Abstract
The TAZ transcription co-activator has been shown to promote cell proliferation and to induce epithelial-mesenchymal transition. Recently we have demonstrated that TAZ is phosphorylated and inhibited by the Hippo tumor suppressor pathway, which is altered in human cancer. The mechanism of TAZ-mediated transcription is unclear. We demonstrate here that TEAD is a key downstream transcription factor mediating the function of TAZ. Disruption of TEAD-TAZ binding or silencing of TEAD expression blocked the function of TAZ to promote cell proliferation and to induce epithelial-mesenchymal transition, demonstrating TEAD as a key downstream effector of TAZ. We also identified CTGF, a gene that regulates cell adhesion, proliferation, and migration, as a direct target of TAZ and TEAD. Our study establishes a functional partnership between TAZ and TEAD under negative regulation by the Hippo signaling pathway.
TAZ (transcriptional co-activator with PDZ binding motif) is a transcription co-activator that was initially identified as a 14-3-3-binding protein (1). TAZ contains a conserved WW domain, a coil-coil domain, a transactivation domain, and a C-terminal PDZ binding motif (2). It is involved in the development of multiple organs such as lung, fat, muscle, bone, limb, and heart tissues (2–5). TAZ also modulates mesenchymal stem cell differentiation by promoting Runx-2-dependent transcription while repressing peroxisome proliferator-activated receptor γ-dependent transcription (3). TAZ knock-out mice have minor skeletal defects, but pathological changes in the kidney and lung resemble polycystic kidney disease and pulmonary emphysema, respectively (6, 7). We recently reported that TAZ is negatively regulated by the Lats tumor suppressor kinase (8), which is a component of the Hippo tumor suppressor pathway initially defined by genetic studies in Drosophila. Phosphorylation of TAZ by Lats leads to 14-3-3 binding and translocation from nucleus to cytoplasm, resulting in functional inactivation of this transcription co-activator (8). This phosphorylation-dependent inactivation of TAZ by Lats is similar to the mechanism of YAP regulation (9), which is a co-activator with similar structure to TAZ, by the Hippo pathway.
Accumulating evidence suggests an important and evolutionary conserved function of the Hippo pathway in control of tissue growth and cancer development. Both genetic and cell biological studies from Drosophila indicated that merlin, a product of the well established human tumor suppressor gene NF2 (10), acts through the Hippo pathway to inhibit cell proliferation (11). Mutations in Sav and Mob, two other components in the Hippo pathway, have also been identified in human tumor cell lines (12, 13), further supporting the importance of the Hippo pathway in human cancer. Interestingly, TAZ is shown to be overexpressed in ∼20% of breast cancer samples and plays an important role in breast tumorigenesis, migration, and invasion (14). Consistent with this observation, TAZ overexpression promotes cell proliferation and induces epithelial-mesenchymal transition (EMT).4 Together, these studies suggest an important role of TAZ in mediating the Hippo pathway signaling to regulate cell growth and tumorigenesis.
TAZ itself has no DNA binding domain; therefore, it must bind to DNA binding transcription factors to stimulate downstream target gene expression. TAZ has been reported to bind many transcription factors such as RUNX family, thyroid TF1 (TTF-1), TBX5, PAX3, PAX8, peroxisome proliferator-activated receptor γ, and TEAD (2, 4, 5, 15). However, the relevance of these putative TAZ target transcription factors in mediating the function of TAZ in promoting cell proliferation and tumorigenesis is not clear.
Here we report that using unbiased biochemical purification, we identified TEAD family transcription factors as the major TAZ interacting transcription factors in HEK293T cells. At the same time, by screening a human transcription factor library, we also identified TEAD family transcription factors as the targets most potently activated by TAZ. We further demonstrated that TEAD is indeed indispensable for TAZ to stimulate cell proliferation, migration, and EMT induction.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum (HyClone) and 100 units/ml penicillin and streptomycin (Invitrogen). MCF10A cells were maintained in Dulbecco's modified Eagle's medium/F-12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 100 ng/ml cholera toxin, 100 units/ml penicillin, and streptomycin (Invitrogen). Cell transfection was performed using Lipofectamine 2000 (Invitrogen) or calcium phosphate methods. Cells were harvested at 24 h post-transfection for protein analysis or luciferase activity assay.
To establish stable TAZ-expressing cells, pBabe-TAZ retroviruses were generated and used to infect MCF10A cells. Stable pools were selected with puromycin for 5 days. TAZ stable pool cells were seeded in 6-well plates with 1 × 105 cells/well in triplicates. Cell growth was counted every day for 7 days.
