Abstract
Disassembly of the yeast V-ATPase into cytosolic V1 and membrane V0 sectors inactivates MgATPase activity of the V1-ATPase. This inactivation requires the V1 H subunit (Parra, K. J., Keenan, K. L., and Kane, P. M. (2000) J. Biol. Chem. 275, 21761–21767), but its mechanism is not fully understood. The H subunit has two domains. Interactions of each domain with V1 and V0 subunits were identified by two-hybrid assay. The B subunit of the V1 catalytic headgroup interacted with the H subunit N-terminal domain (H-NT), and the C-terminal domain (H-CT) interacted with V1 subunits B, E (peripheral stalk), and D (central stalk), and the cytosolic N-terminal domain of V0 subunit Vph1p. V1-ATPase complexes from yeast expressing H-NT are partially inhibited, exhibiting 26% the MgATPase activity of complexes with no H subunit. The H-CT domain does not copurify with V1 when expressed in yeast, but the bacterially expressed and purified H-CT domain inhibits MgATPase activity in V1 lacking H almost as well as the full-length H subunit. Binding of full-length H subunit to V1 was more stable than binding of either H-NT or H-CT, suggesting that both domains contribute to binding and inhibition. Intact H and H-CT can bind to the expressed N-terminal domain of Vph1p, but this fragment of Vph1p does not bind to V1 complexes containing subunit H. We propose that upon disassembly, the H subunit undergoes a conformational change that inhibits V1-ATPase activity and precludes V0 interactions.
V-ATPases are ubiquitous proton pumps responsible for compartment acidification in all eukaryotic cells (1, 2). These pumps couple hydrolysis of cytosolic ATP to proton transport into the lysosome/vacuole, endosomes, Golgi apparatus, clathrin-coated vesicles, and synaptic vesicles. Through their role in organelle acidification, V-ATPases are linked to cellular functions as diverse as protein sorting and targeting, zymogen activation, cytosolic pH homeostasis, and resistance to multiple types of stress (3). They are also recruited to the plasma membrane of certain cells, where they catalyze proton export (4, 5).
V-ATPases are evolutionarily related to ATP synthases of bacteria and mitochondria and consist of two multisubunit complexes, V1 and V0, which contain the sites for ATP hydrolysis and proton transport, respectively. Like the ATP synthase (F-ATPase), V-ATPases utilize a rotational catalytic mechanism. ATP binding and hydrolysis in the three catalytic subunits of the V1 sector generate sequential conformational changes that drive rotation of a central stalk (6–8). The central stalk subunits are connected to a ring of proteolipid subunits in the V0 sector that bind protons to be transported. The actual transport is believed to occur at the interface of the proteolipids and V0 subunit a. Rotational catalysis will be productive in proton transport only if V0 subunit a is held stationary, whereas the proteolipid ring rotates (8). This “stator function” resides in a single peripheral stalk in F-ATPases (9, 10), but is distributed among up to three peripheral stalks in V-ATPases (11–13). The peripheral stator stalks link V0 subunit a to the catalytic headgroup and ensures that there is rotation of the central stalk complex relative to the V0 a subunit and catalytic headgroup.
Eukaryotic V-ATPases are highly conserved in both their overall structure and the sequences of individual subunits. Although homologs of most subunits of eukaryotic V-ATPases are present in archaebacterial V-ATPases (also known as A-ATPases), the C and H subunits are unique to eukaryotes. Both subunits have been localized at the interface of the V1 and V0 sectors, suggesting that they are positioned to play a critical role in structural and functional interaction between the two sectors (14–16). The yeast C and H subunits are the only eukaryotic V-ATPase subunits for which x-ray crystal structures are available (17, 18). The structure of the C subunit revealed an elongated “dumbbell-shaped” molecule, with foot, head, and neck domains (18). The structure of the H subunit indicated two domains. The N-terminal 348 amino acids fold into a series of HEAT repeats and are connected by a 4-amino acid linker to a C-terminal domain containing amino acids 352–478 (17). These two domains have partially separable functions in the context of the assembled V-ATPase (19). Complexes containing only the N-terminal domain of the H subunit (H-NT)2 supported some ATP hydrolysis but little or no proton pumping in isolated vacuolar vesicles (19, 20). The C-terminal domain (H-CT) assembled with the rest of the V-ATPase in the absence of intact subunit H, but supported neither ATPase nor proton pumping activity (19). However, co-expression of the H-NT and H-CT domains results in assembly of both sectors with the V-ATPase and allows increased ATP-driven proton pumping in isolated vacuolar vesicles. These results suggest that the H-NT and H-CT domains play distinct and complementary roles even when the two domains are not covalently attached.
In addition to their role as dedicated proton pumps, eukaryotic V-ATPases are also distinguished from F-ATPases and archaeal V-ATPases in their regulation. Eukaryotic V-ATPases are regulated in part by reversible disassembly of the V1 complex from the V0 complex (1, 21, 22). In yeast, disassembly of previously assembled complexes occurs in response to glucose deprivation, and reassembly is rapidly induced by glucose readdition to glucose-deprived cells. Disassembly down-regulates pump activity, and both the disassembled sectors are inactivated. Inhibition of ATP hydrolysis in free V1 sectors is particularly critical, because release of an active ATPase into the cytosol could deplete cytosolic ATP stores. This inhibition is dependent in part on the H subunit. V1 complexes isolated from vma13Δ mutants, which lack the H subunit gene (V1(-H) complexes) have MgATPase activity. Consistent with a physiological role for H subunit inhibition of V1, heterozygous diploids containing elevated levels of free V1 complexes without subunit H have severe growth defects (23). V1 complexes containing subunit H have no MgATPase activity, but retain some CaATPase activity, suggesting a role for nucleotides in inhibition (24, 25). Consistent with such a role, both the CaATPase activity of native V1 and the MgATPase activity of V1(-H) complexes are lost within a few minutes of nucleotide addition (24).
A number of points of interaction between the H subunit and the V1 and V0 complexes have been identified through two-hybrid assays, binding of expressed proteins, and cross-linking experiments. These experiments have indicated that the H subunit binds to V1 subunits E and G of the V-ATPase peripheral stalks (26, 27), the catalytic subunit (V1 subunit A) (28), regulatory V1 subunit B (15), and the N-terminal domain of subunit a (28). Recently, Jeffries and Forgac (29) have found that cysteines introduced into the C-terminal domain of subunit H can be cross-linked to subunit F in isolated V1 sectors via a 10-Å cross-linking reagent.
In this work, we examine both the subunit-subunit interactions and functional roles of the H-NT and H-CT domains in inhibition of V1-ATPase activity. When expressed in yeast cells lacking subunit H, H-NT can be isolated with cytosolic V1 complexes, but H-CT cannot. We find that both of these domains contribute to inhibition of ATPase activity, but that stable binding to V1 and full inhibition of activity requires both domains. We also find that the H-CT can bind to the cytosolic N-terminal domain of V0 subunit Vph1p (Vph1-NT) in isolation, but does not support tight binding of Vph1-NT to isolated V1 complexes.
