Abstract
APOBEC3G (A3G) is a cellular protein that has been identified as an innate anti-human immunodeficiency virus type 1 (HIV-1) factor. One of the major functions of HIV-1 virion infectivity protein (Vif) protein is to target A3G for ubiquitination/proteasome-mediated degradation and, as a result, evade the host innate defense mechanism. Thus, we wished to devise a strategy to restore the anti-HIV activity of A3G by actively targeting it into HIV-1 virions and countering HIV-1 Vif-targeted degradation. In the current study we performed a series of proof-of-concept experiments for this strategy using as a delivery vehicle of A3G, a derivate of non-pathogenic Nef mutant Nef7 that is capable of being efficiently incorporated into HIV-1 virions. We demonstrate that the Nef7.A3G fusion protein retains several important properties of Nef7; that is, the higher virion incorporation efficiency, no PAK-2 (p21-activated kinase 2) activation, and no CD4 and major histocompatibility complex I down-regulation. Meanwhile, we show that virion incorporated Nef7.A3G possesses the anti-HIV infectivity function of A3G. Moreover, we show that virus-like particle-mediated inverse fusion delivery of Nef7.A3G into HIV-infected CD4+ T lymphocytes leads to potent inhibition of HIV-1 replication in these cells. Taken together, these results indicate that Nef7.A3G can effectively restrict HIV infection and replication by restoring the virion incorporation of A3G, even in the presence of Vif.
Apolipoprotein mRNA editing enzyme catalytic peptide 3G (APOBEC3G, or A3G) is an innate antiviral cellular protein that dramatically reduces human immunodeficiency virus type 1 (HIV-1)2 infectivity when incorporated into virions (1, 2). It is a single-stranded DNA deaminase that functions immediately after viral infection, during the first round of reverse transcription, to deaminate cytidine to uracil on the minus strand of the proviral DNA. This results in hypermutations that lead to loss of genetic stability and protein function (3, 4). The key to the A3G anti-HIV function is virion incorporation. When HIV-1 infects a cell, A3G is incorporated into the progeny virions from that cell through interactions with HIV-1 nucleocapsid protein and RNA. When those virions then infect a second cell, A3G renders them non-replicative. However, lentiviruses such as HIV-1 have evolved a mechanism to prevent the antiviral effects of A3G and other members of APOBEC3 family. The HIV-1 virion infectivity protein (Vif) binds to A3G and targets it for degradation by recruiting the E3 ubiquitin ligase cullin 5-elongin B/C (5), thus preventing A3G encapsidation (6–9).
HIV-1 Nef protein is an accessory protein of ∼27 kDa. It is post-translationally modified by phosphorylation and by the addition of a myristoyl moiety to its second amino acid (glycine), which aids in its membrane targeting and is required for many Nef functions (10–12). Although Nef is dispensable for in vitro HIV-1 replication, it is essential for efficient HIV-1 replication and pathogenesis in vivo (13–16). Nef functions can be broadly separated into three categories: alteration of protein trafficking, especially in the context of the down-regulation of surface receptors (12, 17–21), alterations in cell signaling cascades, including T cell activation (22–25), and enhancement of HIV infectivity (26–29).
Recently a Nef mutant (V153L, E177G) known as Nef7 has been reported to possess some interesting properties. Nef7 is a derivative of F12-HIV, a non-producing HIV-1 strain (30, 31); it is defective for a number of the typical Nef functions, including CD4 and major histocompatibility complex (MHC) class I down-regulation, p21-activated kinase 2 (PAK-2) activation, and binding to the V1H regulatory subunit of the vacuolar ATPase (31). Importantly, although WT Nef is incorporated into the HIV-1 core, Nef7 has the ability to be incorporated into virions at a much higher level. Moreover, the C-terminal fusion of proteins to Nef7 does not significantly alter the high virion incorporation property of Nef7 (32, 33). Collectively, these unique properties offer Nef7 as a potentially novel and non-toxic carrier platform to deliver therapeutic proteins into HIV virions and subsequently block HIV replication.
Thus, we hypothesized that the high virion incorporation of Nef7 in the context of the Nef7.A3G fusion protein would override Vif-targeted A3G degradation and as a result, restore the anti-HIV phenotype of A3G. In the current study we characterized the Nef7.A3G protein for its incorporation into HIV-1 virions, its effects on several well established Nef functions, and its effects on the anti-HIV activity of A3G.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfections—293T and HeLa cells were purchased from American Tissue Culture Collection (ATCC, Manassas, VA). U87.CD4.CXCR4 and U87.CD4.CCR5 cells were obtained from NIH AIDS Reagent Program, which were generously donated by Dr. D. Littman. These cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg/ml penicillin and streptomycin, and 2 mm glutamine at 37 °C with 5% CO2. 293T cells were transfected by the standard calcium phosphate precipitation method. HeLa cells were transfected using the Lipofectamine 2000 system (Invitrogen). pcDNA3 was used throughout the studies to normalize the amounts of DNA for all transfections.