Streptavidin-binding Peptide (SBP) Purification of TAZ Protein Complex—SBP-TAZ stable 293T cells from ten 150-mm plates were collected and lysed in 40 ml of 0.3% Nonidet P-40 buffer (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) containing protease and phosphatase inhibitors. SBP-TAZ in the supernatant was precipitated for 3 h with 100 μl of streptavidin resin (GE HealthCare, 17-5113-01), which was then washed 3 times with 0.3% Nonidet P-40 buffer followed by washing with 50 mm NH4HCO3 3 times. The precipitated protein was digested with trypsin (22). The supernatant was collected, dried, and dissolved in 10% acetonitrile, 0.8% formic acid solution. The peptides were analyzed by MS/MS.
Luciferase Activity Assay—For the luciferase reporter assay, 293T cells were seeded in 24-well plates. A mixture of 5× upstream activating sequence-luciferase reporter, Renilla, and the indicated plasmids was cotransfected. Twenty-four hours after transfection, cells were lysed, and luciferase activity was measured using a dual-luciferase reporter assay system (#E1960; Promega) following the manufacturer's instructions. The luciferase activity was measured by a luminometer (model TD-20/20). Transfection efficiency was normalized to thymidine kinase-driven Renilla luciferase activity as the internal control.
RNA Isolation and Real-time PCR—Total RNA was isolated from cultured cells using Trizol reagent (Invitrogen). cDNA was synthesized by reverse transcription using oligo(dT) as the primer and proceeded to real-time PCR with gene-specific primers in the presence of SYBR Premix Ex Taq (#DRR041A; TaKaRa). The relative abundance of mRNA was calculated by normalization to glyceraldehyde-3-phosphate dehydrogenase mRNA.
Western Blotting Analysis—Protein lysates were prepared from MCF10A, MCF10A TAZ, and mutants stable pool cells in a buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 25 mm NaF, and mixture protease inhibitors (Roche Applied Science). Tissue lysate (40 μg) was resolved by SDS-PAGE followed by Western blotting analysis. Antibodies to FLAG (#A00170 (GenScript) or #A2220 (Sigma)), E-cadherin (monoclonal antibody (mAb) #610405; BD Biosciences), N-cadherin (mAb #610920; BD Biosciences), vimentin (mAb #550513, BD Biosciences), occludin (SC-5562; Santa Cruz Biotechnology), and β-actin (13E5, #4970; Cell Signaling) were purchased commercially.
For immunoprecipitation experiments, 500 μg of cell lysate was incubated with anti-FLAG M2-agarose for 2 h at 4 °C. Beads were washed 3 times with lysis buffer and centrifuged at 5000 × g for 5 min between each wash. Protein was eluted from beads with 50 μl of Laemmli sample buffer (Bio-Rad). Lysates were resolved on 8–10% SDS-PAGE gels and transferred onto nitrocellulose (Bio-Rad) for Western blotting.
Chromatin Immunoprecipitation (ChIP)—ChIP analysis was performed as previously described (23). Briefly, formaldehyde was added directly to cell culture media to a final concentration of 1% at room temperature. Thirty minutes later glycine was added to a final concentration of 0.125 m. The cells were washed with ice-cold phosphate-buffered saline (PBS), scraped, and collected in cold PBS followed by steps of micrococcal nuclease digestion and sonication. After immunoprecipitation with anti-FLAG M2 beads, immunoprecipitated DNA were decross-linked, and proteinase K was digested and purified for PCR amplification. The ChIP-enriched DNA was subjected to PCR using promoter-specific primers (connective tissue growth factor (CTGF) sense, 5′-GGAGTGGTGCGAAGAGGATA-3′; CTGF antisense, 5′-GCCAATGAGCTGAATGGAGT-3′).
Wound-healing Assay—Monolayer cells were wounded with a sterile plastic pipette tip. Cell migration was observed 16 h later by microscopy.
Matrigel Assay—Matrigel assays were performed as reported previously with minor modifications (5). Briefly, cell culture plates were precoated with 100% Matrigel (BD Pharmingen). Cells (5000) were plated onto the Matrigel bed and cultured with mammary epithelial cell growth medium supplemented with hydrocortisone, insulin, epidermal growth factor, fetal bovine serum, and 2% Matrigel. Acinar formation was monitored by microscopy.