EXPERIMENTAL PROCEDURES
Materials and Media—N-Ethylmaleimide (NEM), anti-FLAG M2-agarose beads, and FLAG peptide were all purchased from Sigma. The amylose resin, anti-MBP monocolonal antibody, and all restriction enzymes were purchased from New England BioLabs. Anti-Myc antibody was purchased from Roche Applied Science. Oligonucleotides were synthesized by MWG Biotech, and TaKaRa LA-Taq was purchased from Fisher. pGEM-T Easy TA cloning kit was purchased from Promega. Escherichia coli and yeast media were from Fisher. E. coli cells were grown in LB broth supplemented with 2 g/liter glucose and 100 μg/ml ampicillin.
Strains and Plasmids—Congenic vma13Δ and vma10Δ mutants in the BY4741 background (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) were purchased from Research Genetics/OpenBiosystems. The BY4742 vma10Δ::natR strain (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vma10Δ::natR) was constructed as described (23). These haploid cells were mated, and the resulting diploids were sporulated and tetrads dissected to obtain the vma13Δ::kanR vma10ΔnatR double mutant haploid.
Construction of N-terminal FLAG-tagged VMA10 in pRS315 and N-terminal Myc-tagged NT and CT fragments of VMA13 in pRS316 were described previously (19, 30). These plasmids were introduced into the vma13Δvma10Δ double mutant strain using an overnight lithium acetate protocol (31). The transformants were selected on fully supplemented minimal medium (SC) lacking leucine and/or uracil to select for the presence of one or both plasmids.
Bacterial expression plasmids were constructed by introducing full-length VMA13, a C-terminal fragment of VMA13, or an N-terminal fragment of VPH1 into a modified version of plasmid pMAL c2E (New England Biolabs). (This plasmid, named pMALpAse, has an engineered PreScission protease (New England Biolabs) site downstream of the malE gene at position 2641–2664 and was a generous gift from Gino Cingolani, SUNY Upstate Medical University.) The full-length VMA13 gene and the fragment encoding the H-CT domain were PCR-amplified from wild-type genomic DNA using primers VMA13-BamATG and VMA13-stop Hind (supplemental Table 1) for the full-length gene and VMA13CT-Bam352 and VMA13-stop Hind for the CT fragment. Coding sequence for the N-terminal 406 amino acids of VPH1 was PCR amplified with primers VPH1-BamATG and VPH1–406 Sal. For all the constructs, the resulting PCR fragments were cloned into the pGEM-T Easy TA cloning vector (Promega) according to the manufacturer's instructions. The pieces were then excised and cloned downstream of the malE gene in the pMALpAse vector cleaved with the appropriate enzymes. All PCR-generated constructs were confirmed by DNA sequencing at the Upstate Medical University sequencing core.
For insertion into a two-hybrid plasmid, VMA13-NT was isolated by PCR amplification of the H-NT open reading frame from a yeast shuttle plasmid containing H-NT (19) using primers VMA13/5′2Hyb and VMA13/3′2H. The VMA13-CT was amplified with primers VMA13/CT 5′2H and VMA13/3′2H from a wild-type VMA13 template. Both PCR products were cloned into the pGEM-T Easy vector as described above, then excised and cloned into two-hybrid vector pAS2-1 (32). The VMA13-NT fragment was cloned into BamHI and SalI restriction sites, and the VMA13-CT fragment was cloned into BamHI and PstI sites.
Other V-ATPase subunits were cloned into the pACT2 vector (32) by similar methods (33). VMA7 (encoding subunit F) was PCR amplified with primers VMA7/5′2Hyb and VMA7-C4, VMA8 (encoding subunit D) was amplified with primers VMA8/5′2Hyb and VMA8-C4, and the N-terminal domain of VPH1 was amplified with VPH1 5′2H and VPH13′2H. All primer sequences are listed in supplemental Table 1.
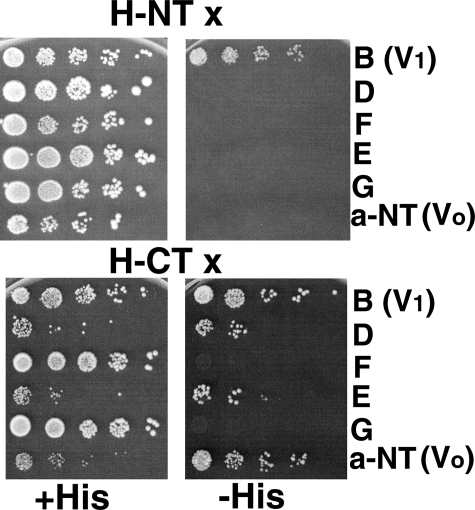
Two-hybrid Assays—Pairs of pACT and pAS-1 plasmids were introduced into yeast strain PJ69-4A (MATa trp1–901 leu2–3,112 ura3–52 his3–200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ (34)) by cotransformation, and transformants were selected on supplemented minimal medium lacking tryptophan and leucine (SC-trp,-leu). Transformants were then grown in liquid medium, serially diluted, and spotted onto: 1) SC-trp,-leu plates (control); 2) SC-trp,-leu,-his plates containing 2.5 mm aminotriazole (to select for expression of the HIS3 gene as a marker of two-hybrid interaction); 3) SC-trp,-leu,-adenine (to select for expression of the ADE2 gene as a marker of two-hybrid interaction); and 4) SC-trp,-leu,-his,-ade (to select for expression of both markers). All of the interactions shown in Fig. 1 were observed on plates 2–4.
FIGURE 1.
Two-hybrid assay for interactions of H-NT and H-CT with other V-ATPase subunits. Amino acids 1–348 (H-NT) and 352–478 (H-CT) of the H subunit were expressed as GAL4 binding domain fusions from the two-hybrid vector pAS2, and genes for V1 subunits B, D, F, E, and G and the N-terminal domain of Vph1p (a-NT) were expressed as GAL4 activation domain proteins from the pACT2 plasmid as described under “Experimental Procedures.” Haploid cells (PJ69-4A) cotransformed with the two plasmids were selected on SC-trp,-leu. Growth of the transformants from serial dilutions on this medium is shown on the left of the figure (+His plates). Panels on the right show growth on SC-trp,-leu medium that also lacks histidine and contains 2.5 mm 3-aminotriazole to strengthen the selection (–His plates). Growth on the –His plates requires two-hybrid interaction (34). None of the plasmids allowed growth under these conditions when mated to a strain containing the empty pAS2 or pACT2 vector (not shown).