Plasmids—pcDNA3.1-APOBEC3G-HA and psPAX2 plasmids were obtained from NIH AIDS Research Reagent Program and were donated by Dr. W. C. Greene and Dr. D. Trono, respectively. pHLA-A2 plasmid was a kind gift from Dr. C. Toloukian. HIV.env–.nef– plasmid was generously provided by Dr. M. Emmerman. HIV.env–vif– plasmid has been previously described (34). HIV-Luc, VSV-G, HXB2.env, YU2.env, pc.CXCR4, and pc.CD4 plasmids were described elsewhere (35). pNef.myc was constructed to express the nef gene from HIV-1 NL4-3 and an Myc epitope at the C terminus of Nef using the standard PCR cloning technique. All mutagenesis was performed using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and appropriate primers. nef point mutations (pNef153.myc, pNef177.myc, and Nef7.myc) were constructed in the context of pNef.Myc with primer pairs 5′-ccc aag ctt atg ggt ggc aag tgg tca-3′ (the HindIII site is underlined) and 5′-ccg gaa ttc tca aga act tca tga ggc-3′ (the EcoRI site is underlined), 5′-ccc aag ctt ctt ctt ctc cgg tta ttt cct ctc ttg tgg-3′ and 5′-ccc aag ctt tag acc gag ttg acc atg atc gaa cat-3′, and 5′-c ctg cat gga atg gat gac ccg ggg aga gaa gtg tta gag tgg ag-3′ and 5′-ct cca ctc taa cac ttc tct ccc cgg gtc atc cat tcc atg cag g-3′. For pNef.HA and pNef7.HA plasmids, the Myc tag in pNef.Myc and pNef7.Myc was replaced with primers 5′-ccc tta cCA TAT GAT GTT CCA GAT TAC GCT tga agc cga att ctg cag ata-3′ and 5′-gg aat tcC ATA TGG GTA ctc tgc gtt ctt gta gta ctc-3′ (NdeI sites are underlined, and the HA tag is capitalized). pNef.GFP and pNef7.GFP plasmids were also constructed in the context of pEGFP.N3 backbone (Clontech, Mountain View, CA) using pNef.Myc and pNef7.Myc as the respective templates and primers 5′-ccg gaa ttc atg ggt ggc aag tgg tca-3′ (EcoRI site underlined) and 5′-ccg act agt gca gtt ctt gaa gta ctc-3′ (BamH1 site underlined). pNef7.A3G fusion vector was constructed by first mutating the stop codon of pNef. Myc with an oligonucleotide 5′-ccg gaa ttc gtt ctt gta gta ctc cgg atg-3′ introducing an EcoR1 site (underlined) and then cloning the APOBEC3G open reading frame at the EcoRI site of Nef. pNef7.A3G/D128K (D128K) and pNef7.A3G/E259Q (E259Q) plasmids were constructed in the context of pNef7.A3G with primer pairs 5′-ctc tact ac ttc tgg AAG cca gat tac cag gag gcg-3′ and 5′-cgc ctc ctg gta atc tgg CTT cca gaa gta gta gag-3′ (D128K mutation capitalized) and pairs 5′-gt ttc ttg aag gcc gcc atg CAC agc tgt gct tcc tg-3′ and 5′-ca gga agc aca gct GTG cat ggc ggc ctt caa gaa ac-3′ (E259Q mutation capitalized). HIV-Luc.vif– was constructed by first subcloning a portion of HIV-Luc containing the vif gene into pBluescript vector followed by site-directed mutagenesis to introduce a stop codon in the Nde1 site of vif using the primer pairs 5′-gta aaa cac cat TAG tat att tca agg aaa gc-3′ and 5′-gc ttt cct tga aat ata CTA atg gtg ttt tac-3′ (stop codon capitalized). The vif– portion was then placed back in HIV-Luc to create HIV-Luc.vif–.
Preparation and Infection of Pseudotyped HIV-1—HIV-1 viruses pseudotyped with different envelope proteins were prepared as previously described (35). Briefly, 293T cells were transfected with plasmids HIV-Luc or HIV-luc.vif–, VSV-G, HXB2.env, or YU2.env and Nef or each of Nef derivatives by the calcium phosphate precipitation method. Cell culture supernatants were collected 72 h after transfection, filtered, and saved as progeny viruses. Progeny viruses were assayed for RT activity. Viruses with an equal level of RT activity were incubated with target cells at 37 °C for 2 h followed by repeated washes with fresh medium to remove the remaining viruses. Infected cells were cultured further in fresh complete medium for 48 h. The cells were analyzed for viral infectivity, as measured by the Luc activity assay. For infection, the amount of VSV-G-pseudotyped viruses used was only one-tenth that of YU2.env and HXB2.env-pesudotyped viruses.
Analysis of Nef Virion Incorporation and Intracellular Delivery—293T cells were transfected with plasmids HIV. env–.nef– or HIV.env–vif– and Nef or each of the Nef derivatives. In some experiments VSV-G, HXB2.env, or YU2.env plasmid was also included. Cell culture supernatants were collected 72 h after transfection, cleared of cell debris, and used as progeny viruses. Progeny viruses were assayed for RT activity as previously described (35). Viruses with an equal level of RT activity were lysed in radioimmune precipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS in phosphate-buffered saline (PBS)), electrophoresed on 12% SDS-PAGE gel, and analyzed by immunoblotting. The blots were probed first with an anti-Myc, anti-HA (Santa Cruz Biotechnologies Inc., Santa Cruz, CA), or anti-HIV p24 antibody (NIH AIDS Reagent Program), then with appropriate horseradish peroxidase-conjugated secondary antibodies and were visualized with a homemade ECL system. Relative levels of protein incorporation into virions were determined by densitometric scanning of the blots and calculated using HIV-1 p24 as an internal standard. To determine delivery of virion-incorporated Nef-GFP fusion proteins, HIV.env–.nef– virions containing Nef-GFP or Nef7-GFP were prepared by transfection as stated above. The viruses were used to infect 293T cells via spinoculation as described (32). Briefly, the viruses were allowed to adsorb onto cells by 1 h of centrifugation at 150 × g at room temperature followed by incubation at 37 °C for 3 h. Cells were then trypsinized to remove cell surface-bound viruses and analyzed for GFP-positive cells using a FACSCalibur II.