RESULTS
TEADs Are Major TAZ Binding Transcription Factors—As a transcription coactivator, TAZ must interact with a downstream transcription factor to induce gene expression. To identify TAZ target transcription factors, we performed TAZ affinity purification. TAZ was expressed as a fusion protein with the SBP, which has high affinity toward streptavidin and can be easily purified by streptavidin beads. To decrease nonspecific interactions caused by high expression level, we used retrovirus to establish 293T stable cell clones with low expression level of SBP-TAZ. SBP-TAZ was purified by streptavidin-Sepharose pulldown, and the purified proteins were detected by Western blotting (Fig. 1A) and silver staining (Fig. 1B). SBP-TAZ and several other proteins were specifically present in cells expressing SBP-TAZ but not in vector control cells (Fig. 1B). After SBP purification, we performed on-beads trypsin digestion, and the entire digestion mixture was analyzed by tandem liquid chromatography and mass spectrometry (LC-MS/MS). This purification strategy identified multiple putative TAZ-interacting proteins. Notably, all four TEAD family members, TEAD1, TEAD2, TEAD3, and TEAD4, were identified with high scores. In fact, multiple peptides for each TEAD protein were identified (Fig. 1C). These peptides were absent from control purification products from cells infected with the SBP vector. Western blotting of the affinity purified SBP-TAZ further confirmed that endogenous TEAD4 was co-purified with SBP-TAZ (Fig. 1D).
FIGURE 1.
TEADs are the major TAZ binding transcription factors. A, SBP-TAZ is stably expressed in 293T cells. Retrovirus was used to establish SBP-TAZ stable cell clones. SBP-TAZ was purified with streptavidin-Sepharose, and the purified proteins were examined by Western blotting. B, silver staining for the streptavidin-Sepharose purified proteins. The streptavidin-Sepharose purified proteins were separated by SDS-PAGE for silver staining. C, identification of multiple TEAD peptides from TAZ immunocomplex. After SBP purification, trypsin digestion was performed on beads, and the total digestion mixture was directly analyzed by liquid chromatography-MS/MS. The identified peptides for each TEAD family member were shown. D, endogenous TEAD4 was coprecipitated by SBP-TAZ. Cell lysate was precipitated with SBP-Sepharose followed by Western blotting with antibodies against TAED4 and TAZ.
TEAD Mediates TAZ-dependent Gene Induction—We also performed an independent screen for TAZ target transcription factors. This screen is based on the ability of TAZ to activate target transcription factors in a luciferase reporter assay. Clones of the Gal4-TF library (a total of 1100 putative human transcription factors fused with Gal4 DNA binding domain) and 5× upstream activating sequence-luciferase reporter driven by five Gal4 binding elements were individually co-transfected with or without TAZ. If a transcription factor is activated by TAZ, co-expression of TAZ should increase luciferase activity. This unbiased screen identified TEAD2, -3, and -4 as the top three transcription factors that are most potently activated by TAZ (Fig. 2A). TEAD1 is not in the library but was also confirmed to be activated by TAZ separately (Fig. 2A). Noteworthy, TEAD3 and TEAD4 were more potently activated by TAZ than TEAD1 and TEAD2 (Fig. 2A). These results complemented the TAZ affinity purification to establish TEADs as major transcription factors activated by TAZ.
FIGURE 2.
TEAD mediates TAZ-dependent gene induction. A, TAZ activates TEAD family transcription factors. 5× upstream activating sequence-Luciferase reporter, the indicated Gal4-fused transcription factors, and renilla were co-transfected with or without TAZ. The renilla activity normalized luciferase activity in the absence of TAZ was set to 1. B, Ser-51 of TAZ disrupts its interaction with TEAD4. The indicated plasmids were cotransfected into 293T cells. FLAG-TAZ was immunoprecipitated (IP) and probed with the indicated antibodies. C, TAZS51A loses the ability to activate TEAD4. The indicated plasmids were co-transfected with a 5× upstream activating sequence-luciferase reporter into 293T cells. Luciferase activity was measured and normalized to renilla activity. WT, wild type. D, dominant-negative TEAD1 blocks TAZ-mediated activation of CTGF promoter. The indicated plasmids were coexpressed in 293T cells. Cells were harvested for measurement of luciferase activity 24 h after transfection. E, TAZ binds to CTGF promoter. A ChIP assay was performed with anti-FLAG antibody using 293T cells stably expressing FLAG-TAZ. The presence of CTGF promoter was detected by PCR. WT, wild type. F, TEAD is required for TAZ-induced CTGF induction. RNA was extracted from the indicated MCF10A stable pools, and CTGF mRNA levels were analyzed by quantitative RT-PCR.
We previously observed that YAPS94A mutation diminished the interaction between YAP and TEAD (9). Interestingly, this residue essential for TEAD binding is conserved in TAZ. We, therefore, generated TAZS51A mutant, which is the equivalent to YAPS94A mutant. We found that TAZS51A or TAZS51D mutation abolished the interaction between TAZ and TEAD4 (Fig. 2B). TAZS51A mutant was also defective in TEAD4 activation (Fig. 2C), indicating that the TAZ-TEAD interaction is functionally important for TAZ to activate TAED4 and, presumably, all TEAD family transcription factors.