Expression and Affinity Purification of MBP- and FLAG-tagged Proteins and Complexes—For bacterial expression of MBP-tagged subunit H and the H-CT fragment, the expression constructs described above were transformed into E. coli (BL21 pLysE) and plated on LB + chloramphenicol (34 μg/ml) + ampicillin (100 μg/ml). MBP-tagged Vph1-NT was transformed into E. coli (BL21) and transformants were selected on LB + ampicillin. Transformed cells were inoculated into LB liquid medium with 125 μg/ml ampicillin and grown overnight at 30 °C. 10 ml of the culture was added to 1 liter of LB containing ampicillin, supplemented with 2 g of glucose. The culture was allowed to grow at 37 °C until it reached A600 of 0.6–0.8, at which point isopropyl 1-thio-β-d-galactopyranoside was added to 300 μm. Cells expressing H and H-CT were then induced for 2 h at 37 °C, whereas the cells expressing Vph1-NT were induced at 30 °C for 6 h. After induction, cells were pelleted by centrifugation, resuspended in TBSE buffer (20 mm Tris-HCl, pH 7.2, 150 mm NaCl, 0.5 mm EDTA), and frozen at –20 °C until use. For Vph1-NT purification, the final suspension was adjusted to 5 mm β-mercaptoethanol. For purification, frozen cell pellets were thawed, and the suspension was sonicated 3 times for 25 s on ice. Lysed cells were centrifuged at 15,000 × g in a J-20 Beckman rotor for 30 min at 4 °C. The supernatant was passed over 1 ml of amylose resin (New England Biolabs) once at the rate of 0.5 ml/min or less. The amylose resin was washed with at least 20 column volumes of cold TBSE and then eluted with 5 fractions of 1 ml of TBSE with 100 mm maltose. 5 ml of eluant was concentrated in a Viva Spin 30-kDa cut off centrifugal filter device (Sartorius) to 300 μl or less. The concentrated sample was then further purified on a SEC250 gel filtration column (Bio-Rad) equilibrated with TBSE. Fractions of the predicted molecular mass of monomeric Vph1-NT were collected, pooled, and the protein concentration was determined from absorption at 280 nm using an extinction coefficient obtained from ProtParam Program (ExPASy). If the experiment required Vph1-NT to be detached from MBP, then 20 μl of Prescission protease at 3 mg/ml was added and left overnight at 4 °C after the concentration step, but before injection into the fast protein liquid chromatography gel filtration column. MBP-tagged H and H-CT were purified similarly except that no β-mercaptoethanol was added to the initial suspension, and the MBP-tagged protein was combined with 1 ml of amylose beads per 50 ml of crude cell extract. The mixture was incubated at 4 °C for 1–2 h with gentle rocking, then the beads were washed three times with 20 ml of TBSE followed by elution in TBSE containing 100 mm maltose. There was no further purification of the eluted proteins; they were used directly, either with or without protease cleavage.
V1 complexes were affinity purified from yeast cells via FLAG-tagged G subunit as described (13) with the following modifications. Cells were not incubated with zymolyase before lysis in the microfluidizer. After ammonium sulfate precipitation and desalting, the cytosolic fraction was centrifuged in a TLA-100 ultracentrifuge for 15 min at 52,000 × g before application of the supernatant to the anti-FLAG M2 affinity column. V1 complexes were eluted with FLAG peptide and used directly in most experiments. An additional gel filtration step using a Sephadex G-250 column on a Bio-Rad BioLogic Duo-Flow fast protein liquid chromatography system was added after affinity purification in a few experiments, as indicated in the figure legends. To test binding of H-CT to V1 pre-bound to anti-FLAG beads (Fig. 4C), V1 complexes were mixed with anti-FLAG beads and incubated for 1 h at 4 °C (∼125 μg of V1 was mixed with 100 μl of beads to give a final volume of 220 μl in TBSE). Purified MBP-H-CT was then added to the mixture (∼9 μg from an 0.1 mg/ml solution). This mixture was incubated for 2–3 additional hours at 4 °C, then the beads were pelleted by centrifugation, rapidly washed twice to remove unbound MBP-H-CT, and the bound material was eluted and subjected to SDS-PAGE and immunoblotting. For experiments with MgATP, 2 mm ATP and 2 mm MgCl2 were added with the MBP-H-CT.
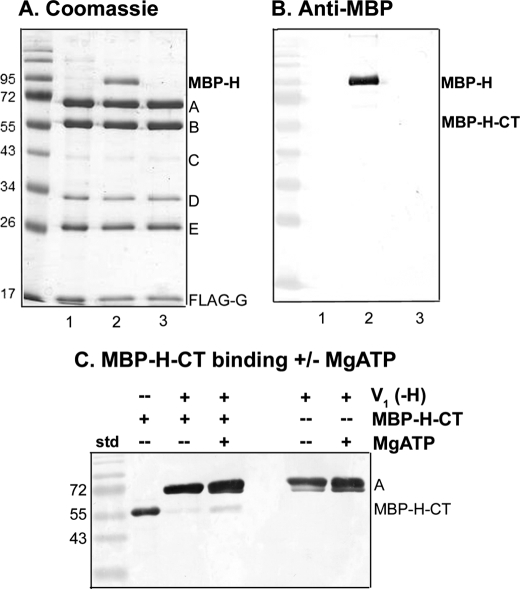
FIGURE 4.
The intact H subunit binds to V1 more tightly than the H-CT fragment. V1(-H) complexes were isolated from vma13Δ yeast cells (lane 1) and incubated with expressed MBP-H (lane 2) or MBP-H-CT (lane 3) followed by reisolation as described in the legend to Fig. 3D. V1(-H) was present at 0.63 μm in each incubation mixture; MBP-H was present at 5.7 μm and MBP-H-CT was present at 19.6 μm. A, Coomassie stained gel of the reisolated V1 complexes is shown. B, an immunoblot with anti-MBP antibody is shown to support identification of MBP-H and the absence of MBP-H-CT after reisolation of complexes as described in Fig. 3D. Note that V1 complexes lacking subunit H were initially isolated by both FLAG affinity chromatography and gel filtration to reduce co-purification of the 95-kDa impurity because it runs near MBP-H. C, V1 complexes were pre-bound to anti-FLAG beads, then incubated with MBP-H-CT (with or without MgATP present) as described under “Experimental Procedures.” After two brief washes to remove unbound MBP-H-CT, the bound material was eluted, and subjected to SDS-PAGE and immunoblotting. The immunoblot was probed with a combination of anti-MBP antibody (1:100,000 dilution) and monoclonal antibody (8B1) against the V1 A subunit (1:10,000 dilution). The MBP-H-CT only sample corresponds to 0.3 μg of purified protein. This blot is representative of four independent experiments.
Enzyme and Binding Assays—ATP hydrolysis activity was measured by a coupled enzyme assay at 37 °C as described (35). V1(-H) complexes were incubated with expressed MBP-H or MBP-H-CT for 1 h in the indicated ratios before addition to the ATPase assay mixture. V1 containing H-NT (V1(-H)/H-NT) was incubated and assayed in the same manner. NEM-sensitive ATPase activity (V1-ATPase activity) was determined by incubating V1 samples for 20 min on ice with 50 μm NEM prior to assay (19).
Immunoprecipitation of the FLAG-tagged proteins was used to assess binding of the bacterially expressed proteins to the purified V1(-H) or V1(-H)/H-NT. 100 μl of anti-FLAG-agarose beads suspended in TBSE were added to a mixture of one part V1 and five parts expressed protein (MBP-H, MBP-H-CT, or the cleaved proteins) to give a final volume of 300 μl. The mixtures were incubated at 4 °C with gentle shaking for 3 h. The resin was washed three times with TBSE to remove nonspecific binding. 100 μg/ml FLAG peptide was used to elute V1 and its binding partners from the beads and eluted protein were concentrated by trichloroacetic acid precipitation. Trichloroacetic acid pellets were solubilized with cracking buffer (50 mm Tris-HCl, pH 6.8, 8 m urea, 5% SDS, 5% β-mercaptoethanol). Samples were separated by SDS-PAGE followed by Coomassie staining. For immunoblotting, samples were transferred to a nitrocellulose membrane. The blots were probed with anti-MBP (1:100,000) and anti-Myc monoclonal antibodies. A colorimetric assay was used for detection after incubation with an anti-mouse secondary antibody linked to alkaline phosphatase.
