attack by electrophilic species and were thought to be important for regulation of its interaction with Nrf2 (25). This notion was strengthened by cell-based studies showing that targeted mutation of Cys151, Cys273, or Cys288 adversely affects Keap1 functional activity (18). Keap1 was found to undergo a yet to be characterized Cys151-dependent modification in cells exposed to t-butylhydroquinone, producing a Keap1 species with reduced mobility on denaturing gels. The significance and nature of this modification were not clear, however, as only a small fraction of the Keap1 protein population was affected by the t-butylhydroquinone treatment (18). In an independent study, mutation of Cys273 or Cys288 was similarly observed to abrogate the repressor activity of Keap1, and this was suggested to be due to the inability of the Keap1 mutant to form an intermolecular disulfide bond needed to release Nrf2 (26). Separately, it has been reported that modifications of Keap1 cysteine residues by electrophilic compounds cause the target substrate for ubiquitylation to be switched from Nrf2 to Keap1 (34, 35). More recently, molecular interaction between Nrf2 and Keap1, whether as wild-type or Cys mutant analogs, was found to be unaffected by different electrophilic inducers (36). This was consistent with an earlier study in which modifications of reactive cysteine residues within the human Keap1 protein were found to have no effects on its binding to the Neh2 domain of Nrf2 (40). Thus, activation of Nrf2 appears to involve mechanisms that are more complex than physical release from an association with Keap1 following modifications of the reactive cysteines.
Nevertheless, data from these studies provide evidence for a mechanism of regulation of Keap1 activity through its reactive cysteine residues. The precise mechanisms leading to Nrf2 stabilization remain to be elucidated, however. Based on our findings showing the nuclear nature of Nrf2 (12), the interaction between Nrf2 and Keap1 is believed to represent a transient event taking place in the nucleus and following the shuttling of Keap1 into this compartment. Control of Nrf2 stability by Keap1 is regulated, spatially and temporally, by mechanisms that restrict their interaction to a specific time and place. It is suggested that stabilization of Nrf2 in response to electrophilic inducers is mediated by mechanisms that either prevent or reduce access of Keap1 to the Nrf2 molecule. In that regard, the reactive cysteines in Keap1 might be important in the regulation of this process. Such mechanisms do not, however, involve regulation of the nucleocytoplasmic shuttling activity of Keap1 (12, 36).
Regulation of Nrf2 activation has also been reported to be mediated by various major protein kinase pathways (27–29, 41–44). Although phosphorylation of Nrf2 has been demonstrated in several studies (27–29), none of the phosphorylations has been conclusively linked to its stabilization. Because these kinase pathways control numerous signaling processes in the cell, additional studies are required to explore whether they are intimately involved in the regulation of Nrf2 protein stability in response to oxidative stress.
Nrf2-ARE Pathway as a Therapeutic Target for Treatment of Neurodegenerative Diseases
The pathogenesis of many neurodegenerative diseases is thought to be associated with oxidative stress due to the accumulation of ROS. Previous work from our laboratory demonstrated that compounds that undergo redox cycling to form ROS as well as oxidants such as H2O2 activate the Nrf2-ARE transcriptional pathway (5). Work from other laboratories has demonstrated that activation of the Nrf2-ARE pathway provides protection from glutamate- and H2O2-induced cell death (45, 46). Furthermore, astrocytes expressing Nrf2 protect neurons from oxidative stress (46). nrf2–/– mice display vacuolar leukoencephalopathy, which is characterized by vascular degeneration involving all major brain regions. In white tracts, the vascular degeneration is accompanied by widespread reactive gliosis (47). Finally, preliminary data suggest that the development of experimental autoimmune encephalomyelitis, which is an experimental model of multiple sclerosis, is exacerbated in nrf2–/–, mice suggesting a possible role of the Nrf2-ARE pathway in the pathogenesis of multiple sclerosis (48).
A compound currently in clinical testing for the treatment of multiple sclerosis is an oral formulation of dimethyl fumarate, which is known as BG12. Preclinical experiments have demonstrated that BG12 can activate the Nrf2-ARE pathway (49–51). It also inhibits the expression of adhesion molecules and cytokines, suggesting potential anti-inflammatory properties. In a randomized, double-blind, placebo-controlled, dose-ranging phase 2b study, BG12 significantly reduced brain MRI activity in patients with relapsing remitting multiple sclerosis (52). These data suggest that BG12 may play a role in the initial therapy of relapsing-remitting multiple sclerosis in patients who cannot tolerate or choose not to start parenteral therapy. A phase 3 clinical study is currently underway in larger patient populations to confirm the findings of the phase 2b study.
In summary, the Nrf2-ARE transcriptional pathway plays an important role in the regulation of genes that control the expression of proteins critical in the detoxication and elimination of ROS and electrophiles. Surprisingly, activation of the Nrf2-ARE pathway may also be important in treatment of multiple sclerosis. Whether patients with other neurodegenerative diseases such as Alzheimer or Parkinson disease might benefit from activation of this pathway remains to be determined.