To further investigate the mechanism of TAZ in gene expression, mRNA expression from vector or TAZ-overexpressing MCF10A cells, which is an immortalized but non-transformed human mammary epithelial cell line, were determined by microarray (supplemental Table 1). Among many genes induced, we observed a robust induction of CTGF, a member of the CCN family of cysteine-rich-secreted proteins that regulates a wide variety of cellular functions, including adhesion, proliferation, migration, and differentiation (16, 17). CTGF has also been implicated in cell growth and cancer. We further examined whether TAZ stimulates CTGF expression and the role of TEAD in this process. We found that TAZ expression strongly activated the luciferase reporter driven by the CTGF promoter (Fig. 2D). Furthermore, co-expression of a dominant negative TEAD1ΔC, which has a deletion of the C-terminal TAZ interacting region but still retains the DNA binding domain, strongly blocked the ability of TAZ to stimulate CTGF promoter in a dose-dependent manner (Fig. 2D). TEAD1ΔC functions as a dominant negative probably by competing with endogenous TEAD for binding to target gene promoters. These observations indicate that TEAD is required for TAZ to activate the CTGF promoter.
To examine a direct interaction of TAZ with CTGF promoter, ChIP was performed in vector or FLAG-TAZ-expressing 293T cells with anti-FLAG antibody. We found that FLAG-TAZ could bind to the CTGF promoter (Fig. 2E). In contrast, the TEAD binding-defective TAZS51A mutant does not bind to CTGF promoter, supporting our model that TEAD mediates the interaction between TAZ and CTGF promoter. These observations are consistent with a previous study demonstrating that the CTGF promoter contains putative TEAD binding sites, and its expression is regulated by TEAD (9). We further determined the effect of TAZ expression on endogenous CTGF mRNA level. CTGF mRNA expression is induced by wild-type TAZ and even more strongly by TAZ4SA, which is a constitutively active mutation because all the Lats inhibitory phosphorylation sites are abolished in this mutant (Fig. 2E). However, the TEAD binding-defective TAZS51A or TAZ4SA-S51A could not induce CTGF expression (Fig. 2E). Together, we conclude that TEAD plays an essential role in the induction of CTGF by TAZ.
TEAD Binding Is Required for TAZ to Stimulate Cell Proliferation—In our previous study, we have established that TAZ acts as a downstream component of the Hippo pathway to promote cell proliferation (8). To examine the role of TEAD in the ability of TAZ to stimulate cell growth, we established MCF10A stable cells expressing TAZ wild-type or mutant proteins. As expected, MCF10A cells expressing TAZ proliferated much faster than the vector control cells (Fig. 3A). Mutation of serine 51 to alanine, which disrupted TEAD binding (Fig. 2B), largely abolished the ability of TAZ to stimulate MCF10A cell proliferation (Fig. 3A), suggesting the importance of TEAD binding for the ability of TAZ to stimulate cell proliferation. Mutation of the four putative Lats phosphorylation sites results in a constitutive active TAZ4SA mutant that is more active in stimulating cell proliferation than the wild-type TAZ (Fig. 3A). TAZ with combined 4SA and S51A mutations (TAZ4SA-S51A) was largely inactive to promote MCF10A proliferation (Fig. 3A), further supporting the notion that TEAD binding is important for TAZ to stimulate cell proliferation and that TEAD is a downstream effector of the Hippo pathway.
FIGURE 3.
Disruption of TAZ-TEAD binding abolishes the function of TAZ in promoting cell proliferation. A, growth curve of MCF10A cells stably expressing wild-type or mutant TAZ as indicated. B, TAZ4SA-S51A is compromised in inducing enlarged MCF10A cell acini. Indicated MCF10A-derived stable cells were cultured in three-dimensions on reconstituted basement membrane for 14 days before the pictures were taken. WT, wild type. C, growth curve of TAZ4SA expressing MCF10A cells. TEAD1/3/4 knockdown by shRNA are indicated. D, TEAD knockdown inhibits MCF10A cell acinar growth. TAZ4SA-expressing MCF10A cells were infected with control or TEAD1/3/4 shRNA lentivirus. Cell growth in Matrigel is shown.
We further investigated the requirement of TEAD binding for TAZ to stimulate MCF10A cell proliferation in a three-dimensional culture on Matrigel. MCF10A cells can form acini in Matrigel with defined size. Such a culture condition is more physiologically relevant to recapitulate the environment and morphogenesis of mammary epithelial tissue. We observed that the size of MCF10A cell acini was significantly increased by expression of TAZ and more potently by expression of TAZ4SA (Fig. 3B). However, the expression of the TAZS51A or TAZ4SA-S51A TEAD binding-defective form did not increase acinar size (Fig. 4B). Therefore, TEAD binding is essential for TAZ to promote cell proliferation.