For immunoprecipitation of Vph1-NT, 250 μl of cultured supernatant containing monoclonal antibody 10D7 (36) was added to a 200-μl mixture of Vph1pNT and a partner protein that had been allowed to stand for ∼3 h on ice. The mixture of the proteins and the antibody was shaken overnight at 4 °C. The next day, 100 μl of a 50% (v/v) Protein A-Sepharose suspension (Sigma) in phosphate-buffered saline buffer (137 mm NaCl, 2.6 mm KCl, 12 mm sodium phosphate, containing 0.2% gelatin) was added to the mixture and incubated on ice for 2 h with occasional shaking. Protein A-Sepharose was gently pelleted by centrifugation and washed twice with the same buffer. The samples were eluted by directly adding cracking buffer to the pellets.
RESULTS
Interactions of the N- and C-terminal Domains of Subunit H with V1 and V0 Subunits—The crystal structure of yeast subunit H indicated that it was comprised of two domains, one consisting of the N-terminal 348 amino acids of the protein (H-NT) and the second consisting of amino acids 352–478 (H-CT) (17). To address the interactions of the two domains with V1 and V0, we introduced each H subunit domain into a two-hybrid vector, and tested its interaction with several V1 subunits and the cytosolic N-terminal domain of V0 subunit a. In this version of the two-hybrid assay, all combinations should grow on supplemented minimal medium lacking leucine and tryptophan, because these two nutritional markers are encoded on the two-hybrid plasmids. As we have seen previously, some combinations of plasmids partially inhibit growth even under the permissive (+His) conditions, possibly reflecting dominant negative interactions as a result of subunit overexpression (13, 27, 37). Two-hybrid interactions are identified by their ability to support growth on medium that also lacks histidine, which requires reconstitution of a transcription factor capable of inducing the HIS3 gene through interaction of the partners (34). As shown in Fig. 1, H-NT displayed an interaction only with the V1 B subunit. H-CT interacted with a number of subunits, including the V1 B, D, and E subunits, and the V0 a-NT domain. Interactions with subunit B of the catalytic headgroup, subunit E, of the peripheral stalks, and the a-NT were expected from previous results. Interaction with the D subunit, of the central rotor stalk was unexpected. This interaction could support a model of H subunit inhibition of V1-ATPase activity in which the H subunit would inhibit V1-ATPase activity by tethering the catalytic headgroup and/or peripheral stalk subunits to the central stalk subunits, preventing rotational catalysis (22, 29).
To further explore this possibility, we isolated V1 sectors from a vma13Δ strain, which lacks subunit H (V1-(H) complexes), with and without a plasmid-borne copy of H-NT or H-CT. As reported previously, V1(-H) complexes had Mg2+-dependent ATPase activity, whereas V1 complexes isolated from strains containing subunit H do not have activity (24). V1 complexes from the strain containing only the H-CT domain (V1(-H)/H-CT) had ATPase activity comparable with those from the strain with no H subunit at all. We have previously shown that H-CT is isolated with the V-ATPase in vacuolar vesicles (19). To determine whether the H-CT was co-purified with V1(-H), we visualized both whole cell lysates and the purified V1 complexes by immunoblotting to detect the Myc epitope present on H-CT. As shown in Fig. 2, A and B, the Myc-tagged H-CT was present in the whole cell lysates, but not in the purified V1 sectors. These results indicate that H-CT does not co-purify with V1(-H) complexes.
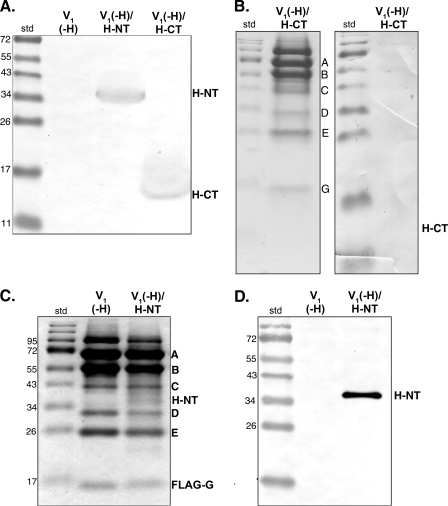
FIGURE 2.
Isolation of V1 sectors from yeast strains containing H-CT and H-NT. A, immunoblot of whole cell lysates from vma13Δ cells, which completely lack subunit H (V1(-H)), vma13Δ cells expressing Myc-tagged H-NT from a plasmid (V1(-H)/H-NT), and vma13Δ cells expressing Myc-tagged H-CT (V1(-H)/H-CT). Lysates were separated by SDS-PAGE and transferred to nitrocellulose, and Myc-tagged proteins were detected with anti-Myc monoclonal antibody. B, V1 sectors were purified from vma13Δ cells expressing Myc-tagged H-CT as described under “Experimental Procedures,” and visualized by Coomassie Blue staining (left) or immunoblot using anti-Myc antibody (right). C and D, V1 sectors were isolated from vma13Δ cells (V1(-H)) and vma13Δ cells containing a plasmid-borne H-NT (V1(-H)/H-NT). Isolated V1 sectors from both strains were separated by SDS-PAGE and visualized by Coomassie Blue staining in C, and by anti-Myc immunoblot in D. The Myc-tagged H-NT runs at ∼40 kDa. The position of molecular mass standards are shown on the left of all panels. Note that in panels B–D, V1 was purified by anti-FLAG affinity chromatography without a final gel filtration step. Under these conditions, varying amounts of a band running near 95 kDa are present in the preparation, along with the expected V1 subunits indicated.
In contrast, the immunoblots in Fig. 2, A and D, show that Myc-tagged H-NT was present in both the whole cell lysate and the purified V1(-H)/H-NT complexes. Coomassie staining of the isolated V1 complexes also supports co-purification of the H-NT with established V1 subunits present in the assembled V1 complexes containing subunit H (Fig. 2C). Comparison of staining in the region corresponding to H-NT to the staining of subunit D, which is also believed to be a single copy subunit, suggests that H-NT levels are substoichiometric. We measured NEM-sensitive MgATPase activity in the isolated V1(-H)/H-NT complexes. The average NEM-sensitive ATPase activity in these complexes was 0.49 ± 0.04 μmol/min/mg protein (mean ± S.E., n = 3), whereas the average V1 activity from strains lacking any subunit H was 1.92 ± 0.57 μmol/min/mg (n = 5). (NEM is a specific inhibitor of the V1 ATPase that binds at the catalytic site.) These results indicate that the H-NT domain can be co-purified with V1, and that MgATPase activity of V1(-H) complexes is partially inhibited by the presence of H-NT. These results are summarized in Table 1.
TABLE 1.
Summary of V1-ATPase inhibition experiments
| V1 complex expressed in yeast | Bacterially expressed (H subunit added) | H subunit bound? | % V1 (-H) (ATPase activity) |
|---|---|---|---|
| V1(-H) | None | None | 100 |
| V1(-H)/H-NT | None | H-NT co-isolates with V1 | 26 |
| V1(-H)/H-CT | None | None | 100 |
| V1(-H) | Full-length H | H binds tightly through repurification | 20 |
| V1(-H) | H-CT | Very little detected after repurification | 38 |
| V1(-H) | H-CT (+MgATP) | More H-CT retained after repurification | NDa |
| V1(-H)/H-NT | Full-length H | H subunit partially displaces H-NT | 5 (20)b |
| V1(-H)/H-NT | H-CT | ND | 14 (55)b |
ND, not determined.
