Abstract
Mutation of CCM2 predisposes individuals to cerebral cavernous malformations, vascular abnormalities that cause seizures and hemorrhagic stroke. CCM2 has been proposed to regulate the activity of RhoA for maintenance of vascular integrity. Herein, we define a novel mechanism where the CCM2 phosphotyrosine binding (PTB) domain binds the ubiquitin ligase (E3) Smurf1, controlling RhoA degradation. Brain endothelial cells with knockdown of CCM2 have increased RhoA protein and display impaired directed cell migration. CCM2 binding of Smurf1 increases Smurf1-mediated degradation of RhoA. CCM2 does not significantly alter the catalytic activity of Smurf1, nor is CCM2 a Smurf1 substrate. Rather the CCM2-Smurf1 interaction functions to localize Smurf1 for RhoA degradation. These findings provide a molecular mechanism for the pathogenesis of cerebral cavernous malformations (CCM) resulting from loss of CCM2-mediated localization of Smurf1, which controls RhoA degradation required for maintenance of normal endothelial cell physiology.
We previously characterized a scaffold-like protein named osmosensing scaffold for MEKK3 (OSM) for its ability to bind actin and localize to Rac-containing membrane ruffles and its obligate requirement for p38 activation in response to hyperosmotic stress (1). Subsequently, the gene encoding OSM, CCM2, was found to be mutated in the human disease cerebral cavernous malformations (CCM)2 (2). Cerebral cavernous malformations are vascular lesions of the central nervous system characterized as clusters of dilated, thin walled blood vessels. CCM lesions are fragile and prone to vascular leakiness and rupture, leading to hemorrhages that cause seizure and stroke (3, 4).
Recently, CCM2 knockdown endothelial cells were shown to have increased activation of RhoA (5), although the mechanism was not defined. Herein, we demonstrate a molecular mechanism for activation of this pathway. Through a novel CCM2 PTB domain interaction with the Smurf1 homologous to the E6-AP C terminus (HECT) domain, we now show that CCM2 binds the E3 ligase Smurf1 for the control of RhoA degradation.
EXPERIMENTAL PROCEDURES
RNAi—bEND.3 cells were purchased from ATCC. pLKO.1 and pLKO.1 CCM2 shRNA plasmids were purchased from Open Biosystems. Lentiviral infection was done according to the RNAi consortium protocol.
In Vitro Ubiquitination Assay—The in vitro ubiquitination assay was done as described (6).
In Vitro Wound Healing Assay—bEND.3 cells stably expressing pLKO.1 or CCM2 shRNAs were transduced with pLL5.0 for expression of green fluorescent protein. Cells were grown in 24-well dishes to a monolayer and imaged on the BD Biosciences Pathway high content imager. Wounds were imaged every 15 min for 16 h on a ×10 objective. The percentage of wound healing was determined as the area of the wound remaining after 16 h divided by the initial area of the wound (ImageJ).
RESULTS AND DISCUSSION
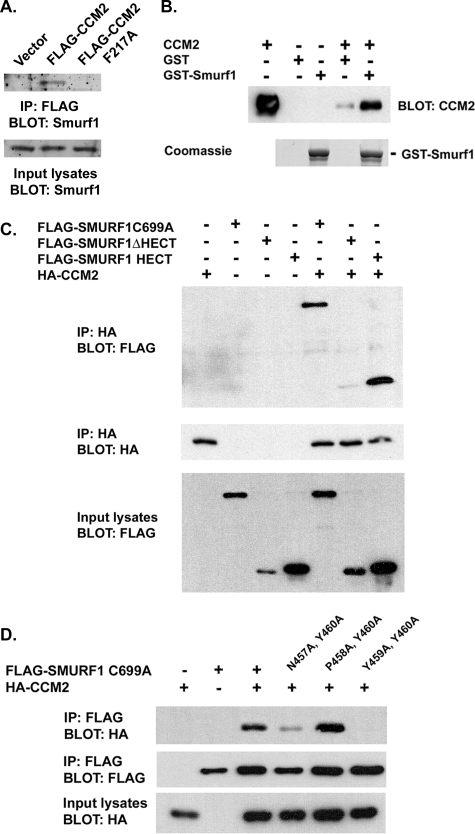
CCM2 Binds Smurf1—We have previously demonstrated that MEKK3, a Ste11-like MAP3K, regulates the p38 pathway in mammalian cells in response to hyperosmolarity (1). MEKK3 is highly homologous to MEKK2, and both have a conserved proline-rich motif that, in MEKK2, has been shown to bind the E3 ubiquitin ligase Smurf1 (7). Therefore, we asked whether Smurf1 was found in the CCM protein complex (8), possibly by binding to the PY motif of MEKK3. Cells stably expressing FLAG-tagged wild type CCM2 or a CCM2 PTB domain mutant (CCM2-F217A) that is unable to bind NPXY motifs in target proteins were used in co-immunoprecipitation assays to define endogenous proteins that bind to the CCM protein complex (8). Fig. 1A shows that endogenous Smurf1 co-immunoprecipitated with FLAG-CCM2 but not CCM2-F217A, indicating that Smurf1 associates with the CCM2 protein complex in a PTB domain-dependent manner.