Preparation of Cell Lysates, Immunoprecipitation/Western Blot Analysis, and in Vitro Kinase Assay—Cells were washed twice with ice-cold PBS. Cells were collected and lysed in radioimmune precipitation assay buffer and 1× protease inhibitor mixture (Roche Diagnostics), and cell lysates were obtained by centrifugation and removal of the cell debris. For immunoprecipitation/Western blot analysis, cell lysates were subjected to immunoprecipitation in a buffer (137 mm NaCl, 50 mm Tris·HCl, pH 8.0, 2 mm EDTA, 0.56% Nonidet P-40, and 2 mm Na3VO4) using an anti-Nef antibody (NIH AIDS Reagent Program) followed by Western blot analysis using an anti-PAK-2 antibody (Santa Cruz). For in vitro kinase assay, the immunoprecipitates were suspended in 30 μl of kinase assay buffer (50 mm HEPES, pH 8.0, 150 mm NaCl, 5 mm EDTA, 10 mm MgCl2, 0.02% Triton X-100). Ten μCi of [γ-32P]ATP was added to the reaction. After incubation at room temperature for 5 min, the reaction was separated by SDS-PAGE. The gel was dried and exposed to x-ray film.
Immunofluorescence Staining—Transfected HeLa cells were washed with cold PBS and detached using 0.5 mm EDTA in PBS. An anti-CD4 antibody (NIH AIDS Reagent Program) or an anti-MHC I antibody (a gift from Dr. J. Blum) was added to the cell suspension and incubated on ice for 30 min. An appropriate phycoerythrin-conjugated secondary antibody was then added, and the cells were incubated on ice in the dark for another 30 min. The cells were then fixed in 1% paraformaldehyde at 4 °C for 4 h, then washed with PBS and analyzed using a FACSCalibur II. Cells were washed 3 times with cold HBSS (1× Hanks' salt solution, 4 mm NaHCO3, 1% bovine serum albumin, and 0.02% NaN3) between the primary antibody staining, secondary antibody staining, and fixation steps.
Preparation and Transduction of Virus-like Particles (VLPs)—293T cells were transfected with psPAX2 packaging vector (36), pc.CXCR4 pc.CD4, and Nef, Nef7, Nef7.A3G or A3G. Cell culture supernatants were collected 72 h after transfection, cleared of cell debris, and used as VLPs. The VLPs were assayed for RT activity. Jurkat cells were infected at 37 °C for 3 h with replication-competent HIV-1 HXB2 viruses of an RT activity of 10,000 cpm, which was estimated to give rise to about 20–25% infection efficiency using a replication-defective HIV-green fluorescence protein reporter virus (HIV.env–.GFP) (35, 38) and washed to remove the remaining viruses. Infected cells were then transduced twice with appropriate VLPs, 50,000 cpm after HIV-1 infection and another 50,000 cpm the next day. One-third of cell culture supernatants was collected every other day and assayed for RT activity, whereas fresh medium was added to the cultures. The RT activity was normalized to the cell counts.
Data Analysis—All values were expressed as the mean ± S.E. All comparisons were made based on wild-type Nef (WT Nef) using two-tailed Student's t test. A p value of <0.05 was considered statistically significant (*), and p < 0.01 highly significant (**).
RESULTS
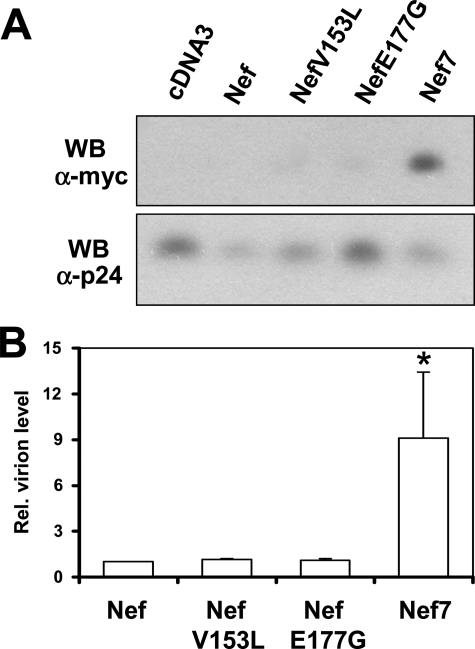
Enhanced Nef7 Incorporation into HIV-1 Virions—To test our hypothesis that A3G fusion to Nef7 (Nef7.3AG) can restore the virion incorporation of A3G and inactivate HIV-1 infectivity in the presence of HIV-1 Vif expression, we first decided to ascertain the high virion incorporation property of Nef7. We constructed Nef and Nef7 mutants in which V152L and E177G point mutations were introduced and compared their ability to be incorporated into HIV-1 virions. We then transfected 293T cells with an env- and nef-defective HIV-1 (HIV.env–.nef–) and wild-type Nef (WT Nef) or Nef 7. We also constructed and transfected two single point Nef7 mutants, V153L and E177G, in these experiments. Viruses were collected 72 h post-transfection and assayed for RT activity. Viruses with equal levels of RT activity were pelleted and lysed in radioimmune precipitation assay buffer then subjected to Western blot analysis using an antibody against a Myc epitope that was engineered to the C terminus of all Nefs and an anti-HIV p24 antibody (Fig. 1A). Relative virion Nef incorporation was normalized to HIV-1 p24. Compared with WT Nef, Nef7 showed ∼9-fold higher virion incorporation (Fig. 1B). There was little virion incorporation of WT Nef, V153L, and E177G over the cDNA background control. These results confirm that Nef7 is capable of being incorporated into HIV-1 virions at a higher level than WT Nef.