Supplementary Material
This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: ARE, antioxidant response element; ROS, reactive oxygen species; XRE, xenobiotic response element; AhR, aryl hydrocarbon receptor.
T. Nguyen, P. Nioi, and C. B. Pickett, unpublished data.
References
- 1.Rushmore, T. H., and Pickett, C. B. (1990) J. Biol. Chem. 265 14648–14653 [PubMed] [Google Scholar]
- 2.Friling, R. S., Bensimon, A., Tichauer, Y., and Daniel, V. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6258–6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favreau, L. V., and Pickett, C. B. (1991) J. Biol. Chem. 266 4556–4561 [PubMed] [Google Scholar]
- 4.Li, Y., and Jaiswal, A. K. (1992) J. Biol. Chem. 267 15097–15104 [PubMed] [Google Scholar]
- 5.Rushmore, T. H., Morton, M. R., and Pickett, C. B. (1991) J. Biol. Chem. 266 11632–11639 [PubMed] [Google Scholar]
- 6.Rushmore, T. H., King, R. G., Paulson, K. E., and Pickett, C. B. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3826–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moi, P., Chan, K., Asunis, I., Cao, A., and Kan, Y. W. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9926–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venugopal, R., and Jaiswal, A. K. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen, T., Sherratt, P. J., and Pickett, C. B. (2003) Annu. Rev. Pharmacol. Toxicol. 43 233–260 [DOI] [PubMed] [Google Scholar]
- 10.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., Yamamoto, M., and Nabeshima, Y. I. (1997) Biochem. Biophys. Res. Commun. 236 313–322 [DOI] [PubMed] [Google Scholar]
- 11.McMahon, M., Itoh, K., Yamamoto, M., Chanas, S. A., Henderson, C. J., McLellan, L. I., Wolf, C. R., Cavin, C., and Hayes, J. D. (2001) Cancer Res. 61 3299–3307 [PubMed] [Google Scholar]
- 12.Nguyen, T., Sherratt, P. J., Nioi, P., Yang, C. S., and Pickett, C. B. (2005) J. Biol. Chem. 280 32485–32492 [DOI] [PubMed] [Google Scholar]
- 13.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., Igarashi, K., Engel, J. D., and Yamamoto, M. (1999) Genes Dev. 13 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen, T., Sherratt, P. J., Huang, H.-C., Yang, C. S., and Pickett, C. B. (2003) J. Biol. Chem. 278 4536–4541 [DOI] [PubMed] [Google Scholar]
- 15.Stewart, D., Killeen, E., Naquin, R., Alam, S., and Alam, J. (2003) J. Biol. Chem. 278 2396–2402 [DOI] [PubMed] [Google Scholar]
- 16.McMahon, M., Itoh, K., Yamamoto, M., and Hayes, J. D. (2003) J. Biol. Chem. 278 21592–21600 [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., O'Connor, T., and Yamamoto, M. (2003) Genes Cells 8 379–391 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, D. D., and Hannink, M. (2003) Mol. Cell. Biol. 23 8137–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K., and Yamamoto, M. (2004) Mol. Cell. Biol. 24 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W., and Diehl, J. A. (2004) Mol. Cell. Biol. 24 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, D. D., Lo, S.-C., Cross, J. V., Templeton, D. J., and Hannink, M. (2004) Mol. Cell. Biol. 24 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furukawa, M., and Xiong, Y. (2005) Mol. Cell. Biol. 25 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakabayashi, N., Itoh, K., Wakabayashi, J., Motohashi, H., Noda, S., Takahashi, S., Imakado, S., Kotsuji, T., Otsuka, F., Roop, D. R., Harada, T., Engel, J. D., and Yamamoto, M. (2003) Nat. Genet. 35 238–245 [DOI] [PubMed] [Google Scholar]
- 24.Devling, T. W., Lindsay, C. D., McLellan, L. I., McMahon, M., and Hayes, J. D. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinkova-Kostova, A. T., Holtzclaw, W. D., Cole, R. N., Itoh, K., Wakabayashi, N., Katoh, Y., Yamamoto, M., and Talalay, P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakabayashi, N., Dinkova-Kostova, A. T., Holtzclaw, W. D., Kang, M. I., Kobayashi, A., Yamamoto, M., Kensler, T. W., and Talalay, P. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, H.-C., Nguyen, T., and Pickett, C. B. (2002) J. Biol. Chem. 277 42769–42774 [DOI] [PubMed] [Google Scholar]