FIGURE 1.
Smurf1 binds CCM2 via a novel PTB domain-HECT domain interaction. A, RAW264.7 stable cell lines expressing empty vector, FLAG-CCM2, or FLAG-CCM2 F217A were lysed and incubated with anti-FLAG-coated Sepharose beads. The associated endogenous Smurf1 was examined by Western blot using anti-Smurf1 antibody. IP, immunoprecipitation. B, the Smurf1-CCM2 interaction is direct. Purified GST or GST-Smurf1 was incubated with purified His-CCM2, and associating CCM2 was determined by Western blot using anti-CCM2 antibody. C, cells expressing HA-CCM2 with FLAG-Smurf1 C699A (a catalytically inactive mutant of Smurf1 used to examine protein-protein interactions), FLAG-Smurf1ΔHECT, or FLAG-Smurf1 HECT were lysed and immunoprecipitated with anti-HA antibody. The associating proteins were determined by Western blot with anti-FLAG antibody. D,an NPYY motif within the HECT domain of Smurf1 is responsible for interaction with CCM2. Cells expressing HA-CCM2 and FLAG-Smurf1 C699A, FLAG-Smurf1 N457A/Y460A/C699A, FLAG-Smurf1 P458A/Y460A/C699A, or FLAG-Smurf1 Y459A/Y460A/C699A were lysed and immunoprecipitated with anti-FLAG antibody. Associated HA-CCM2 was determined by Western blot using anti-HA antibody.
MEKK3 binds CCM2 independent of the CCM2 PTB domain (8), suggesting that Smurf1 association with the CCM complex is independent of MEKK3. However, we found that Smurf1 was also able to associate with MEKK3 by co-immunoprecipitation (supplemental Fig. 1A). Using purified proteins, CCM2 and MEKK3 both directly bind GST-Smurf1 (Fig. 1B and supplemental Fig. 1B). From this we conclude that Smurf1 and CCM2 are binding partners both in vivo and in vitro.
CCM2 Binds Smurf1 via a PTB Domain-HECT Domain Interaction—Smurf1 is a member of the NEDD4 family of E3 ligases, which have a common domain structure of an N-terminal C2 domain followed by multiple WW domains and a C-terminal HECT domain (9). Smurf1 constructs were generated that encoded either the N-terminal moiety of Smurf1 (FLAG-Smurf1 ΔHECT containing the C2 and WW domains) or the C-terminal moiety of Smurf1 (FLAG-Smurf1 HECT encoding only the HECT domain). Co-expression of the Smurf1 moieties with CCM2 indicated that CCM2 predominantly bound the HECT domain with little binding to the N-terminal portion of Smurf1 (Fig. 1C), indicating that CCM2 was binding specifically to the HECT domain. Consistent with this, the HECT domain of Smurf1 was sufficient to bind CCM2 by co-immunoprecipitation (supplemental Fig. 1C). To our knowledge, this is the first demonstration of a PTB domain-HECT domain interaction.
