Abstract
Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) are essential for detecting viral RNA and triggering antiviral responses, including production of type I interferon. We analyzed the phenotype of non-synonymous mutants of human RIG-I and MDA5 reported in databases by functional complementation in cell cultures. Of seven missense mutations of RIG-I, S183I, which occurs within the second caspase recruitment domain repeat, inactivated this domain and conferred a dominant inhibitory function. Of 10 mutants of MDA5, two exhibited loss of function. A nonsense mutation, E627*, resulted in deletion of the C-terminal region and double-stranded RNA (dsRNA) binding activity. Another loss of function mutation, I923V, which occurs within the C-terminal domain, did not affect dsRNA binding activity, suggesting a novel and essential role for this residue in the signaling. Remarkably, these mutations are implicated in resistance to type I diabetes. However, the A946T mutation of MDA5, which has been implicated in type I diabetes by previous genetic analyses, affected neither dsRNA binding nor IFN gene activation. These results provide new insights into the structure-function relationship of RIG-I-like receptors as well as into human RIG-I-like receptor polymorphisms, antiviral innate immunity, and autoimmune diseases.
Innate and adaptive immune systems constitute the defense against infections by pathogens. Immediately after an infection occurs, various cells in the body sense the virus and initiate antiviral responses in which type I IFN2 plays a critical role, both in viral inhibition and in the subsequent adaptive immune response (1). The production of IFN is initiated when sensor molecules such as Toll-like receptors (TLRs) and RLRs detect virus-associated molecules. TLRs detect pathogen-associated molecular patterns (PAMPs) at the cell surface or in the endosome in immune cells such as dendritic cells and macrophages (2). RLRs sense viral RNA in the cytoplasm of most cell types and induce antiviral responses, including the activation of IFN genes (3). RLRs include RIG-I, MDA5, and laboratory of genetics and physiology 2 (LGP2).
It is proposed that RLRs sense and activate antiviral signals through the coordination of their functional domains (4). The N-terminal region of RIG-I and MDA5 is characterized by two repeats of CARD and functions as an activation domain (3). This domain is responsible for the transduction of signals downstream to IFN-β promoter stimulator 1 (IPS-1) (also known as MAVS, VISA, and Cardif). The primary sequence of the CTD, consisting of ∼140 amino acids, is conserved among RLRs. The CTD of RIG-I functions as a viral RNA-sensing domain as revealed by biochemical and structural analyses (5, 6). Both dsRNA and 5′-ppp-ssRNA, which are generated in the cytoplasm of virus-infected cells, are recognized by a basic cleft structure of RIG-I CTD. In addition to its RNA recognition function, the CTD of RIG-I and LGP2 functions as a repression domain through interaction with the activation domain. The repression domain is responsible for keeping RIG-I inactive in non-stimulated cells (3, 7). The helicase domain, with DEXD/H box-containing RNA helicase motifs, is the largest domain found in RLRs. Once dsRNA or 5′-ppp-ssRNA is recognized by the CTD, the helicase domain causes structural changes to release the activation domain. ATP binding and/or its hydrolysis is essential for the conformational change because Walker's ATP-binding site within the helicase domain is essential for signaling by RIG-I and MDA5.
Analyses of knock-out mice have revealed that RIG-I and MDA5 recognize distinct RNA viruses (8, 9). Picornaviruses are detected by MDA5, but many other viruses such as influenza A, Sendai, vesicular stomatitis, and Japanese encephalitis are detected by RIG-I. The difference is based on the distinct non-self RNA patterns generated by viruses, as demonstrated by the finding that RIG-I is selectively activated by dsRNA or 5′-ppp ssRNA, whereas MDA5 is activated by long dsRNA (10–12).
Single nucleotide polymorphisms (SNPs) of the human RIG-I and MDA5 genes including several non-synonymous SNPs (nsSNPs), which potentially alter the function of the proteins encoded, are reported in databases. In this report, we investigated the functions of nsSNPs of RIG-I and MDA5 by functional complementation using respective knock-out cells. We identified loss of function mutations of RIG-I and MDA5. Notably, two MDA5 mutations, E627* and I923V, recently reported to have a strong association with resistance to T1D (13), were severely inactive. The results suggest a novel molecular mechanism for the activation of RLRs and will contribute to our understanding of the functional effects of RLR polymorphisms and the critical relationship between RLR nsSNPs and diseases.