FIGURE 4.
Disruption of TEAD-TAZ binding blocks the ability of TAZ to promote EMT. A, S51A mutation in TAZ blocks TAZ induced EMT-like morphological change. Phase-contrast images of MCF10A cells stably expressing TAZ and its mutants are shown. B, Ser-51 mutation abolishes actin organization remodeling induced by TAZ. Cells were stained with rhodamine-conjugated phalloidin. C, Ser-51 mutation abolishes TAZ-induced EMT. Cell lysates from MCF10A cells stably expressing the wild-type (WT) and mutant TAZ were analyzed with the indicated epithelial and mesenchymal marker antibodies. D, Ser-51 mutation abolishes TAZ-induced change of E-cadherin and N-cadherin mRNA expression. Total RNA was extracted from MCF10A cells stably expressing the wild-type and mutant TAZ, and quantitative RT-PCR was employed to determine E-cadherin and N-cadherin mRNA levels. E, TEAD1/3/4 knockdown blocks TAZ-induced EMT-like morphological change. TEAD1/3/4 were knocked down by shRNA. Indicated MCF10A stable cells were cultured, and the pictures were taken.
To provide further evidence for the functional importance of TEAD-TAZ interaction, we utilized shRNA to knock down TEADs in MCF10A cells. We have previously generated a lentiviral construct, shTEAD1/3/4, which can successfully knock down TEAD1, TEAD3, and TEAD4 simultaneously (9). We examined the effect of TEAD1/3/4 knockdown on TAZ-induced cell proliferation. Our data further support an important role of TEAD in TAZ-mediated stimulation of cell proliferation as TEAD knockdown dramatically retarded proliferation of the TAZ4SA-expressing cells (Fig. 3, C and D). Collectively, these results provide compelling evidence that TEAD is required for the function of TAZ in promoting cell proliferation.
TEAD Binding Is Essential for TAZ to Induce EMT—We previously reported that expression of wild-type TAZ or the active TAZ4SA mutant in MCF10A cells induces EMT. We examined whether TEAD binding is required for TAZ to promote EMT in MCF10A cells. TAZ or TAZ4SA expression induces dramatic EMT-like morphological changes, including a loss of cell-cell contact and cell scattering (Fig. 4A). However, both TEAD binding-deficient mutant TAZS51A and TAZ4SA-S51A were unable to promote such morphological alterations (Fig. 4A). We also performed F-actin staining to examine the organization of cytoskeleton. Cells expressing TAZ exhibited disorganization of cell junction, and the TAZ4SA-expressing cells displayed a dramatic increase of stress fibers along with disorganization of cell junction, consistent with the EMT phenotypes (Fig. 4B). In contrast, the TAZS51A- and TAZ4SA-S51A-expressing cells exhibited normal structures of cytoskeleton as observed in the MCF10A vector control cells (Fig. 4, A and B). These data indicate that TEAD binding is important for TAZ to induce mesenchymal-like morphological changes in MCF10A cells.
Western blotting for EMT markers was performed to further confirm whether TEAD-TAZ binding plays a role in TAZ-induced EMT. Epithelial marker E-cadherin and occludin were indeed down-regulated in cells expressing TAZ and more significantly in cells expressing TAZ4SA (Fig. 4C). Consistent with the down-regulation of epithelial markers, expression of mesenchymal markers N-cadherin and vimentin were up-regulated in TAZ4SA-expressing cells but not in the TEAD binding-defective TAZ4SA-S51A-expressing cells (Fig. 4C). The levels of N-cadherin and E-cadherin mRNA were also regulated by TAZ but not by the TEAD binding-defective mutants in a manner similar to their corresponding protein levels (Fig. 4D). Furthermore, infection of MCF10A/TAZ4SA cells with this shTEAD1/3/4 lentivirus inhibited the ability of TAZ4SA in promoting cell proliferation and EMT-like morphological alterations (Fig. 4E). Together, our data indicate that TEAD-TAZ binding is required for TAZ to induce EMT in MCF10A cells.