The consensus PTB-binding motif is Asn-Pro-Xaa-Tyr (NPXY), where Xaa and X represent any amino acid (10). Examination of the amino acid sequence of the Smurf1 HECT domain identified an NPYY motif (amino acids 457–460) that is also conserved in the Smurf2 HECT domain. As would be predicted, CCM2 can bind Smurf1 or Smurf2 (supplemental Fig. 1D). The tyrosine of the NPXY motif is critical for interaction of binding partners with many PTB domains (10). Mutation of Tyr-460 alone within the Smurf1 NPYY motif to alanine had little effect on interaction with CCM2 by co-immunoprecipitation (data not shown). Engineering combinations of mutations involving two mutations within the NPYY motif of Smurf1 determined that N457A/Y460A (APYA) had a partial inhibition of association, whereas mutation at residues Tyr-459 and Tyr-460 (NPAA, referred to as Smurf1 YY-AA below) caused a total loss of association with CCM2 (Fig. 1D). These results indicate that the PTB domain of CCM2 requires the NPYY motif within the Smurf1 HECT domain for binding.
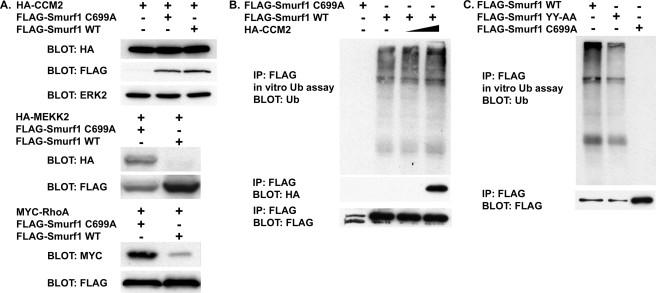
CCM2 Is Not a Smurf1 Substrate, nor Does It Affect Smurf1 Catalytic Activity—To determine the function of the CCM2-Smurf1 interaction, we first examined whether CCM2 was a Smurf1 substrate. In cells expressing Smurf1 WT or the catalytically inactive Smurf1 C699A, there was no change in abundance of CCM2 relative to control transfected cells, although we did detect loss of known Smurf1 substrates MEKK2 and RhoA (Fig. 2A) (7, 11). As the HECT domain catalyzes the ubiquitination of Smurf1 substrates, binding of CCM2 to the Smurf1 HECT domain could therefore regulate the catalytic activity of Smurf1. Based on in vitro ubiquitination assays, we were not able to detect any substantial change in Smurf1 catalytic activity in the presence of CCM2 (Fig. 2, B and C). These results indicate that CCM2 is not a substrate for Smurf1, nor does it regulate Smurf1 catalytic activity.
FIGURE 2.
CCM2 is not a Smurf1 substrate, nor does it impact Smurf1 catalytic activity. A, cells expressing HA-CCM2, HA-MEKK2, Myc-RhoA, FLAG-Smurf1 C699A, or FLAG-Smurf1 WT were lysed, and steady-state levels of HA-CCM2, HA-MEKK2, or Myc-RhoA were determined by Western blot. B, cells expressing FLAG-Smurf1 C699A, FLAG-Smurf1 WT, or HA-CCM2 were lysed, and FLAG-Smurf1 proteins were immunopurified (IP). The immunopurified FLAG-Smurf1 or FLAG-Smurf1 C699A was subsequently used for an in vitro ubiquitination assay. C, cells expressing FLAG-Smurf1 C699A, FLAG-Smurf1 WT, or FLAG-Smurf1 YY-AA were lysed, and immunopurified FLAG-Smurf1 was used for in vitro ubiquitination.
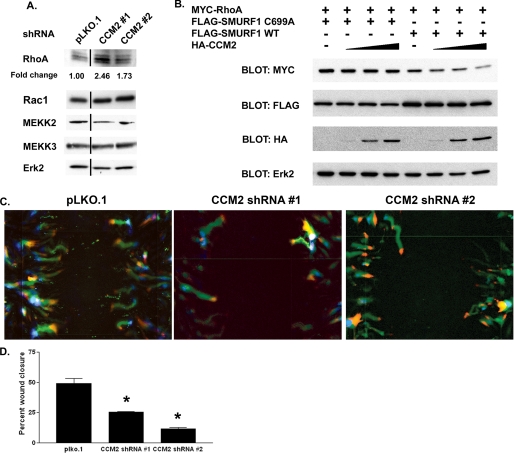
CCM2 Regulates RhoA Degradation—To determine whether CCM2 regulates degradation of Smurf1 substrates, we utilized RNAi. Using a lentivirus-based shRNA system, brain microvascular endothelial cells (bEND.3) were infected with either a control vector (pLKO.1) or two independent CCM2 shRNA vectors. These shRNA vectors were found to be effective at knocking down CCM2 expression by 90% (supplemental Fig. 2A). Cells expressing shRNA for CCM2 had increased levels of RhoA as determined by immunoblotting (Fig. 3A). These cells did not show a significant increase in RhoA message (supplemental Fig. 2B), suggesting that the increased RhoA detected was occurring post-transcriptionally. Importantly, knockdown of CCM2 did not affect abundance of the Smurf1 substrate MEKK2, the Smurf1-interacting protein MEKK3, or the GTPase Rac1 (Fig. 3A). These findings indicate that loss of CCM2 expression selectively facilitates degradation of RhoA but not other Smurf1 substrates such as MEKK2.