EXPERIMENTAL PROCEDURES
Cells, DNA Transfection, and Preparation of Cell Extracts—L929 cells were maintained in minimum essential medium Eagle (Sigma) with 5% fetal bovine serum and penicillin/streptomycin. Mouse embryonic fibroblasts (MEFs) were obtained from Dr. S. Akira (Osaka University). MEFs and 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin/streptomycin. L929 cells were transiently transfected with the DEAE-dextran method. MEFs and 293T cells were transiently transfected by FuGENE 6 (Roche Applied Science). For the preparation of cell extracts, cells were lysed with lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.1 mg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride, and 1 mm sodium orthovanadate) and centrifuged at 245,000 × g for 10 min. The supernatant was used for SDS-PAGE and electrophoretic mobility shift assays (EMSAs).
Oligonucleotides—Oligonucleotides (800 bp) corresponding to the coding sequence of green fluorescent protein were amplified by PCR using T7-primer (5′-CGTAATACGACTCACTATAGGGGATATCAGCAAAGGAGAAGAACTTTT-3′) and T3-primer (5′-GCAATTAACCCTCACTAAAGGGAGGCCTAGGGAGAAGACAGTGAGCTC-3′). Long dsRNA (ds800) was prepared by annealing complementary strands separately synthesized by in vitro transcription using the AmpliScribe T7 flash transcription kit (EPICENTRE Biotechnologies) and the AmpliScribe SP6 high yield transcription kit (EPICENTRE Biotechnologies). The annealed dsRNA was treated with S1 nuclease (Takara Bio) to generate a blunt end and alkaline phosphatase (Takara Bio) to remove 5′-phosphate. 32P-ds800 was prepared by labeling the 800-bp dsRNA using T4 polynucleotide kinase and [γ-32P]ATP.
Reporter Assay—MEFs were transfected with p55-C1Bluc, pRLtK, and expression plasmids for RIG-I mutants or MDA5 mutants. Cells were split into two aliquots, stimulated with RNA (5′-pppGG25) (6) or poly(I-C) transfection or Sendai virus (SeV) infection for 12 h, and harvested at 48 h after DNA transfection. L929 cells were transfected similar to MEFs but were stimulated by Newcastle disease virus infection. The infections were performed as described previously (14). The RNA transfection and poly(I-C) transfection were performed using Lipofectamine™ RNAiMAX (Invitrogen). The luciferase assay was performed with a Dual-Luciferase reporter assay system (Promega). Luciferase activity was normalized using Renilla luciferase activity (pRLtk) as a reference.
Quantitative PCR Assay—Quantitative PCR was performed as described previously (3).
Plasmid Constructs—p-55C1Bluc and pRLtk, pEF-FLAG-RIG-I, pEF-FLAG-RIG-ICARD, pEF-FLAG-MDA5, and pEF-FLAG-MDA5CARD were described previously (14). Data on SNPs for RIG-I and MDA5 were obtained from the NCBI (www.ncbi.nlm.nih.gov) and HapMap databases. The expression plasmids for RIG-I mutants (pEF-FLAG-RIG-IR7C, pEF-FLAG-RIG-IS144F, pEF-FLAG-RIG-IS183I, pEF-FLAG-RIG-IT260P, pEF-FLAG-RIG-II406T, pEF-FLAG-RIG-ID580E, pEF-FLAG-RIG-IF789L, and pEF-FLAG-RIG-ICARDS183I) and MDA5 mutants (pEF-FLAG-MDA5T260S, pEF-FLAG-MDA5L274I, pEF-FLAG-MDA5K351E, pEF-FLAG-MDA5I442V, pEF-FLAG-MDA5H460R, pEF-FLAG-MDA5E627*, pEF-FLAG-MDA5H843R, pEF-FLAG-MDA5I923V, pEF-FLAG-MDA5A946T, and pEF-FLAG-MDA5D1014E) were generated using a GeneEditor in vitro site-directed mutagenesis system (Promega). The mutations were confirmed by sequencing.