FIGURE 1.
Virion incorporation of Nef and its mutants. 293T cells were transfected with HIV.env–.nef– and WT Nef, NefV153L, NefE177G, or Nef7 with pcDNA3 included as a control. Progeny viruses were collected 72 h post-transfection and quantified for their RT activity. Viruses with an equal level of RT activity were pelleted and analyzed for Western blot (WB) analysis using an antibody against the Myc epitope that was added to the C terminus of all Nefs and an antibody against HIV-1 p24 antigen (A). The relative (Rel.) level of virion protein from three independent experiments was determined by densitometry and normalized to the virion p24 level (B). The WT Nef value was set as 1.
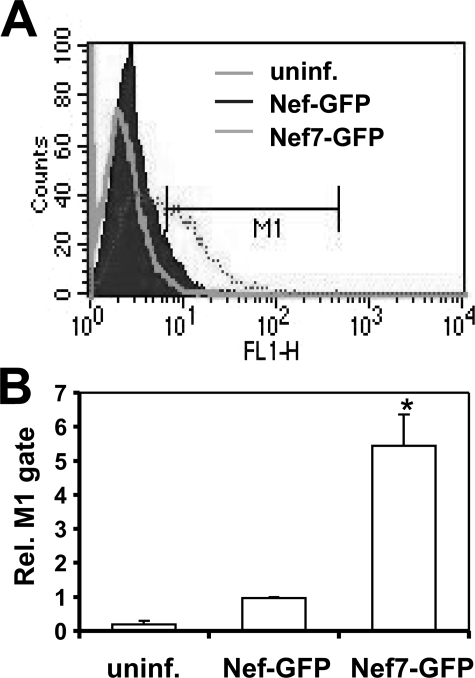
Efficient Delivery of Virion Nef7 into Cells—We next determined whether virion incorporated Nef would be efficiently delivered into target cells. We constructed Nef-GFP and Nef7-GFP fusion plasmids and transfected them into 293T cells along with HIV.env–.nef– and VSV-G to produce GFP-labeled VSV-G-pseudotyped HIV-1. GFP expression in trans and incorporation into HIV-1 virions would allow accurate quantitation of the delivery efficiency of virion proteins into host cells. Thus, we infected fresh 293T cells with these newly pseudotyped viruses containing an equal level of RT activity by spinoculation and analyzed the percentage of GFP+ cells in each infection (Fig. 2A). In agreement with the virion incorporation, about 5–6-fold more GFP+ cells were detected in the infections by viruses carrying Nef7-GFP than those of viruses carrying WT Nef-GFP (Fig. 2B), suggesting that virion Nef7 protein can be delivered into target cells through infection.
FIGURE 2.
Delivery of virion WT Nef and Nef7 into cells. 293T cells were transfected with HIV.env–.nef– and VSV-G and with WT Nef-GFP or Nef7-GFP expression plasmids. Progeny viruses were collected and quantitated as stated above. Viruses with an equal level of RT activity were used to infect fresh 293T by spinoculation. Uninfected cells (uninf.) were used as a control. After 3 h cells were washed to remove unbound viruses and then trypsinized to remove cell surface-bound virus. These cells were then analyzed for GFP-positive cells by fluorescence-activated cell sorter to detect intracellular delivery of virion Nef (A). FL1-H, increasing GFP intensity. The relative level of GFP-positive cells from three independent experiments was calculated with the number of GFP-positive cells in WT Nef-GFP infection set to 1 (B).