FIGURE 3.
CCM2 regulates Smurf1-medated RhoA abundance and migration of brain endothelial cells. A, bEND.3 cells expressing pLKO.1 or CCM2 shRNAs were lysed, and the abundance of RhoA, Rac1, MEKK2, and MEKK3 was analyzed by Western blot. Erk2 was measured as a loading control. -Fold change was calculated by determining the RhoA intensity relative to the Erk2 intensity for that sample standardized to the RhoA/Erk2 ratio for cells expressing pLKO.1. The line denotes where lanes were removed for clarity. B, HEK293 cells were transfected with Myc-RhoA, FLAG-Smurf1 WT, FLAG-Smurf1 C699A, and increasing amounts of HA-CCM2. Cells were subsequently lysed and analyzed by Western blot. C, bEND.3 cells grown to confluency were subjected to an in vitro wound healing assay. Cells were imaged every 15 min for 16 h. The paths taken by cells (green) expressing either pLKO.1 or CCM2 shRNAs demonstrates decreased migration in CCM2 knockdown cells. The starting point of the cell is shown in blue, and its final location is shown in red. D, quantitation of in vitro wound healing assays. Cells lacking CCM2 expression are less able to close a wound as measured by the percentage of wound closure. Error bars,*, p < 0.02.
To determine whether CCM2 facilitates Smurf1-mediated degradation of RhoA, we expressed increasing amounts of CCM2 in combination with Smurf1 and RhoA (Fig. 3B). Cells expressing Smurf1 C699A and increasing amounts of CCM2 had little effect on RhoA levels. In contrast, increasing amounts of CCM2 in the presence of wild type Smurf1 led to significant loss of RhoA protein, consistent with CCM2 regulating Smurf1-dependent RhoA degradation.
CCM2 Regulates Endothelial Cell Migration—Consistent with our observation that CCM2 was capable of regulating RhoA levels, knockdown of CCM2 led to increased stress fiber formation in brain endothelial cells. CCM2 knockdown cells had increased association of phosphorylated myosin light chain 2 with these stress fibers. Treatment of CCM2 knockdown cells with the Rho kinase (ROCK) inhibitor Y-27632 was able to reverse this effect (supplemental Fig. 2, C–E). To determine whether these cytoskeletal changes impacted cell migration, we performed wound healing assays. bEND.3 cells expressing a control vector were capable of healing 50% of the wound in 16 h; however, cells lacking CCM2 were only capable of closing 25% of the wound in the same period of time (Fig. 3, C and D). This finding is consistent with increased RhoA abundance as RhoA degradation is necessary for cytoskeletal turnover and migration. The defects we observed in CCM2 knockdown cells were not limited to migration. We observed increased permeability of endothelial cell monolayers and decreased tubule formation in CCM2 knockdown cells (supplemental Fig. 3). Therefore, in addition to migration, endothelial tubule formation and maintenance of a permeability barrier require CCM2.
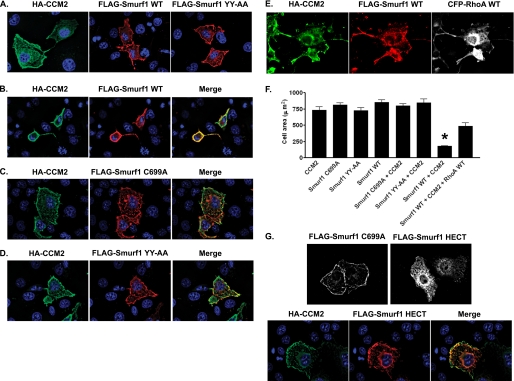
Co-expression of CCM2 and Smurf1 Leads to Cell Rounding—When expressed in COS-7 cells, CCM2 was localized in the cytoplasm with enhanced localization at the plasma membrane (Fig. 4A). Smurf1 was also localized at the cell periphery, a function of the C2 domain of NEDD4 family members (12). When co-expressed, Smurf1 and CCM2 co-localized to the plasma membrane and induced a dramatic rounding morphology (Fig. 4B). Cell rounding was not observed in cells expressing CCM2 or Smurf1 alone nor in cells expressing CCM2 and Smurf1 C699A (Fig. 4, A and C). Furthermore, cells co-expressing CCM2 and Smurf1 YY-AA, which cannot bind CCM2, did not show a cell-rounding phenotype (Fig. 4D), suggesting that the CCM2-Smurf1 complex is regulating the cytoskeleton. Importantly, cell spreading was rescued with expression of exogenous RhoA (Fig. 4, E and F), signifying that cell rounding was due to loss of RhoA protein.