Activity relative to isolated V1(-H)/H-NT complexes (from Fig. 3) is shown in parentheses. To compare the overall activity and inhibition of these complexes to V1(-H) complexes, the % activity in the V1(-H)/H-NT complexes as isolated (26%) was multiplied by the proportion of the activity remaining after addition of expressed H or H-CT.
Inhibitory Activity of the Expressed H-CT Domain—The absence of H-CT from isolated V1 sectors could reflect either poor binding to V1 sectors or preferential binding to other partners in the cell. To assess the ability of the H-CT domain to bind V1(-H) and inhibit V1-ATPase activity, we first expressed both the intact H subunit and H-CT as N-terminal MBP fusions in E. coli, and purified the proteins by amylose affinity chromatography. As shown in Fig. 3A, both proteins were obtained in good purity, although some fragments were present with the MBP-H fusion even without addition of protease. Cleavage with protease generated MBP and untagged H or H-CT. We next assessed the ability of both H and H-CT to bind and inhibit V1. As shown in Fig. 3B, the expressed, intact H subunit efficiently inhibited MgATPase activity in V1(-H) complexes, even when present at only a 2-fold molar excess over V1(-H). Somewhat more surprisingly, the H-CT inhibited almost as efficiently, although the level of inhibition reached was never as high (average of 80% inhibition for intact H and 62% for H-CT at 5-fold excess). There was no difference in inhibition by MBP-tagged H and H-CT versus H and H-CT that had been proteolytically cleaved from MBP. We also expressed and purified MBP alone and tested for inhibition of V1-ATPase activity. There was no inhibition by MBP alone (data not shown).
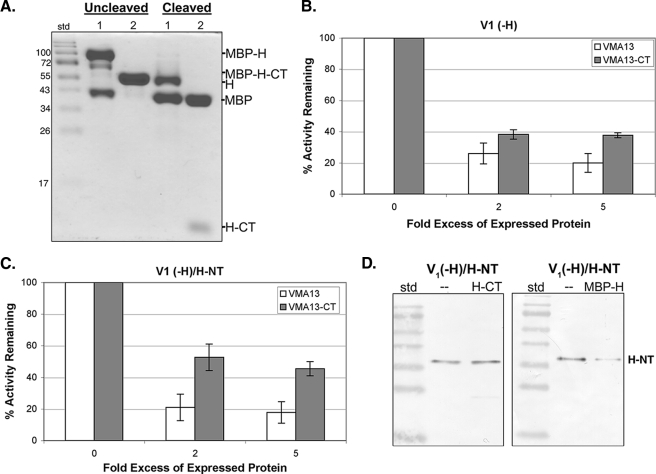
FIGURE 3.
Inhibition of V1-ATPase activity by intact H subunit and H-CT fragment expressed in E. coli. A, the intact H subunit and H-CT fragment were fused to MBP, expressed in E. coli, and purified by amylose affinity chromatography as described under “Experimental Procedures.” A Coomassie-stained gel of the intact fusion proteins (uncleaved) and products of cleavage by PreScission protease (cleaved). Sample 1 is MBP-H and sample 2 is MBP-H-CT. B and C, ATPase activity of V1(-H) (B) and V1(-H)/H-NT (C) was assayed in the absence of any bacterially expressed protein and in the presence of a 2- or 5-fold molar excess of the intact H subunit (VMA13, open bars) or H-CT fragment (VMA13-CT, shaded bars). The percentage of the ATPase activity remaining was calculated relative to the same V1 preparation and is shown as mean ± S.E. (error bars) for three to four separate purifications. Both the H and the H-CT fragment gave nearly identical levels of inhibition whether or not the MBP tag was removed, and inhibition with and without cleavage to remove MBP is combined in the graphs. D, V1(-H)/H-NT was repurified by FLAG affinity chromatography after a mock incubation (—) or incubation with a 5-fold excess of H-CT or MBP-H. An immunoblot probed with anti-Myc antibody to assess the level of Myc-tagged H-NT that remains associated with reisolated V1 is shown.
When expressed together in yeast, the H-NT and H-CT domains show increased activation of ATP-driven proton transport by the V-ATPase in vacuolar vesicles over either domain alone (19). We next tested whether the H-CT would give further inhibition of ATPase activity in V1(-H)/H-NT. We found that addition of H-CT to H-NT-containing V1 complexes inhibited an average of 55% of the MgATPase activity present in these complexes (Fig. 3C). The uncleaved MBP-H-CT inhibited both V1(-H) and V1(-H)/H-NT complexes to a level comparable with the cleaved H-CT. This result suggests that steric hindrance between the MBP and H-NT does not significantly affect inhibition. We also tested whether the expressed, intact H subunit could further inhibit V1(-H)/H-NT complexes. As shown in Fig. 3C, the intact H subunit did inhibit V1(-H)/H-NT containing complexes as efficiently as it inhibited V1(-H) complexes. These results are summarized in Table 1.
Inhibition by intact H in the presence of H-NT was somewhat surprising, but one explanation might be displacement of H-NT from V1(-H)/H-NT complexes by the intact subunit. We tested this by incubating V1(-H)/H-NT complexes with both intact H and H-CT, as for the inhibition assays, and then re-isolating V1 by affinity chromatography via the FLAG-tagged G subunits. As shown in Fig. 3D, whereas comparable levels of H-NT are present after the re-isolation whether or not H-CT is present, incubation with intact H partially displaced H-NT, so that less Myc-tagged H-NT is present after re-isolation of V1 complexes. This suggests that H-NT binds to V1 less tightly than the intact H subunit and thus can be displaced.
To address whether H-CT was easily removed from V1, possibly accounting for the loss of H-CT expressed in yeast during V1 isolation, we incubated V1(-H) complexes with the expressed MBP-H subunit or MBP-H-CT under the same preincubation conditions as for the inhibition experiments, re-isolated V1, and assessed binding of the expressed subunits. Intact MBP-H subunit was isolated in near-stoichiometric amounts with V1, as suggested by the prominent band at ∼95 kDa in the Coomassie-stained gel (Fig. 4A), and confirmed by MBP immunoblot in Fig. 4B. MBP-tagged H-CT, which inhibited ATPase activity as well as the cleaved H-CT, was not present in the re-isolated V1. This result suggests that although the inhibition of V1-ATPase activity by H-CT indicates that it must be able to bind, there may be a relatively low affinity/high off-rate that could account for its absence in V1 isolated from yeast cells expressing H-CT. To further address this issue, we concentrated V1 by pre-binding to anti-FLAG beads, and then incubated with MBP-tagged H-CT, followed by rapid washing to elute unbound MBP-H-CT. A small amount of MBP-H-CT was bound under these conditions, as shown by the anti-MBP Western blot in Fig. 4C. However, if MgATP was included in the incubation, more MBP-H-CT was bound to V1 on the beads. Although the levels of bound MBP-H-CT, even with MgATP, were low, they were reproducibly higher in the presence of MgATP in four independent experiments. This result suggests that the substrate MgATP, or a product of its hydrolysis, may promote binding of H-CT and inhibition of V1-ATPase activity. Consistent with the high level of binding in the absence of MgATP shown in Fig. 4A, inclusion of MgATP did not increase the binding of MBP-tagged H subunit to V1 (data not shown).