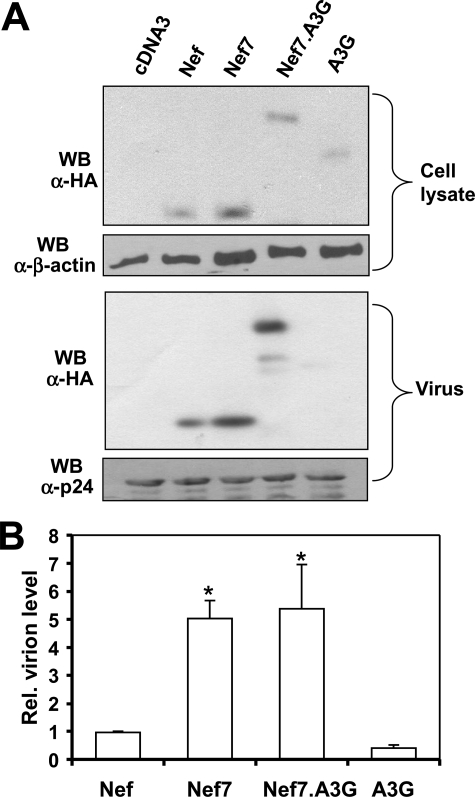
Higher Virion Incorporation of the Nef7.A3G Fusion Protein—Next, we determined whether Nef7 in the form of fusion with A3G (Nef7.A3G) retained the higher virion incorporation property of Nef7. We constructed the Nef7.A3G fusion expression plasmid. We then prepared HIV.env–.nef– viruses carrying WT Nef, Nef7, Nef7.A3G, or A3G, as described above and compared Nef7 and Nef7.A3G for their virion incorporation by Western blot analysis. We also included WT Nef and A3G as controls in these experiments. We performed Western blot analysis to ensure a comparable expression level of all proteins (Fig. 3A). Quantitation of virion protein incorporation showed that Nef7 and Nef7.A3G had comparable levels in HIV-1 virions (Fig. 3B). As Nef is a virion protein, it was detected in the virus blot. On the other hand, Vif expression greatly diminishes encapsidation of A3G in the virus particles, and as a result, only a very faint band of A3G appeared in the virus blot. These results indicate that Nef7.A3G fusion does not alter the higher incorporation of Nef7 into HIV-1 virions.
FIGURE 3.
Virion incorporation of the Nef7.A3G fusion protein. 293T cells were transfected with HIV.env–.nef– and WT Nef, Nef7, Nef7.A3G, or A3G expression plasmids with pcDNA3 included as a control. Progeny viruses were collected and subjected to Western blot (WB) analysis along with whole cell lysate using an antibody against an HA epitope that was added to the C terminus of all Nefs, Nef7.A3G, and A3G and antibodies against HIV-1 p24 or β-actin as loading controls (A) followed by quantitation of the protein level in virions from three independent experiments, as stated above (B).
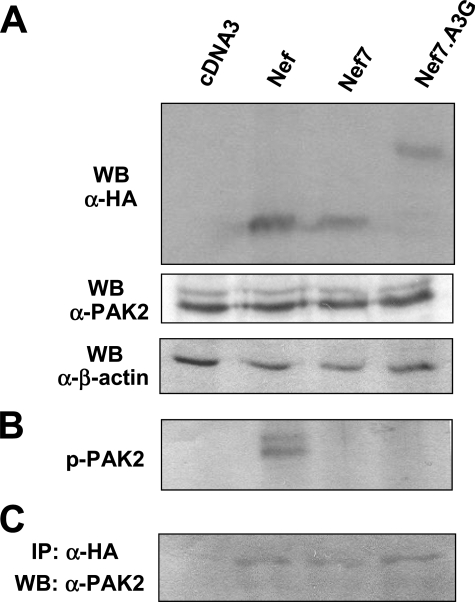
No PAK-2 Activation by the Nef7.A3G Fusion Protein—Activation of PAK-2 is an important mechanism whereby Nef exerts its pathogenic function on the host (39). Nef7 does not activate PAK-2 (31). Therefore, we determined whether the Nef7.A3G fusion led to PAK-2 reactivation. We transfected 293T cells with HIV.env–.nef– and WT Nef, Nef7, or Nef7.A3G. We first performed Western blot analysis for endogenous PAK-2 expression in all transfections using an anti-PAK-2 antibody. We also performed Western blot analysis using an anti-HA antibody to ensure a comparable level of Nef expression using an anti-β-actin antibody to ensure equal loading of the cell lysates. There was little change in endogenous PAK-2 levels among all transfections (Fig. 4A). We then performed immunoprecipitation of the cell lysates using an anti-HA antibody followed by an in vitro kinase assay for PAK-2 phosphorylation. PAK-2 phosphorylation, an indicator of PAK-2 activation, was only detected in cells that were transfected with WT Nef (Fig. 4B). There was no phosphorylation signal in cells that were transfected with Nef7, Nef7.A3G, and pcDNA3 control. To ascertain that the inability of Nef7 and Nef7.A3G to inactivate PAK-2 is not due to altered association between PAK-2 and these Nef derivatives, we performed immunoblotting of the immunoprecipitates using an anti-PAK-2 antibody. Compared with the pcDNA3 control, Nef7 and Nef7.A3G exhibited a similar level of immunoprecipitated PAK-2 to that of WT Nef (Fig. 4C). Taken together, these results show that, like Nef7, Nef7.A3G does not activate PAK-2 and that the failure of these two Nef derivatives to activate PAK-2 is not due to changes in the level of endogenous PAK-2 expression by these Nefs or to changes in the binding affinity of these Nefs to PAK-2.
FIGURE 4.
Effects of Nef, Nef7, and Nef7.A3G on PAK-2 activation. 293T cells were transfected with HIV.env–.nef– and WT Nef, Nef7, or Nef7.A3G expression plasmids with pcDNA3 included as a control. Cells were harvested for whole cell lysates 72 h after transfection for Western blot (WB) analysis against anti-HA and anti-PAK-2 (A), with β-actin used to ensure comparable loading of cell lysates. B, cell lysates were also subjected to immunoprecipitation with anti-HA antibodies followed by an in vitro kinase assay using [γ-32P]ATP to detect PAK-2 activation. C, cell lysates were first immunoprecipitated (IP) with anti-Nef antibody and Western-blotted with anti-PAK-2 antibody.