Antibodies and Immunoblotting—The anti-FLAG (M2; Sigma) antibody is a commercial product. SDS-PAGE and immunoblotting were performed as described previously (14).
EMSA—293T cells (1 × 106/6-cm dish) were transfected with 1 μg of expression vector. At 24 h after transfection, cell extract was prepared and mixed with anti-FLAG beads (Sigma) to adsorb FLAG-tagged proteins. The beads were washed, and bound protein was eluted with FLAG peptide (Sigma). The method of EMSA was described previously (6).
RESULTS
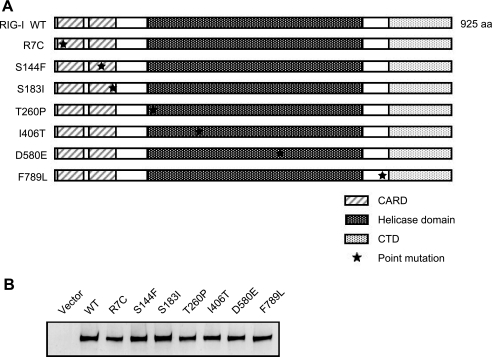
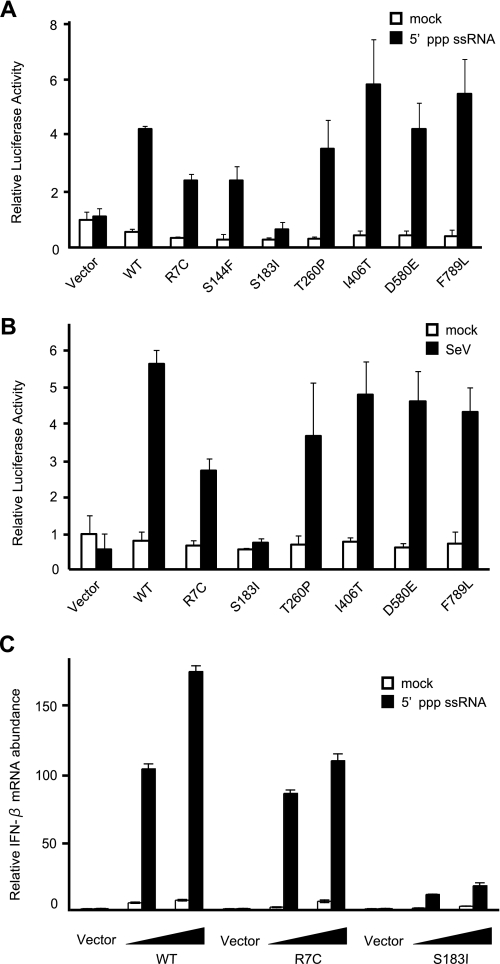
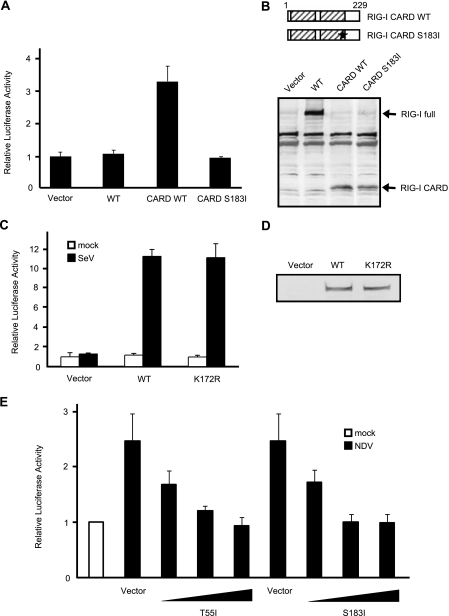
Construction of RIG-I Mutants and Their Biological Activities in MEFs Derived from RIG-I Knock-out Mice—nsSNPs were selected from nucleotide sequence polymorphisms of human RIG-I reported in databases and introduced into the RIG-I expression vector by site-directed mutagenesis. Domain structure and locations of the mutations are indicated in Fig. 1A. In RIG-I–/– MEFs transiently transfected with these vectors, the wild type and seven human RIG-I mutants were expressed at comparable levels (Fig. 1B). These results indicate that none of the amino acid substitutions significantly affect the synthesis and/or stability of RIG-I. The signaling function of the mutants was analyzed by temporarily complementing the function of RIG-I in RIG-I–/– MEFs using a virus-responsive luciferase reporter gene (Fig. 2A). Although the transfection of 5′-pppRNA did not activate the reporter gene significantly in RIG-I–/– MEFs (Fig. 2A, Vector), the ectopic expression of WT RIG-I restored the responsiveness to the ligand. All the mutant constructs exhibited functional complementation except the R7C, S144F, and S183I mutants, which exhibited a reduced response to 5′-pppRNA, particularly the S183I mutant, which exhibited a severe defect. Because the activity of S144F was not reproducible, we did not investigate this mutant further. It has been shown that RIG-I senses SeV and activates the IFN promoter. Therefore, the ability of the mutants to respond to this viral inducer was tested (Fig. 2B). SeV efficiently activated the reporter gene when WT RIG-I was expressed; however, S183I was virtually inactive and R7C exhibited partial activity. Other mutants sensed the virus as efficiently as the WT. We further confirmed the effect of the R7C and S183I mutations by monitoring the expression of endogenous mouse IFN-β mRNA (Fig. 2C). The results clearly demonstrate that S183I is barely active and that R7C is partially active. The above results strongly suggest that Ser-183 is critical