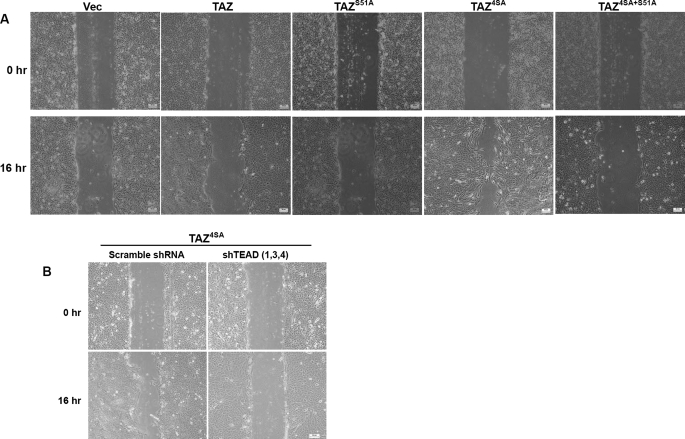
TEAD Is Required for TAZ to Promote Cell Migration—Cell migration plays an important role in cancer development. To examine the requirement of TEAD binding in mediating TAZ function in promoting cell migration, we performed wound-healing assays. Expressing of TAZ could promote the wound healing in cultured MCF10A cells (Fig. 4A). Mutation of the Lats phosphorylation sites dramatically increased the effect of TAZ to promote wound closure. However, cells expressing TAZS51A and TAZ4SA-S51A exhibited minimal wound closure, indicating that TEAD binding is required for TAZ to promote cell migration (Fig. 5A). This observation is consistent with a defect of TAZS51A or TAZ4SA-S51A in inducing EMT.
FIGURE 5.
TEAD is required for TAZ function in promoting cell migration. A, Ser-51 mutation abolishes TAZ-induced cell migration. Migration of MCF10A cells stably expressing the wild-type and mutant TAZ was determined by a wound-healing assay. B, TEAD1/3/4 knockdown blocks TAZ induced cell migration. Migration ability of indicated MCF10A stable cells was determined by a wound-healing assay.
We also examined the effect of TEAD knockdown on the activity of TAZ to promote cell migration. Knockdown of TEAD significantly compromised the ability of TAZ4SA in wound healing when compared with cells infected with scramble shRNA viruses (Fig. 5B). Collectively, these results support the conclusion that TEAD is required for the function of TAZ in promoting cell migration.
DISCUSSION
Breast cancer is the second-leading cause of cancer-related death in females in United States(18). Elucidating the molecular mechanisms underlying the development and progression of breast cancer is critical for its prevention, diagnosis, and therapy. TAZ has been reported to promote cell proliferation, EMT, and cell migration in normal mammary epithelial cells (8). TAZ expression level has also been reported to correlate with breast cancer cell invasiveness (14). Elevated TAZ expression is observed in more than 20% of breast cancers especially invasive ductal carcinomas (14). Consistently, TAZ has also been shown to be inhibited by the Hippo tumor suppressor pathway, which contains well established human tumor suppressor NF2 and other genes mutated in human cancer, such as Sav and Mob.
However, the key transcription factors mediating TAZ function in promoting cell proliferation, migration, and EMT have not been defined, although several transcription factors, such as RUNX, peroxisome proliferator-activated receptor γ, Tbx5, and Pax3, have been implicated to act downstream of TAZ (3–5). The lack of systemic study on TAZ transcription factor targets is certainly a reason for the absence of this information; therefore, we addressed this question by a combination of biochemical purification and functional screening. Interestingly, TAZ affinity purification identified all four of the TEAD family transcription factors as TAZ-binding proteins. Besides the four TEAD proteins, only two additional transcription factors were identified in our affinity purification but with lower mass spectrometry scores (data not shown). This strongly suggests TEADs as the major TAZ transcription factor partners at least in HEK293T cells. Consistently, an independent screen of a Gal4 fusion human transcription factor library identified three of the four TEAD family transcription factors as the most potent targets of TAZ determined by the reporter activation assay, whereas the other TEAD family member, TEAD1, was not present in the library. Together, these data support the notion that TEADs are bona fide TAZ target transcription factors, although other transcription factors may be used for the diverse functions of TAZ in other cell types.
We demonstrated in this report that TEAD mediates TAZ function in promoting cell proliferation, EMT, and cell migration, which are all involved in cancer initiation and progression. We manipulated TAZ-TEAD interaction mainly by two approaches; that is, a point mutation on TAZ (S51A) that abolishes TAZ-TEAD interaction and shRNA knockdown of TEAD. Both approaches support that perturbation of TAZ-TEAD interaction aborts or attenuates the effect of TAZ on promoting cell proliferation in two-dimensional or three-dimensional culture, inducing EMT-like morphological changes and EMT molecular markers and promoting wound closure in wound healing cell migration assay. Therefore, TAZ-TEAD interaction might be a target of therapy for cancers showing elevated TAZ expression or Hippo pathway dysfunction.