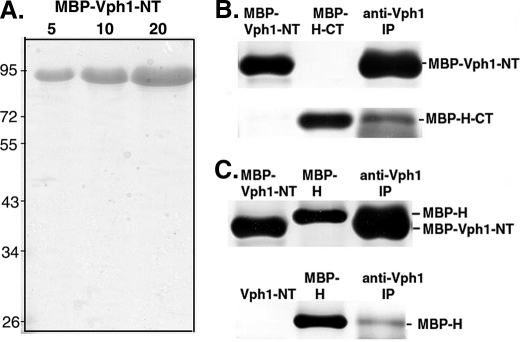
Binding to the N-terminal Domain of Vph1p—The two-hybrid assay shown in Fig. 1 indicates that the H-CT domain interacts with the cytosolic N-terminal domain of Vph1p (Vph1-NT). Binding of both intact H and a fragment of the H subunit consisting of amino acids 160–478 to Vph1-NT was documented previously (28). (The latter fragment contains both the entire H-CT and approximately half of the H-NT domain and is capable of complementing the growth and biochemical phenotypes of a vma13Δ mutant.) To determine whether the H-CT domain alone is able to bind Vph1p-NT in vitro, we first expressed the first 406 amino acids of Vph1p as an N-terminal MBP fusion protein, and purified the protein on amylose resin. Purified MBP-Vph1-NT is shown in Fig. 5A. We then incubated MBP-Vph1-NT with MBP-tagged H-CT, followed by immunoprecipitation with monoclonal antibody (10D7) against Vph1-NT. As shown in Fig. 5B, the antibody was able to co-precipitate H-CT with Vph1-NT. These results support the interaction between the H-CT domain and Vph1-NT suggested by the two-hybrid data in Fig. 1. We also confirmed binding of the intact H subunits to both the expressed MBP-Vph1-NT, which is very close in molecular mass to the MBP-tagged H subunit, and to cleaved Vph1-NT, which is well separated (Fig. 5C).
FIGURE 5.
Binding of expressed Vph1-NT to expressed H subunit and H-CT fragment. A, amino acids 1–406 of Vph1p were expressed as an MBP fusion protein in E. coli and purified by amylose affinity chromatography. 5, 10, and 20 μg of the purified MBP-Vph1-NT were separated by SDS-PAGE and visualized with Coomassie Blue. B and C, immunoblots probed with anti-MBP antibody. The indicated MBP fusion proteins were loaded singly (MBP-Vph1-NT, MBP-H-CT, or MBP-H) or incubated together for 3 h at 4 °C followed by immunoprecipitation with anti-Vph1p antibody to isolate complexes as described under “Experimental Procedures.” In B, final concentrations in the mixture used for immunoprecipitation were 10.3 μm MBP-Vph1-NT and 47.6 μm MBP-H-CT. In A, final concentrations were 5.9μm MBP-Vph1-NT and 7.5μm MBP-H. In C, MBP-tagged H subunit was incubated with protease-cleaved Vph1-NT before immunoprecipitation because the two MBP-tagged proteins have similar molecular masses.
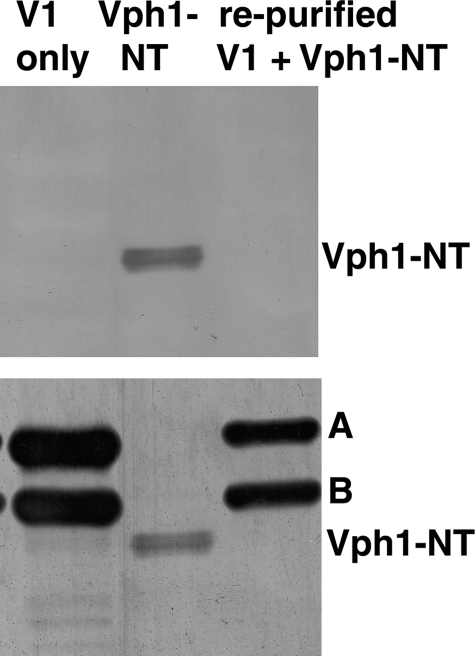
V1 sectors isolated from glucose-deprived cells contain the intact H subunit, but have been released from the V0 sector. Given the evidence that Vph1-NT can bind to the H subunit and H-CT, we tested whether expressed Vph1p-NT can bind to isolated V1 complexes, because these complexes contain the H subunit. We combined either MBP-tagged Vph1-NT or cleaved Vph1-NT with isolated, FLAG-tagged V1 containing subunit H (to give final concentrations of 2.2 μm V1 and 8.6 μm Vph1-NT), then re-purified V1 complexes on an anti-FLAG column. As shown in Fig. 6 (top panel), no Vph1-NT binding was detected. We confirmed that V1 had been re-purified by reprobing the same Western blot for V1 subunits A and B (Fig. 6, bottom panel); both were present at high levels in the reisolated material. These results suggest that even though they contain subunit H, V1 sectors do not bind Vph1-NT tightly.
FIGURE 6.
Vph1-NT does not bind stably to purified V1 sectors. V1 sectors containing subunit H were isolated, then incubated with Vph1-NT followed by repurification by FLAG-affinity chromatography as described in the legend to Fig. 4. Purified V1 only, expressed Vph1-NT cleaved from MBP, and V1 complexes incubated with Vph1-NT and repurified were separated by SDS-PAGE and transferred to nitrocellulose. The same blot was probed first with monoclonal antibody (10D7) to Vph1 (top) and then reprobed with monoclonal antibodies to the A and B subunits (bottom) to confirm that V1 complexes were present after re-isolation, but did not contain bound Vph1p-NT.
DISCUSSION
Potential Mechanisms for Inhibition of V1-ATPase Activity by Subunit H—The experiments described here build on previous data indicating that the H subunit is largely responsible for MgATPase inhibition of V1-ATPase activity (24). We find that expressed and purified H subunit binds tightly and stoichiometrically to isolated V1(-H) complexes and is a potent inhibitor of MgATPase activity. Inhibition of MgATPase activity in cytosolic V1 sectors is likely to be critical, but the mechanism of this inhibition is incompletely understood. Recent evidence of cross-linking between an introduced cysteine in the H-CT domain and the F subunit in free V1 suggested a tethering mechanism of inhibition (29), in which the H subunit prevents ATP hydrolysis by tethering the rotor and stator domains and thus prohibiting rotation. However, the cross-linking approach does not actually test for inhibition of MgATPase activity and did not employ a zero-length cross-link so direct interaction cannot be inferred. Notably, the two-hybrid interaction between the H-CT domain and subunit D of the central stalk shown here is also consistent with the arrangement of H-CT and the central stalk suggested by the cross-linking experiment, because the D and F subunits are adjacent in the central stalk (38).