for RIG-I to sense transfected 5′-pppRNA as well as SeV-derived PAMPs. Ser-183 resides within the second CARD, and this prompted us to explore the impact of the S183I substitution on the signaling function of the isolated RIG-I CARD. Unlike that of the full-length RIG-I, overexpression of the truncated RIG-I (1–229), which encompasses the two repeats of CARD, constitutively activated the reporter p-55C1B without a viral stimulus (Fig. 3A). However, RIG-I (1–229) with S183I failed to activate the reporter gene. In these cells, levels of RIG-I (1–229) with or without the S183I mutation were comparable, suggesting that Ser-183 is critical for the signaling function but does not affect protein levels of RIG-I CARD (Fig. 3B). It was reported that human RIG-I undergoes ubiquitination at Lys-172 in the second CARD, and this process is essential for RIG-I signaling (15). To compare the mutation at Ser-183, we generated a K172R mutant and tested its activity (Fig. 3, C and D). Surprisingly, the mutant exhibited virus-induced signaling activity comparable with the WT, suggesting that the ubiquitination of Lys-172 plays a minor role in the regulation of RIG-I and that the phenotype of the S183I mutant is unlikely due to a failure of ubiquitination.
FIGURE 1.
RIG-I nsSNP mutants and their expression in MEFs. A, schematic representation of the RIG-I wild type and nsSNP-containing mutants. RIG-I has a tandem CARD, RNA helicase domain, and CTD. Positions of the mutations are indicated by asterisks. aa, amino acids. B, FLAG-tagged WT RIG-I and SNPs were produced in RIG-I–/– MEFs and detected by immunoblotting using an anti-FLAG antibody.
FIGURE 2.
Functional analysis of RIG-I nsSNP mutants. A and B, RIG-I–/– MEF cells were transiently transfected with p-55CIBluc together with empty vector (Vector) or the indicated constructs. The cells were subjected to a Dual-Luciferase assay after stimulation with 5′-ppp-ssRNA (12 h) (A) or SeV (12 h) (B). The relative firefly luciferase activity, normalized to the Renilla luciferase activity, is shown. Error bars show the SDs for triplicate transfections. mock, mock-treated. C, RIG-I–/– MEFs were transfected with empty vector (Vector) or expression vectors for WT RIG-I or mutants as indicated (the total amount of plasmid was kept at 6μgby adding empty vector). To observe the dose response, cells were transfected with 3 or 6 μg of the expression plasmid. Cells were mock-treated or transfected with 5′-ppp-ssRNA for 12 h, and IFN-β mRNA was quantified by quantitative PCR by using the Applied Biosystems primer set for mouse interferon-β1: Mm00439546_S1.