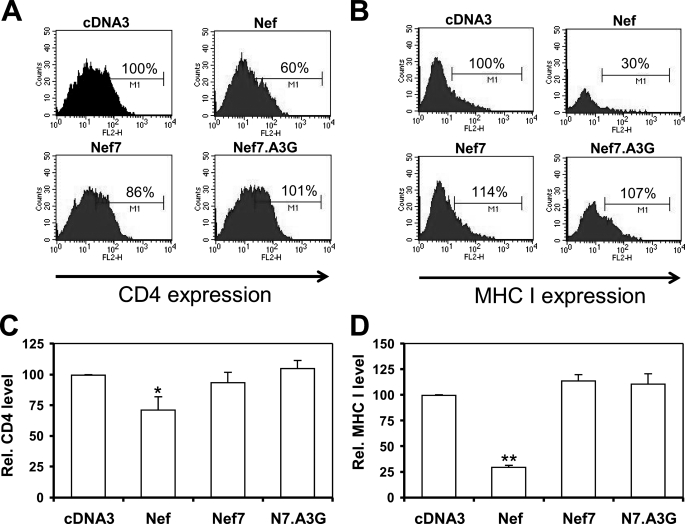
Down-regulation of CD4 and MHC I by Nef7.A3G—Another key function of Nef is the down-regulation of cell surface expression of various membrane proteins, the most well known being CD4 and MHC I (40). Nef7 is reported to be defective in both CD4 and MHC I down-regulation (31). We then attempted to confirm the diminished pathogenic phenotype of Nef7 and to determine whether this activity was present in the Nef7.A3G fusion protein by taking advantage of a system that was previously validated to study Nef-mediated MHC I down-regulation (41). We adapted this system to determine the relationship between cell surface expression of CD4 and MHC I and expression of Nef or its derivatives. We transfected HeLa cells with plasmids expressing CD4, GFP, and each of the Nefs using Lipofectamine. After 24 h we performed cell surface immunofluorescence staining using an anti-CD antibody and a phycoerythrin-conjugated secondary antibody followed by fluorescence-activated cell sorter for CD4+ cells among all GFP+ cells. We also performed similar experiments with MHC I (Fig. 5B). As expected, WT Nef down-regulated both CD4 and MHC I expression (Fig. 5, C and D). In contrast, Nef7 and Nef7.A3G showed no significant changes in the surface expression of these two receptors. These results show that both Nef7 and Nef7.A3G do not alter the cell surface expression of CD4 and MHC I.
FIGURE 5.
Effects of Nef, Nef7, and Nef7.A3G on cell surface expression of CD4 or MHC I. HeLa cells were transfected with CD4, GFP, and Nef, Nef7, or Nef7.A3G expression plasmids with pcDNA3 included as a control. At 24 h cells were harvested and stained with an anti-CD4 antibody followed by a phycoerythrin-conjugated secondary antibody. The cells were then gated for the GFP+ cells by fluorescence-activated cell sorter, and only the GFP+ cells were analyzed for cell surface CD4 expression (A). WB, Western blot. Similarly, HeLa cells were transfected with HLA-A2, GFP, and Nef, Nef7, or Nef7.A3G expression plasmids and analyzed for cell surface MHC I expression (B). Cell staining without anti-CD4 or anti-MHC I antibodies was included as the negative control to ensure that these secondary antibodies did not have nonspecific staining (data not shown). The relative CD4 and MHC I levels from three independent experiments were calculated with the CD4 or MHC I level in pcDNA3-transfected cells set to 100% (C and D).
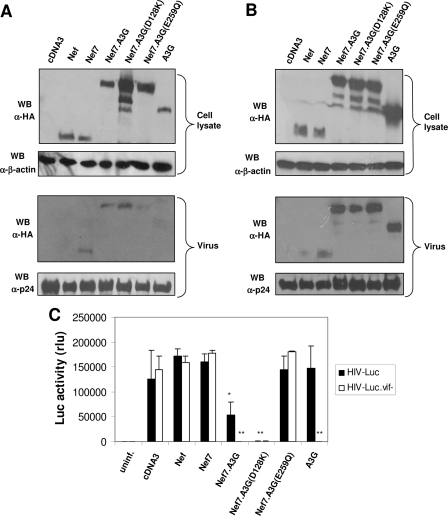
Impaired HIV-1 Infectivity by Nef7.A3G Virion Incorporation—Nef7.A3G was detected in HIV-1 virions in the context of HIV-1 Vif expression (Fig. 3). It did not activate PAK-2 (Fig. 4) or down-regulate CD4 and MHC I expression (Fig. 5). These findings led us to determine the effect of Nef7.A3G on the HIV-1 infectivity. We transfected 293T cells with HIV-Luc or HIVΔvif, VSV-G, and Nef or each of the Nef derivatives to produce VSV-G-pseudotyped HIV-Luc viruses carrying each of the Nefs. HIVΔvif has no functional env or vif gene, whereas HIV-Luc contains no functional HIV-1 env or nef gene and has the Luc reporter gene inserted at the 5′ end of the nonfunctional nef gene; it allows in trans complementation of other viral envelopes to determine the viral tropism and only single round infection for accurate determination of HIV-1 entry (35). VSV-G envelope was used to facilitate HIV-1 infection of cells that do not usually express HIV-1 receptor CD4 and chemokine receptors CXCR4 or CCR5. To ensure the specificity of Nef7.A3G, we also constructed and included two Nef7.A3G point mutants as controls, Nef7.A3G/D128K (D128K) and Nef7.A3G/E259Q (E259Q), in these experiments. Both point mutations were made in the A3G portion of the fusion protein. D128K mutation does not bind to HIV-1 Vif protein and renders the A3G protein Vif-resistant (6, 9), whereas E259Q mutation within the second deaminase domain leads to enzymatically inactive A3G (42). In addition, we also included A3G alone in the transfection, and we performed