However, tethering of the catalytic domain/peripheral stalk to the central stalk as a means of blocking rotation does not necessarily account for all characteristics of V1 MgATPase inhibition. First, both yeast and Manduca sexta V1 complexes retain CaATPase activity even under conditions where MgATPase activity is completely inhibited and the H subunit has presumably assumed its inhibitory conformation (24, 25). The similarity between these V1-ATPases and chloroplast F1, which exhibits CaATP hydrolysis without coupling to proton transport, has been noted previously (25). In an active F1 hybrid ATPase consisting of α and β subunits from Rhodospirillum rubrum and a γ subunit from spinach chloroplast, CaATP was shown to induce rotation of the central stalk with characteristics similar to MgATP-induced rotation (39). It seems unlikely that V1-ATPases could achieve the observed rates of CaATP hydrolysis, which are similar to the rates of MgATP hydrolysis in the absence of subunit H, without rotation (24), although it is possible that CaATP binding perturbs inhibitory interactions between subunit H with the central stalk. In addition, the experiments shown here demonstrate that both the H-NT and H-CT domains individually have some inhibitory activity. Inhibition of MgATPase activity by the expressed H-CT domain could be accommodated by a tethering model for V1 inhibition because Fig. 1 indicates two-hybrid interactions of this domain with multiple V1 subunits, including a subunit of the peripheral stalk (E subunit), the catalytic headgroup (subunit B), and the central stalk (subunit D). However, inhibition of V1-ATPase activity by the H-NT domain alone does not necessarily support tethering to the central stalk as important for inhibition. There is no evidence that this domain of the H subunit approaches the central stalk, yet complexes isolated from cells containing only this domain have only 26% of the NEM-sensitive activity of complexes with no subunit H (Table 1).
It could be argued that the V1 sector does not assemble normally in the presence of H-NT only. However, the ability of the intact H subunit to mediate further inhibition and even partially displace H-NT suggests that the V1(-H)/H-NT complexes are both capable of activity and susceptible to inhibition. In fact, as summarized in Table 1, if the lower level of MgATPase activity in V1(-H)/H-NT complexes isolated from yeast is considered along with the levels of inhibition achieved by addition of expressed H and H-CT, the V1(-H)/H-NT complexes are inhibited even more completely than V1(-H) complexes. This result is initially surprising. However, one intriguing possibility is that the ATPase activity of the V1(-H)/H-NT complexes shown in Fig. 3C arises largely from a population of V1(-H) complexes without H-NT bound and thus is inhibited to a similar extent as the V1(-H) complexes in Fig. 3B. Consistent with this explanation, H-NT appears to be substoichiometric in V1(-H)/H-NT complexes based on Coomassie staining (Fig. 2C). This interpretation would suggest that there is strong inhibition of ATPase activity in V1 (-H)/H-NT complexes. Further experiments will be necessary to confirm this.
In our experiments, binding of H-CT to V1(-H) complexes is notably weak, unable to survive isolation from yeast cells expressing H-CT or reisolation of purified V1(-H) complexes incubated with bacterially expressed H-CT (Table 1). It is likely that the H-NT domain provides further binding energy in the intact subunit. Our data do not conclusively demonstrate whether the H-NT and H-CT domains have an additive inhibitory effect. MgATPase activity is further inhibited when H-CT is added to V1(-H)/H-NT (Fig. 3), but as described above, occupancy of these complexes with H-NT may not be complete.
Some insights into potential mechanisms for inhibition may be provided by biochemical and structural experiments with the F1-ATPase. The Walker lab has identified several modes of action that account for mechanisms of multiple inhibitors (40). One class of inhibitor, characterized by the regulatory IF1 inhibitor for the F1-ATPase, appears to inhibit by binding to an area of the α and β subunits that would be oriented toward the membrane domain in the assembled ATP synthase. The binding site also places IF1 in proximity to the γ subunit of the central stalk (41). However, it is believed that the inhibitory activity of IF1 derives from tight binding to the α and β subunits that blocks conformational changes essential for rotational catalysis rather than interaction with the γ-subunit (41, 42). Given the established position of the H subunit at the membrane-proximal side of V1, it is tempting to speculate that binding to the B subunits (and possibly the A subunits as well (28)), which may involve areas of both H-NT and H-CT, is a more important source of inhibition than tethering of the central stalk. Further experiments will be necessary to explore this possibility.
It should also be noted that binding of the H subunit is unlikely to be the sole mechanism for inhibition of MgATPase activity in V1. We found previously that V1 complexes lacking subunit H showed a burst of ATPase activity upon addition of MgATP (24). In the experiments described here, we measured activity in the presence of an ATP regenerating system; the initial activity was still ultimately silenced, even in the absence of any subunit H (not shown). Huss and Wieczorek (43) recently showed that the M. sexta V1 contains 1.7 mol of ADP/mol of enzyme when isolated and proposed that MgADP generated from hydrolysis contributes to both release of V1 from the membrane and inhibition of its activity. Although yeast V1 complexes isolated by an earlier protocol contained no bound nucleotide (24), it would not be surprising if MgADP inhibition contributes to the eventual loss of hydrolytic activity even in the absence of subunit H. It is also entirely possible that nucleotide binding is entwined with the mechanism of inhibition by subunit H. The small enhancement of H-CT binding in the presence of MgATP (Fig. 4C) may support such a mechanism but requires further investigation. An interplay between nucleotide binding, directional rotation, and inhibitor binding is proposed in the mechanism of IF1 (41).
Does the H Subunit Contribute to Vph1p Binding to the V1 Complex?—Previous experiments demonstrated binding of subunit H to the N-terminal cytosolic domain of Vph1p (28). The experiments described here extend those studies by identifying the C-terminal domain of subunit H as the probable site of binding. Binding of H-CT to Vph1p, along with its rather weak affinity for V1, may help to account for the absence of H-CT in isolated V1. In addition, improved coupling of ATP hydrolysis and proton pumping in vacuolar vesicles from a strain containing both H-NT and H-CT, relative to the strain with H-NT alone, argued that H-CT provided functionally important interactions with the V0 sector (19). It is initially surprising, then, that the expressed Vph1-NT did not bind to V1 containing the intact H subunit. Landolt-Marticorena et al. (28) coprecipitated Vph1-NT with V1 subunits, including subunit H, from yeast cells lacking an intact Vph1 subunit. Vph1p-NT has been shown to interact with the E, G, and A subunits, all of which are present in purified V1, as well as subunit H (28, 44). It is probable that V1 purified from a cytosolic fraction used in our experiments is in an inhibited conformation that disfavors binding to the membrane sector. The structural basis of this conformation is not understood, but it is tempting to speculate that H-CT binds at a position in free V1 that does not allow Vph1p binding. Interestingly, the relatively large number of interactions with H-CT suggested by Fig. 1 may reflect binding partners that interact under different circumstances. There must be similar mechanisms for masking Vph1-NT and other V0 binding sites in V1 subunits when V1 is released from the membrane, and it is in fact likely that there are post-translational modifications or additional protein-protein interactions in the cells that modulate binding of V1 to Vph1-NT (33, 45, 46). These cellular factors may impart glucose dependence on interactions between V1 and Vph1-NT as well.
Supplementary Material
Acknowledgments
We thank Stephan Wilkens for helpful comments on the project and the manuscript, and the Wilkens and Cingolani laboratories for helpful advice on protein expression.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM50322 (to P. M. K.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: H-NT, N-terminal domain (amino acids 1–348) of the yeast H subunit; H-CT, C-terminal domain (amino acids 352–478) of the yeast H subunit; V1(-H), V1 complexes lacking subunit H isolated from a vma13Δ mutant yeast strain; V1(-H)/H-NT, V1 complexes containing H-NT isolated from a vma13Δ strain expressing H-NT from a plasmid; Vph1-NT, N-terminal domain of Vph1p (amino acids 1–406); MBP, maltose-binding protein; SC, synthetic complete medium (fully supplemented minimal medium); NEM, N-ethylmaleimide.