FIGURE 3.
Characterization of RIG-I S183I. A, RIG-I–/– MEF cells were transfected with reporter genes together with empty vector (Vector) or plasmid expressing FLAG-tagged WT RIG-I, RIG-I CARD (the N-terminal region, amino acid 1–229), or RIG-I CARD S183I. After transfection (24 h), the cells were subjected to a Dual-Luciferase assay. Error bars show the S.D. values for triplicate transfections. B, each protein expressed in RIG-I–/– MEF cells was detected by immunoblotting using an anti-FLAG antibody. RIG-I full, full-length RIG-I. C, reporter assay of the K172R mutant was performed as in Fig. 2B. D, protein levels were determined by immunoblotting. RIG-I K172R and WT RIG-I were expressed at comparable levels. E, empty vector or expression vectors for full-length RIG-I with the T55I or S183I mutation were introduced into L929 cells (the total amount of plasmid was kept at 9 μg by adding empty vector) and infected with Newcastle disease virus, and reporter activity was analyzed as in panel A. To observe the dose response, cells received 1, 5, or 9 μg of the expression plasmid for T55I and S183I as indicated. NDV, Newcastle disease virus.
It is known that human RIG-I with the amino acid substitution T55I acts as a dominant inhibitor (7). This mutation within the first CARD inactivates the signaling function of the isolated tandem CARDs. We next tested whether RIG-I S183I exhibits a dominant negative phenotype. L929 cells were transfected with control vector or RIG-I mutants and then activated by infection of Newcastle disease virus. Cells transfected with the vector exhibited IFN promoter activity due to endogenous RIG-I (Fig. 3E). Expression of T55I as well as S183I significantly reduced the promoter activity in a dose-dependent manner, suggesting that S183I causes a dominant negative phenotype similar to T55I.
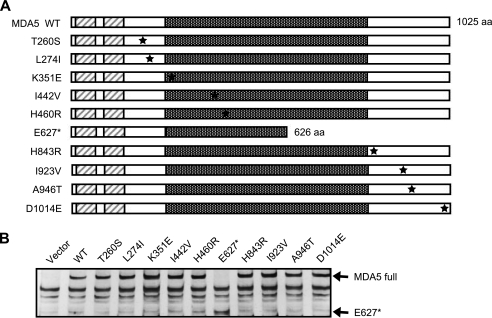
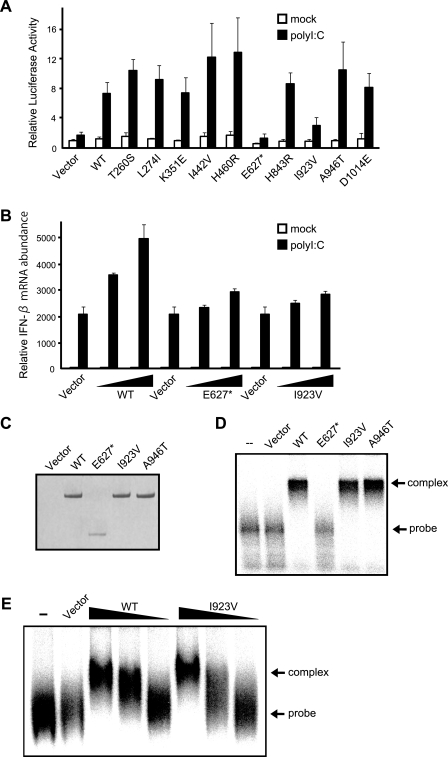
Construction of MDA5 Mutants and Their Biological Activities in MEFs Derived from MDA5 Knock-out Mouse—Next, the biological activity of human MDA5 mutants was analyzed similarly using MDA5-deficient MEFs. Ten nsSNPs including A946T identified in familial T1D (16) were introduced into the MDA5 expression vector (Fig. 4A). The wild type and mutants were expressed in MDA5–/– MEFs at comparable levels (Fig. 4B), showing that the mutations, including the E627* truncation, did not affect MDA5 protein levels. The biological activity of the mutants was assayed similarly to that of the RIG-I mutants. As a MDA5 agonist, a commercial poly(I-C) with an average length of 2 kbp was used, which selectively activates MDA5 (6, 11). Wild-type MDA5 clearly conferred responsiveness to the poly(I-C) (Fig. 5). Eight MDA5 mutants, including A946T, which was implicated in human T1D (16), exhibited complementing activity comparable with the wild type. However, E627* and I923V showed significantly low levels of activity. The phenotypes of these mutations were further tested by transient expression in MDA5–/– MEFs and monitoring endogenous IFN-β mRNA (Fig. 5B). Although MDA5 is absent, intracellular poly(I-C) induced IFN-β gene expression in the control cells (Vector). This is presumably