Another interesting aspect of the function of TAZ-TEAD in inducing EMT is its implication in stem cell self-renewal. A recent report suggested a correlation between mesenchymal traits and the gain of epithelial stem cell properties using immortalized human mammary epithelial cells as a model system (19). Interestingly, TAZ has recently been implicated in maintaining stem cell self-renewal ability, although this function of TAZ has been attributed to its ability to regulate Smad subcellular localization (20). It will be interesting to test whether TEAD also contributes to the ability of TAZ to maintain stem cell self-renewal or cancer stem cell population in breast cancer.
To explore the regulation of gene expression by TAZ-TEAD, we used gene expression microarray to examine genes induced by TAZ expression. As expected for a transcription co-activator, ectopic expression of TAZ induces expression of many genes. We compared the gene expression profile of TAZ- or YAP-overexpressing MCF10A cells. Interestingly, although TAZ and YAP are highly homologous and there are significant portion of genes commonly induced by both TAZ and YAP, there are also many genes uniquely induced by TAZ or YAP (supplemental Fig. 1 and Table 1). We previously reported that many YAP target genes are TEAD-dependent. Importantly, by looking at the distribution of 55 high confidence TEAD-dependent YAP target genes (those induced by YAP-wild type and active 5SA protein but not YAP-S94A TEAD binding-deficient protein), we found that 32 of them are common among YAP- and TAZ-inducible genes, whereas 23 of them are uniquely induced by YAP. Therefore, TAZ and YAP can use TEAD family transcription factors to induce sets of common as well as distinct target genes, which might be responsible for the shared and unique functions of TAZ and YAP. To further confirm the function of TEAD in TAZ-dependent gene induction, we showed that CTGF is a direct target gene of TAZ-TEAD in mammalian cells and TEAD is required for TAZ to induce CTGF expression. Interestingly, CTGF has been implicated in multiple human cancer development and progression, consistent with the positive role of TAZ in tumorigenesis (16, 17, 21).
In summary, our study identified TEAD family transcription factors as major TAZ target transcription factors and established a functional partnership between TAZ and TEAD in cell proliferation, EMT, and cell migration. These findings advance our understanding of the role of TAZ in cancer development and provide a potential therapeutic target for human breast cancer treatment.
Supplementary Material
Acknowledgments
We thank the members of the Fudan Molecular and Cell Biology laboratory for discussions throughout this study and the Institutes of Biomedical Sciences for support.
This work was supported, in whole or in part, by National Institutes of Health grants RO1CA65572 and RO1CA132809 (to Y. X. and K.-L. G.). This work was also supported by 985 Program from the Chinese Ministry of Education, National High Technology Research and Development Program of China Grants 2004BA711A18 and 2006AA02A308, State Key Development Program for Basic Research of China Grants 2006CB806700 and 2009CB918401, National Natural Science Foundation of China Grants 30600112 and 30871255, Shanghai Key Project Grant 06JC14086, Pufa Talent Grant 07pj14011, Shanghai Leading Academic Discipline Project, project number B110, and a University of Michigan Rackham predoctoral fellowship (to B. Z.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
Footnotes
The abbreviations used are: EMT, epithelial-mesenchymal transition; SBP, streptavidin-binding peptide; ChIP, chromatin immunoprecipitation; CTGF, connective tissue growth factor; MS, mass spectroscopy; shRNA, short hairpin RNA; YAP, Yes-associated protein; TEAD, TEA domain.