References
- 1.Forgac, M. (2007) Nat. Rev. Mol. Cell Biol. 8 917–929 [DOI] [PubMed] [Google Scholar]
- 2.Kane, P. M. (2006) Microbiol. Mol. Biol. Rev. 70 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, S. C., and Kane, P. M. (2009) Biochim. Biophys. Acta 1793 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breton, S., and Brown, D. (2007) Am. J. Physiol. 292 F1–F10 [DOI] [PubMed] [Google Scholar]
- 5.Nelson, N., and Harvey, W. R. (1999) Physiol. Rev. 79 361–385 [DOI] [PubMed] [Google Scholar]
- 6.Imamura, H., Nakano, M., Noji, H., Muneyuki, E., Ohkuma, S., Yoshida, M., and Yokoyama, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirata, T., Iwamoto-Kihara, A., Sun-Wada, G. H., Okajima, T., Wada, Y., and Futai, M. (2003) J. Biol. Chem. 278 23714–23719 [DOI] [PubMed] [Google Scholar]
- 8.Cross, R. L. (2000) Biochim. Biophys. Acta 1458 270–275 [DOI] [PubMed] [Google Scholar]
- 9.Walker, J. E., and Dickson, V. K. (2006) Biochim. Biophys. Acta 1757 286–296 [DOI] [PubMed] [Google Scholar]
- 10.Dickson, V. K., Silvester, J. A., Fearnley, I. M., Leslie, A. G., and Walker, J. E. (2006) EMBO J. 25 2911–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lolkema, J. S., Chaban, Y., and Boekema, E. J. (2003) J. Bioenerg. Biomembr. 35 323–335 [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa, N., Mazon, H., Heck, A. J., and Wilkens, S. (2008) J. Biol. Chem. 283 3329–3337 [DOI] [PubMed] [Google Scholar]
- 13.Ohira, M., Smardon, A. M., Charsky, C. M., Liu, J., Tarsio, M., and Kane, P. M. (2006) J. Biol. Chem. 281 22752–22760 [DOI] [PubMed] [Google Scholar]
- 14.Venzke, D., Domgall, I., Kocher, T., Fethiere, J., Fischer, S., and Bottcher, B. (2005) J. Mol. Biol. 349 659–669 [DOI] [PubMed] [Google Scholar]
- 15.Wilkens, S., Inoue, T., and Forgac, M. (2004) J. Biol. Chem. 279 41942–41949 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Z., Inoue, T., Forgac, M., and Wilkens, S. (2006) FEBS Lett. 580 2006–2010 [DOI] [PubMed] [Google Scholar]
- 17.Sagermann, M., Stevens, T. H., and Matthews, B. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7134–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drory, O., Frolow, F., and Nelson, N. (2004) EMBO Rep 5 1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, M., Tarsio, M., Charsky, C. M., and Kane, P. M. (2005) J. Biol. Chem. 280 36978–36985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannery, A. R., and Stevens, T. H. (2008) J. Biol. Chem. 283 29099–29108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyenbach, K. W., and Wieczorek, H. (2006) J. Exp. Biol. 209 577–589 [DOI] [PubMed] [Google Scholar]
- 22.Kane, P. M., and Smardon, A. M. (2003) J. Bioenerg. Biomembr. 35 313–321 [DOI] [PubMed] [Google Scholar]
- 23.Rizzo, J. M., Tarsio, M., Martinez-Munoz, G. A., and Kane, P. M. (2007) J. Biol. Chem. 282 8521–8532 [DOI] [PubMed] [Google Scholar]
- 24.Parra, K. J., Keenan, K. L., and Kane, P. M. (2000) J. Biol. Chem. 275 21761–21767 [DOI] [PubMed] [Google Scholar]
- 25.Graf, R., Harvey, W. R., and Wieczorek, H. (1996) J. Biol. Chem. 271 20908–20913 [DOI] [PubMed] [Google Scholar]
- 26.Fethiere, J., Venzke, D., Madden, D. R., and Bottcher, B. (2005) Biochemistry 44 15906–15914 [DOI] [PubMed] [Google Scholar]
- 27.Lu, M., Vergara, S., Zhang, L., Holliday, L. S., Aris, J., and Gluck, S. L. (2002) J. Biol. Chem. 277 38409–38415 [DOI] [PubMed] [Google Scholar]
- 28.Landolt-Marticorena, C., Williams, K. M., Correa, J., Chen, W., and Manolson, M. F. (2000) J. Biol. Chem. 275 15449–15457 [DOI] [PubMed] [Google Scholar]
- 29.Jefferies, K. C., and Forgac, M. (2008) J. Biol. Chem. 283 4512–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Z., Charsky, C., Kane, P. M., and Wilkens, S. (2003) J. Biol. Chem. 278 47299–47306 [DOI] [PubMed] [Google Scholar]
- 31.Elble, R. (1992) BioTechniques 13 18–20 [PubMed] [Google Scholar]
- 32.Bai, C., and Elledge, S. J. (1997) Methods Enzymol. 283 141–156 [DOI] [PubMed] [Google Scholar]
- 33.Smardon, A. M., and Kane, P. M. (2007) J. Biol. Chem. 282 26185–26194 [DOI] [PubMed] [Google Scholar]
- 34.James, P., Halladay, J., and Craig, E. A. (1996) Genetics 144 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lotscher, H. R., deJong, C., and Capaldi, R. A. (1984) Biochemistry 23 4128–4134 [DOI] [PubMed] [Google Scholar]
- 36.Kane, P. M., Kuehn, M. C., Howald-Stevenson, I., and Stevens, T. H. (1992) J. Biol. Chem. 267 447–454 [PubMed] [Google Scholar]
- 37.Keenan Curtis, K., and Kane, P. M. (2002) J. Biol. Chem. 277 2716–2724 [DOI] [PubMed] [Google Scholar]
- 38.Makyio, H., Iino, R., Ikeda, C., Imamura, H., Tamakoshi, M., Iwata, M., Stock, D., Bernal, R. A., Carpenter, E. P., Yoshida, M., Yokoyama, K., and Iwata, S. (2005) EMBO J. 24 3974–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker, W. C., Schwarz, A., Levine, T., Du, Z., Gromet-Elhanan, Z., Richter, M. L., and Haran, G. (2004) J. Biol. Chem. 279 47415–47418 [DOI] [PubMed] [Google Scholar]
- 40.Gledhill, J. R., and Walker, J. E. (2005) Biochem. J. 386 591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gledhill, J. R., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15671–15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cabezon, E., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2003) Nat. Struct. Biol. 10 744–750 [DOI] [PubMed] [Google Scholar]
- 43.Huss, M., and Wieczorek, H. (2007) FEBS Lett. 581 5566–5572 [DOI] [PubMed] [Google Scholar]
- 44.Qi, J., and Forgac, M. (2008) J. Biol. Chem. 283 19274–19282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, M., Sautin, Y. Y., Holliday, L. S., and Gluck, S. L. (2003) J. Biol. Chem. 279 8732–8739 [DOI] [PubMed] [Google Scholar]
- 46.Voss, M., Vitavska, O., Walz, B., Wieczorek, H., and Baumann, O. (2007) J. Biol. Chem. 282 33735–33742 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.