due to the activation of RIG-I by a short poly(I-C) present in the preparation we used. Unlike the reporter assay, which monitors only transfected cells, this quantitative PCR assay detects IFN-β transcripts from both transfected and non-transfected cells, increasing the background signal. Irrespective of the background, overexpression of WT MDA5 resulted in an enhancement of IFN-β expression by poly(I-C), and this induction was not observed with E627* and I923V. Further, the effect was observed at different levels of MDA5 expression. The E627* mutant lacks a part of the helicase domain and the entire CTD in which Ile-923 resides.
FIGURE 4.
MDA5 nsSNP mutants and their expression in MEFs. A, schematic representation of WT MDA5 and its nsSNPs. Point mutations are indicated by asterisks. E627* is the nonsense mutant. aa, amino acids. B, FLAG-tagged MDA5 SNPs were produced in MDA5–/– MEFs and detected by immunoblotting using an anti-FLAG antibody. Vector, empty vector; MDA5 full, full-length MDA5.
FIGURE 5.
Functional analysis of MDA5 nsSNP mutants. A, MDA5–/– MEFs were transfected with reporter genes together with the indicated constructs as in Fig. 2A. After stimulation with poly(I-C) (12 h), the cells were subjected to a Dual-Luciferase assay. Error bars show the S.D. values for triplicate transfections. mock, mock-treated. Vector, empty vector. B, MDA5–/– MEFs were transfected with expression vectors for WT MDA5 or E627* or Ile-923 mutants (the total amount of plasmid was kept at 6 μg by adding empty vector). To observe the dose response, cells were transfected with 3 or 5.7 μg of the expression plasmid. Cells were mock-treated or transfected with poly(I-C) for 12 h, and IFN-β mRNA was quantified by quantitative PCR as in Fig. 2C. C, 293T cells were transfected with empty vector, WT MDA5, E627*, I923V, or A946T, and the produced proteins were purified using anti-FLAG (“Experimental Procedures”). The purified proteins were separated by SDS-PAGE and stained by Coomassie Brilliant Blue. D, EMSA of the purified MDA5 proteins (500 ng) using 32P-labeled poly(I-C) as a probe. complex, probe protein complex; probe, free probe. E, dose response of RNA binding by WT MDA5 and the I923V mutant. EMSA was performed using 500, 300, and 100 ng of MDA5 and Ile-923 protein.
Because in the case of RIG-I, the CTD determines RNA recognition specificity, we investigated the RNA binding activity of these mutants by EMSA using 32P-labeled poly(I-C). Wild-type and recombinant MDA5 proteins were expressed in 293T cells and purified. The recombinant proteins were virtually free of other cellular proteins as analyzed by Coomassie Brilliant Blue staining (Fig. 5C). Wild-type MDA5 clearly formed a complex with poly(I-C) (Fig. 5D), but E627* did not exhibit detectable binding activity. I923V and A946T exhibited activity to bind poly(I-C) as strongly as the wild type under these conditions. We further compared the RNA binding at different protein concentrations (Fig. 5E) and confirmed that MDA5 WT and the I923V mutant bind to dsRNA in a comparable fashion. These results suggest that the E627* mutant is biologically inactive due to its failure to recognize its agonist.