all infections with both HIV-Luc. We collected viruses from transfected cells, harvested these transfected cells for cell lysates, and performed Western blot analysis using an anti-HA antibody on the cell lysate and virus of both the HIV-Luc (Fig. 6A) and HIVΔvif (Fig. 6B) transfections. As expected, due to differential sensitivity to Vif-mediated degradation, A3G had the lowest level in the cells, whereas Nef7-A3G(D128K) had the highest level in the cells. The expression levels of both Nef7.A3G and Nef7.A3G(E259Q) were found to be between A3G and Nef7.A3G(D128K), suggesting that Nef fusion had made A3G less sensitive to Vif-mediated degradation. As a result, Nef7.A3G(D128K) was incorporated slightly more into viruses than Nef7, Nef7.A3G, or Nef7A3G(E259Q). Compared with the above findings, lack of Vif expression (and, therefore, Vif-mediated degradation) resulted in a similar level of detection of A3G and the three A3G fusion proteins in both the cells and the viruses. On the other hand, lack of Vif expression did not alter the fact that Nef7 was always detected in virions much more than its wild-type counterpart Nef. Taken together, these results indicate that Nef7-A3G is partially Vif-resistant but not to the same extent as Nef7-A3G(D128K), possibly due to structural constraints on the binding of Vif to the fusion protein. However, the extent to which the levels of Nef7-A3G and Nef7-A3G(D128K) differ between cells and viruses suggests that the slight resistance of Nef7-A3G to Vif is not solely responsible for its higher virion incorporation. Meanwhile, we used virus collected from the HIV-Luc transfections to infect fresh 293T cells. As expected, viruses carrying WT Nef and Nef7 and HIV-Luc viruses collected from A3G-transfected cells showed a level of infectivity similar to that of pcDNA3 control viruses, as measured by the Luc activity (Fig. 6C). Compared with viruses carrying WT Nef or Nef7, HIV-Luc viruses carrying Nef7.A3G showed a marked decrease in infectivity. Moreover, the infectivity of the viruses carrying the Vif-resistant D128K mutant was further decreased over that of its counterpart Nef7.A3G, whereas the viruses carrying the nonfunctional A3G mutant E259Q showed little inhibition. To ensure the functionality of all the A3G derivatives and the non-functionality of the E259Q mutant, we constructed HIV-Luc.vif–, which contains a stop codon at the beginning of the vif gene, rendering it nonfunctional, and included it as a control in the infectivity experiments. The HIV-Luc.vif– viruses containing Nef7.A3G, Nef7.A3G(D128K) and A3G showed complete inhibition of infection, similar to that observed in the HIV-Luc viruses containing the Vif-resistant Nef7.A3G(D128K) mutant. On the other hand, the inactive Nef7.A3G(E259Q) mutant had little effect on infectivity regardless of Vif expression. These results show that although Nef7.A3G virion incorporation does not restrict HIV-1 infection to the same extent as A3G does in the absence of Vif, as demonstrated by the Nef7.A3G(D128K) mutation, it does significantly impair HIV-1 infectivity, even in the presence of Vif.
FIGURE 6.
Effects of virion incorporation of Nef and its derivatives on the infectivity of VSG-pseudotyped HIV-Luc reporter viruses. 293T cells were transfected with HIV-Luc, HIV-Luc.vif–, or HIVΔvif, VSV-G, and WT Nef, Nef7, Nef7.A3G, Nef7.A3G(D128K), Nef7.A3G(E259Q), or A3G with pcDNA3 included as a control. At 72 h progeny viruses were collected and quantitated for RT activity. Meanwhile, HIV-Luc (A) or HIVΔvif (B) whole cell lysates and progeny viruses were analyzed by Western blot using an anti-HA antibody with a β-actin or p24 Western blot (WB) analysis to ensure comparable gel loading. HIV-Luc or HIV-Luc.vif– progeny viruses with an equal level of RT activity were then used to infect fresh 293T cells. Infected cells were harvested 48 h post-infection and analyzed for Luc activity (C). The data were mean ± S.E. of triplicate experiments. rlu, relative light units.