References
- 1.Kanai, F., Marignani, P. A., Sarbassova, D., Yagi, R., Hall, R. A., Donowitz, M., Hisaminato, A., Fujiwara, T., Ito, Y., Cantley, L. C., and Yaffe, M. B. (2000) EMBO J. 19 6778–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong, J. H., and Yaffe, M. B. (2006) Cell Cycle 5 176–179 [DOI] [PubMed] [Google Scholar]
- 3.Hong, J. H., Hwang, E. S., McManus, M. T., Amsterdam, A., Tian, Y., Kalmukova, R., Mueller, E., Benjamin, T., Spiegelman, B. M., Sharp, P. A., Hopkins, N., and Yaffe, M. B. (2005) Science 309 1074–1078 [DOI] [PubMed] [Google Scholar]
- 4.Murakami, M., Tominaga, J., Makita, R., Uchijima, Y., Kurihara, Y., Nakagawa, O., Asano, T., and Kurihara, H. (2006) Biochem. Biophys. Res. Commun. 339 533–539 [DOI] [PubMed] [Google Scholar]
- 5.Murakami, M., Nakagawa, M., Olson, E. N., and Nakagawa, O. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18034–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makita, R., Uchijima, Y., Nishiyama, K., Amano, T., Chen, Q., Takeuchi, T., Mitani, A., Nagase, T., Yatomi, Y., Aburatani, H., Nakagawa, O., Small, E. V., Cobo-Stark, P., Igarashi, P., Murakami, M., Tominaga, J., Sato, T., Asano, T., Kurihara, Y., and Kurihara, H. (2008) Am. J. Physiol. Renal Physiol. 294 542–553 [DOI] [PubMed] [Google Scholar]
- 7.Hossain, Z., Ali, S. M., Ko, H. L., Xu, J., Ng, C. P., Guo, K., Qi, Z., Ponniah, S., Hong, W., and Hunziker, W. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei, Q. Y., Zhang, H., Zhao, B., Zha, Z. Y., Bai, F., Pei, X. H., Zhao, S., Xiong, Y., and Guan, K. L. (2008) Mol. Cell. Biol. 28 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao, B., Ye, X., Yu, J., Li, L., Li, W., Li, S., Yu, J., Lin, J. D., Wang, C. Y., Chinnaiyan, A. M., Lai, Z. C., and Guan, K. L. (2008) Genes Dev. 22 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClatchey, A. I., and Giovannini, M. (2005) Genes Dev. 19 2265–2277 [DOI] [PubMed] [Google Scholar]
- 11.Hamaratoglu, F., Willecke, M., Kango-Singh, M., Nolo, R., Hyun, E., Tao, C., Jafar-Nejad, H., and Halder, G. (2006) Nat. Cell Biol. 8 27–36 [DOI] [PubMed] [Google Scholar]
- 12.Lai, Z. C., Wei, X., Shimizu, T., Ramos, E., Rohrbaugh, M., Nikolaidis, N., Ho, L. L., and Li, Y. (2005) Cell 120 675–685 [DOI] [PubMed] [Google Scholar]
- 13.Tapon, N., Harvey, K. F., Bell, D. W., Wahrer, D. C., Schiripo, T. A., Haber, D. A., and Hariharan, I. K. (2002) Cell 110 467–478 [DOI] [PubMed] [Google Scholar]
- 14.Chan, S. W., Lim, C. J., Guo, K., Ng, C. P., Lee, I., Hunziker, W., Zeng, Q., and Hong, W. J. (2008) Cancer Res. 68 2592–2598 [DOI] [PubMed] [Google Scholar]
- 15.Park, K. S., Whitsett, J. A., Di Palma, T., Hong, J. H., Yaffe, M. B., and Zannini, M. (2004) J. Biol. Chem. 279 17384–17390 [DOI] [PubMed] [Google Scholar]
- 16.Chu, C. Y., Chang, C. C., Prakash, E., and Kuo, M. L. (2008) J. Biomed. Sci. 15 675–685 [DOI] [PubMed] [Google Scholar]
- 17.Sala-Torra, O., Gundacker, H. M., Stirewalt, D. L., Ladne, P. A., Pogosova-Agadjanyan, E. L., Slovak, M. L., Willman, C. L., Heimfeld, S., Boldt, D. H., and Radich, J. P. (2007) Blood 109 3080–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinestrosa, M. C., Dickersin, K., Klein, P., Mayer, M., Noss, K., Slamon, D., Sledge, G., and Visco, F. M. (2007) Nat. Rev. Cancer 7 309–315 [DOI] [PubMed] [Google Scholar]
- 19.Mani, S. A., Guo, W., Liao, M. J., Eaton, E.N., Ayyanan, A., Zhou, A. Y., Brooks, M., Reinhard, F., Zhang, C. C., Shipitsin, M., Campbell, L. L., Polyak, K., Brisken, C., Yang, J., and Weinberg, R. A. (2008) Cell 133 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varelas, X., Sakuma, R., Samavarchi-Tehrani, P., Peerani, R., Rao, B. M., Dembowy, J., Yaffe, M. B., Zandstra, P. W., and Wrana, J. L. (2008) Nat. Cell Biol. 10 837–848 [DOI] [PubMed] [Google Scholar]
- 21.Dornhöfer, N., Spong, S., Bennewith, K., Salim, A., Klaus, S., Kambham, N., Wong, C., Kaper, F., Sutphin, P., Nacamuli, R., Höckel, M., Le, Q., Longaker, M., Yang, G., Koong, A., and Giaccia, A. (2006) Cancer Res. 66 5816–5827 [DOI] [PubMed] [Google Scholar]
- 22.Rybak, J. N., Ettorre, A., Kaissling, B., Giavazzi, R., Neri, D., and Elia, G. (2005) Nat. Methods 2 291–298 [DOI] [PubMed] [Google Scholar]
- 23.Wang, G., Balamotis, M. A., Stevens, J. L., Yamaguchi, Y., Handa, H., and Berk, A. J. (2005) Mol. Cell 17 683–694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.