DISCUSSION
Human RIG-I Polymorphism—We identified S183I as a loss of function mutation of RIG-I. This serine residue is conserved in human, monkey, cow, and pig RIG-I. The mutation apparently inactivates the tandem CARD, which relays signals downstream. Furthermore, S183I exhibits a dominant inhibitory phenotype, suggesting that individuals retaining this mutation as a heterozygote would exhibit hypersensitivity to viral infections. It has been shown that RIG-I mutants with a CARD deletion (RIG-IC), CARD point mutation (T55I), and ATP-binding site mutation (RIG-I K270A) all function as a dominant inhibitor (3, 7). It has been reported that the repression domain present in the C-terminal region of RIG-I and LGP2 dominantly suppresses the activation of RIG-I in trans through interaction with CARD and the helicase loop region (7). Because the repression domain encompasses the RNA-binding domain of RIG-I (6), one mechanism is likely competition of RNA binding with WT RNA. However, the RNA binding-deficient RIG-I mutants, K888A/K907A and K858A/K861A (6), functioned as a dominant negative inhibitor.3 Interestingly, LGP2 mutants, K643E and K651E, which correspond to Lys-888 and Lys-907 of RIG-I, lost RNA binding activity but retain repression function (17). These results strongly suggest an RNA-independent mechanism.
One proposed function of the second CARD is to conjugate to ubiquitin (at Lys-172), which may be essential for signaling activity. However, human RIG-I with K172R, which is resistant to ubiquitination, did not affect phenotype (Fig. 3, C and D). Moreover, the corresponding amino acid in mouse RIG-I is glutamine, and the corresponding position of MDA5 is glutamic acid (human, monkey, mouse, cow, and pig), suggesting that the ubiquitination of Lys-172 has a minor impact on signaling activity. The precise function of the second CARD is not clear, but a comparative study of Ser-183 and WT RIG-I will elucidate the molecular function of this residue.
Human MDA5 Polymorphism—We identified two loss of function mutations, E627* and I923V, in human MDA5. Each of these mutations is actually present in the human population (13). It is worth investigating the phenotype of the homozygote because MDA5 knock-out mice clearly exhibit hypersusceptibility to the family Picornaviridae, genus Cardiovirus. E627*, which lacks CTD and a part of the helicase domain, lost its dsRNA binding activity; hence there was no signaling activity. Although the I923V mutation occurs in the CTD, this mutant exhibited intact dsRNA binding, suggesting a novel function of Ile-923 other than the recognition of RNA. It is worth noting that this isoleucine is conserved in human, monkey, mouse, cow, and pig MDA5. It is tempting to speculate that Ile-923 participates in an interaction with some other domain of MDA5 or other unknown regulatory protein(s).
A new report by Nejentsev et al. (13) describing the relationship between susceptibility to T1D and MDA5 polymorphism was published. The report describes that four rare mutations (two mutations in the exon and two mutations in the intron) in the human MDA5 gene are associated with protection against T1D. Our analysis included the two exon mutations (E627* and I923V), which exhibited a loss of function phenotype. The genetic analysis included another rare nsSNP, H460R, shown to be independent of T1D resistance. The phenotype of this mutant was normal in our analysis. On the other hand, A946T, which was suggested to associate with T1D in previous reports (13, 16), did not exhibit a loss of function phenotype. However, because Nejentsev et al. (13) suggested that the association of A946R with T1D was due to the effect of another nsSNP, R843H, a combination of these mutations might confer loss of function on MDA5. In summary, our analysis strongly suggests that loss of function mutations of MDA5 have a causative role in resistance to T1D. Although T1D has a complex pathology, these findings may provide a new strategy for establishing an animal model for T1D.
The human genome encodes multiple sensors, including TLRs and RLRs for viral PAMPs. Some of these may act redundantly to secure defense against viral infections. In the case of RLRs, RIG-I and MDA5 detect a distinct spectrum of viruses, as suggested from the phenotype of respective knock-out mice. There is variation within human populations in susceptibility to a particular viral infection. We argue for a possible contribution of the genetic diversity of RLRs, including those identified in the current investigation, to susceptibility. In addition to the impact of the infection itself, secondary effects such as autoimmunity, which is remotely triggered by certain viral infections, may be influenced at least in part through the functional diversity of RLRs. In summary, our results suggest a critical relationship between RLR polymorphisms and diseases including viral infections and autoimmunity.