FIGURE 4.
Expression of Smurf1 WT and CCM2 leads to cell morphology changes. A, COS-7 cells expressing HA-CCM2, FLAG-Smurf1 WT, or FLAG-Smurf1 YY-AA were fixed and stained with anti-HA antibody or anti-FLAG antibody. B, COS-7 cells expressing HA-CCM2 and Smurf WT display cell rounding, whereas cells expressing HA-CCM2 and FLAG-Smurf1 C699A (C) or HA-CCM2 and FLAG-Smurf1 YY-AA (D) do not. E, expression of exogenous RhoA (CFP-RhoA WT, where CFP indicates cyan fluorescent protein) rescues cell spreading in cells expressing CCM2, Smurf1 WT, and RhoA WT. F, quantitation of cell spreading. The area of cell spreading was measured using ImageJ, n = 20. Error bars represent S.E. *, p < 0.0001 relative to cells expressing CCM2 alone. G, the Smurf1 HECT domain does not localize to the plasma membrane, and CCM2 recruits the Smurf1 HECT domain to the plasma membrane. COS-7 cells expressing HA-CCM2, FLAG-Smurf1 C699A, or FLAG-Smurf1 HECT were fixed and stained with anti-FLAG and anti-HA antibodies.
CCM2 Localizes Smurf1 by Binding the HECT Domain—We have shown that CCM2 regulates localization of the MEKK3-MKK3-p38 signaling module and localization of KRIT1 (1, 13). Therefore, we hypothesized that CCM2 regulates Smurf1 localization. The C2 domain of Smurf1 localizes Smurf1 to the plasma membrane. Therefore, CCM2 binding is not required for membrane localization of Smurf1 but would be predicted to localize Smurf1 to CCM2 protein complexes. As expected, the Smurf1 HECT domain, when expressed alone, was not able to localize to the cell membrane because it lacks a C2 domain (Fig. 4G). Expression of CCM2 with the Smurf1 HECT domain resulted in relocalization of the Smurf1 HECT domain to the plasma membrane, where it co-localized with CCM2 (Fig. 4G). Thus, the CCM2 PTB-Smurf1 HECT domain interaction localizes Smurf1 to CCM2 complexes primarily at the cell periphery.
The significance of our work is, in part, the knockdown of CCM2 in a brain microvascular endothelial cell line, bEND.3. Endothelial cells lacking CCM2 expression are defective in cell migration, permeability, and tubule formation. bEND.3 cells display an increase in RhoA abundance, predictably leading to increased signaling via the ROCK pathway. Indeed, the importance of ROCK signaling in endothelial cells has been recognized. Subsequent phosphorylation of MLC2 results in actomyosin contractility, loss of cell-to-cell contact, and increased permeability (14). We show that CCM2 knockdown leads to increased abundance of RhoA and stress fibers as well as increased phosphorylated MLC2 associated with stress fibers. Further, co-expression of CCM2 and Smurf1 led to cell rounding, likely due to dramatic loss of RhoA and cytoskeletal collapse.
Recent work has shown that CCM2 is a negative regulator of RhoA signaling in human umbilical vein endothelial cells (5); however, the molecular mechanism was not determined. We also provide evidence for activation of this pathway, although we observed a decrease in directed cell migration in bEND.3 cells, a response not observed with human umbilical vein endothelial cells. This suggests variability in endothelial cell physiology between these two cell types. Taken together, these studies strongly suggest that defective RhoA regulation is a significant contributor to the pathogenesis of CCM.