- 28.Bloom, D. A., and Jaiswal, A. K. (2003) J. Biol. Chem. 278 44675–44682 [DOI] [PubMed] [Google Scholar]
- 29.Cullinan, S. B., Zhang, D., Hannink, M., Arvisais, E., Kaufman, R. J., and Diehl, J. A. (2003) Mol. Cell. Biol. 23 7198–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karapetian, R. N., Evstafieva, A. G., Abaeva, I. S., Chichkova, N. V., Filonov, G. S., Rubtsov, Y. P., Sukhacheva, E. A., Melnikov, S. V., Schneider, U., Wanker, E. E., and Vartapetian, A. B. (2005) Mol. Cell. Biol. 25 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velichkova, M., and Hasson, T. (2005) Mol. Cell. Biol. 25 4501–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, Z., Zhang, S., Chan, J. Y., and Zhang, D. D. (2007) Mol. Cell. Biol. 27 6334–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watai, Y., Kobayashi, A., Nagase, H., Mizukami, M., McEvoy, J., Singer, J. D., Itoh, K., and Yamamoto, M. (2007) Genes Cells 12 1163–1178 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, D. D., Lo, S.-C., Sun, Z., Habib, G. M., Lieberman, M. W., and Hannink, M. (2005) J. Biol. Chem. 280 30091–30099 [DOI] [PubMed] [Google Scholar]
- 35.Hong, F., Sekhar, K. R., Freeman, M. L., and Liebler, D. C. (2005) J. Biol. Chem. 280 31768–31775 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi, A., Kang, M. I., Watai, Y., Tong, K. I., Shibata, T., Uchida, K., and Yamamoto, M. (2006) Mol. Cell. Biol. 26 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muratani, M., and Tansey, W. P. (2003) Nat. Rev. Mol. Cell Biol. 4 1–10 [DOI] [PubMed] [Google Scholar]
- 38.Katoh, Y., Itoh, K., Yoshida, E., Miyagishi, M., Fukamizu, A., and Yamamoto, M. (2001) Genes Cells 6 857–868 [DOI] [PubMed] [Google Scholar]
- 39.Shen, G., Hebbar, V., Nair, S., Xu, C., Li, W., Lin, W., Keum, Y. S., Han, J., Gallo, M. A., and Kong, A. N. (2004) J. Biol. Chem. 279 23052–23060 [DOI] [PubMed] [Google Scholar]
- 40.Eggler, A. L., Liu, G., Pezzuto, J. M., van Breemen, R. B., and Mesecar, A. D. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10070–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Numazawa, S., Ishikawa, M., Yoshida, A., Tanaka, S., and Yoshida, T. (2003) Am. J. Physiol. 285 C334–C342 [DOI] [PubMed] [Google Scholar]
- 42.Kang, K. W., Cho, M. K., Lee, C. H., and Kim, S. G. (2001) Mol. Pharmacol. 59 1147–1156 [DOI] [PubMed] [Google Scholar]
- 43.Lee, J.-M., Hanson, J. M., Chu, W. A., and Johnson, J. A. (2001) J. Biol. Chem. 276 20011–20016 [DOI] [PubMed] [Google Scholar]
- 44.Nakaso, K., Yano, H., Fukuhara, Y., Takeshima, T., Wada-Isoe, K., and Nakashima, K. (2003) FEBS Lett. 546 181–184 [DOI] [PubMed] [Google Scholar]
- 45.Shih, A. Y., Johhnson, D. A., Wong, G., Kraft, A. D., Jiang, L., Erb, H., Johnson, J. A., and Murphy, T. H. (2003) J. Neurosci. 23 3394–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraft, A. D., Johnson, D. A., and Johnson, J. A. (2004) J. Neurosci. 24 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbs, A. F., Benkovic, S. A., Miller, D. B., O'Callaghan, J. P., Battelli, L., Schwegler-Berry, D., and Ma, Q. (2007) Am. J. Pathol. 170 2068–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson, D. A., Resch, J. M., Fabry, Z., and Johnson, J. A. (2007) SfN Neuroscience Meeting Planner, Program No. 901.26, Society for Neuroscience, San Diego, CA
- 49.Gröger, M., Holnthoner, W., Wolff, K., Wiegrebe, W., Jirovsky, D., and Petzelbauer, P. J. Investig. Dermatol. 117 1363–1368 [DOI] [PubMed]
- 50.Wierinckx, A., Breve, J., Mercier, D., Schultzberg, M., Drukarch, B., and Van Dam, A.-M. (2005) J. Neuroimmunol. 166 132–143 [DOI] [PubMed] [Google Scholar]
- 51.Schilling, S., Goelz, S., Linker, R., and Luehder, R. (2006) Clin. Exp. Immunol. 145 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmierer, K., Yousry, T. A., Yang, M., Eraksoy, M., Meluzinova, E., Rektor, I., Dawson, K. T., Sandrock, A. W., and O'Neill, G. N. (2008) Lancet 372 1463–147218970976 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.