Acknowledgments
We greatly appreciate the gift of MEFs from Dr. S. Akira (Research Institute for Microbial Diseases, Osaka University, Osaka). We give our thanks to Dr. F. Matsuda (Center for Genomic Medicine, Kyoto University, Kyoto) for discussion.
Author's Choice—Final version full access.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Science and Technology Agency, The Mochida Memorial Foundation for Medical Pharmaceutical Research, and Nippon Boehringer Ingelheim.
Footnotes
The abbreviations used are: IFN, interferon; IPS-1, IFN-β promoter stimulator 1; RIG-I, retinoic acid-inducible gene I; MDA5, melanoma differentiation-associated gene 5; LGP2, laboratory of genetics and physiology 2; TLR, Toll-like receptor; RLR, RIG-I-like receptor; PAMP, pathogen-associated molecular pattern; CARD, caspase recruitment domain; CTD, C-terminal domain; SNP, single nucleotide polymorphisms; nsSNP, non-synonymous SNP; MEF, mouse embryonic fibroblast; SeV, Sendai virus; T1D, type I diabetes; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; EMSA, electrophoretic mobility shift assay; WT, wild type.
J.-P. Zheng and T. Fujita, unpublished observation.
References
- 1.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immun. 5 730–737 [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama, M., and Fujita, T. (2008) Immunity 29 178–181 [DOI] [PubMed] [Google Scholar]
- 5.Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A., and Hopfner, K. P. (2008) Mol. Cell 29 169–179 [DOI] [PubMed] [Google Scholar]
- 6.Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M., Jr., Inagaki, F., and Fujita, T. (2008) Mol. Cell 29 428–440 [DOI] [PubMed] [Google Scholar]
- 7.Saito, T., Hirai, R., Loo, Y. M., Owen, D., Johnson, C. L., Sinha, S. C., Akira, S., Fujita, T., and Gale, M., Jr. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gitlin, L., Barchet, W., Gilfillan, S., Cella, M., Beutler, B., Flavell, R. A., Diamond, M. S., and Colonna, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101–105 [DOI] [PubMed] [Google Scholar]
- 10.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994–997 [DOI] [PubMed] [Google Scholar]
- 11.Kato, H., Takeuchi, O., Mikamo-Satoh, E., Hirai, R., Kawai, T., Matsushita, K., Hiiragi, A., Dermody, T. S., Fujita, T., and Akira, S. (2008) J. Exp. Med. 205 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Reis e Sousa, C. (2006) Science 314 997–1001 [DOI] [PubMed] [Google Scholar]
- 13.Nejentsev, S., Walker, N., Riches, D., Egholm, M., and Todd, J. A. (March 5, 2009) Science 10.1126/science.1167728 [DOI] [PMC free article] [PubMed]
- 14.Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M., Jr., Akira, S., Yonehara, S., Kato, A., and Fujita, T. (2005) J. Immunol. 175 2851–2858 [DOI] [PubMed] [Google Scholar]
- 15.Gack, M. U., Shin, Y. C., Joo, C. H., Urano, T., Liang, C., Sun, L., Takeuchi, O., Akira, S., Chen, Z., Inoue, S., and Jung, J. U. (2007) Nature 446 916–920 [DOI] [PubMed] [Google Scholar]
- 16.Smyth, D. J., Cooper, J. D., Bailey, R., Field, S., Burren, O., Smink, L. J., Guja, C., Ionescu-Tirgoviste, C., Widmer, B., Dunger, D. B., Savage, D. A., Walker, N. M., Clayton, D. G., and Todd, J. A. (2006) Nat. Genet. 38 617–619 [DOI] [PubMed] [Google Scholar]
- 17.Li, X., Ranjith-Kumar, C. T., Brooks, M. T., Dharmaiah, S., Herr, A. B., Kao, C., and Li, P. (March 28, 2009) J. Biol. Chem. 10.1074/jbc.M900818200 [DOI] [PMC free article] [PubMed]