We now provide the mechanism of increased RhoA signaling associated with loss of CCM2. For the first time, we show that CCM2 associates with Smurf1 to regulate the degradation of the GTPase RhoA. Further, we have characterized a novel interaction between the CCM2 PTB domain and the Smurf1 HECT domain. Although the CCM2 interaction with Smurf1 does not regulate the catalytic activity of Smurf1 nor act to recruit CCM2 as a Smurf1 substrate, this interaction plays a role in localization of Smurf1 to promote degradation of RhoA.
Supplementary Material
The on-line version of this article (available at http://www.jbc.org) contains supplemental methods and three supplemental figures.
Footnotes
The abbreviations used are: CCM, cerebral cavernous malformations; E3, ubiquitin-protein isopeptide ligase; RNAi, RNA interference; shRNA, short hairpin RNA; ROCK, Rho kinase; WT, wild type; HA, hemagglutinin; GST, glutathione S-transferase; PTB, phosphotyrosine binding; HECT, homologous to the E6-AP C terminus.
References
- 1.Uhlik, M. T., Abell, A. N., Johnson, N. L., Sun, W., Cuevas, B. D., Lobel-Rice, K. E., Horne, E. A., Dell'Acqua, M. L., and Johnson, G. L. (2003) Nat. Cell Biol. 5 1104–1110 [DOI] [PubMed] [Google Scholar]
- 2.Liquori, C. L., Berg, M. J., Siegel, A. M., Huang, E., Zawistowski, J. S., Stoffer, T., Verlaan, D., Balogun, F., Hughes, L., Leedom, T. P., Plummer, N. W., Cannella, M., Maglione, V., Squitieri, F., Johnson, E. W., Rouleau, G. A., Ptacek, L., and Marchuk, D. A. (2003) Am. J. Hum. Genet. 73 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchuk, D. A., Srinivasan, S., Squire, T. L., and Zawistowski, J. S. (2003) Hum. Mol. Genet. 12 Spec. No. 1, R97–R112 [DOI] [PubMed] [Google Scholar]
- 4.Plummer, N. W., Zawistowski, J. S., and Marchuk, D. A. (2005) Curr. Neurol. Neurosci. Rep. 5 391–396 [DOI] [PubMed] [Google Scholar]
- 5.Whitehead, K. J., Chan, A. C., Navankasattusas, S., Koh, W., London, N. R., Ling, J., Mayo, A. H., Drakos, S. G., Marchuk, D. A., Davis, G. E., and Li, D. Y. (2009) Nat. Med. 15 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, H. R., Ogunjimi, A. A., Zhang, Y., Ozdamar, B., Bose, R., and Wrana, J. L. (2006) Methods Enzymol. 406 437–447 [DOI] [PubMed] [Google Scholar]
- 7.Yamashita, M., Ying, S. X., Zhang, G. M., Li, C., Cheng, S. Y., Deng, C. X., and Zhang, Y. E. (2005) Cell 121 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilder, T. L., Malone, M. H., Bencharit, S., Colicelli, J., Haystead, T. A., Johnson, G. L., and Wu, C. C. (2007) J. Proteome Res. 6 4343–4355 [DOI] [PubMed] [Google Scholar]
- 9.Ingham, R. J., Gish, G., and Pawson, T. (2004) Oncogene 23 1972–1984 [DOI] [PubMed] [Google Scholar]
- 10.Smith, M. J., Hardy, W. R., Murphy, J. M., Jones, N., and Pawson, T. (2006) Mol. Cell Biol. 26 8461–8474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, H.-R., Zhang, Y., Ozdamar, B., Ogunjimi, A. A., Alexandrova, E., Thomsen, G. H., and Wrana, J. L. (2003) Science 302 1775–1779 [DOI] [PubMed] [Google Scholar]
- 12.Plant, P. J., Yeger, H., Staub, O., Howard, P., and Rotin, D. (1997) J. Biol. Chem. 272 32329–32336 [DOI] [PubMed] [Google Scholar]
- 13.Zawistowski, J. S., Stalheim, L., Uhlik, M. T., Abell, A. N., Ancrile, B. B., Johnson, G. L., and Marchuk, D. A. (2005) Hum. Mol. Genet. 14 2521–2531 [DOI] [PubMed] [Google Scholar]
- 14.Wojciak-Stothard, B., and Ridley, A. J. (2002) Vasc. Pharmacol. 39 187–199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.