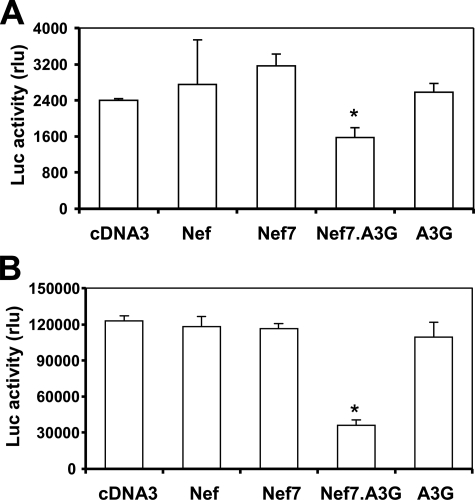
To further corroborate these findings, we prepared HXB2. env- and YU2.env-pseudotyped HIV-Luc viruses carrying WT Nef, Nef7, Nef7.A3G, or A3G, used HXB2.env pseudotyped viruses to infect U87.CD4.CXCR4 cells or YU2.env-pseudotyped viruses to infect U87.CD4.CCR5 cells, and determined the infectivity. Similar to the results obtained using VSV-G-pseudotyped HIV-Luc viruses, Nef7.A3G-containing viruses showed a significantly lower level of viral infection than all others (Fig. 7). Moreover, we also infected U87.CD4.CXCR4 and U87.CD4.CCR5 cells with VSV-G-pseudotyped viruses carrying the respective proteins and obtained similar results (data not shown).
FIGURE 7.
Effects of virion incorporation of Nef and its derivatives on the infectivity of HXB2. env- and YU2.env-pseudotyped HIV-Luc reporter viruses. 293T cells were transfected with HIV-Luc, HXB2.env, and WT Nef, Nef7, Nef7.A3G, or A3G expression plasmids with pcDNA3 included as a control. At 72 h the viruses were collected and quantitated for RT activity. Viruses with an equal level of RT activity were used to infect U87.CD4.CXCR4 cells, and the infected cells were harvested for the Luc activity assay 48 h post-infection (A). Similarly, various YU2.env-pseudotyped HIV-Luc viruses carrying Nef and each of its derivatives were prepared and used to infect U87.CD4.CCR5 cells (B). The data are the mean ± S.E. of triplicate experiments. rlu, relative light units.
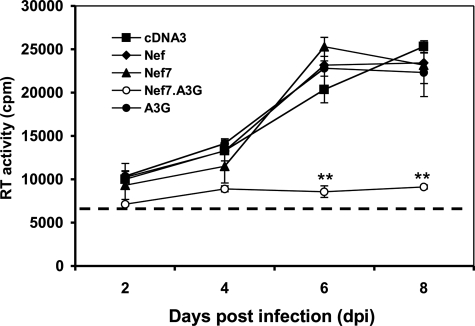
Block of HIV-1 Replication by Nef7.A3G-containing VLPs—CD4+ T lymphocytes are natural target cells for HIV-1 infection. Thus, we next determined the feasibility of using Nef7.A3G to target and inhibit HIV-1 replication in Jurkat cells, a CD4+ human T lymphocyte cell line commonly used for HIV-1 infection. To ensure efficient delivery of Nef7.A3G into HIV-1-infected cells, we took advantage of the VLPs-based inverse fusion strategy (43). We transfected 293T cells with an HIV-based packaging vector psPAX2 (36), Nef7.A3G, CD4, and CXCR4 and collected the cell culture supernatants as the VLPs. We also included WT Nef, Nef7, and A3G to produce control VLPs. CD4 and CXCR4 expression and presentation on the outer viral membrane allows recognition and binding to the gp120 that is expressed on the cell surface of HIV-1-infected cells, resulting in fusion of VLPs with these cells and delivery of virion components within VLPs into these cells (43). We infected Jurkat cells with replication-competent HIV-1 HXB2 viruses and exposed the infected cells to each of the VLPs. We then monitored HIV-1 infection and replication in these cells. Notably, treatment of HIV-infected Jurkat cells with VLPs that were derived from Nef7.A3G transfection gave rise to little viral replication (Fig. 8). In contrast, treatment of VLPs derived from cells expressing WT Nef, Nef7, and A3G all showed similar viral replication kinetics to that of VLPs from pcDNA3 control. Based on the number of cells that were inoculated with HXB2 virus (1 × 106) and the estimated initial infection efficiency (20–25%), the antiviral activity of the Nef7.A3G-containing VLP was in the range of 2–2.5 × 105-fold. These results suggest that Nef7.A3G-containing VLPs expressing CD4 and CXCR4 can successfully target HIV-infected cells and inhibit virus replication in HIV-1-infected Jurkat cells.
FIGURE 8.
Effects of Nef-, Nef7-, Nef7.A3G-, and A3G-containing VLPs on HIV-1 replication. 293T cells were transfected with psPAX2, CD4, CXCR4, and WT Nef, Nef7, Nef7.A3G, or A3G expression plasmids. At 72 h post-transfection culture supernatants containing VLPs were collected, cleared of cell debris, and quantitated for the RT activity. CD4+ Jurkat T lymphocytes were first infected with HIV-1 HXB2 strain. At 3 h the remaining viruses were removed from the cells by repeated washes with fresh medium and then treated with each of the VLPs. The cells were cultured for various lengths of time as indicated. At each time point, cell culture supernatant was collected and quantitated for RT activity. The RT activity of input VLPs in the culture medium was marked as a dotted line.
DISCUSSION
In this study we demonstrate that Nef7.A3G incorporated into HIV-1 virions (Fig. 3). We further show that the Nef7.A3G fusion protein fails to activate PAK2 (Fig. 4) or to down-regulate CD4 or MHC I (Fig. 5). Importantly, we show that the Nef7.A3G fusion protein retains the anti-HIV activity of A3G even in the context of HIV-1 Vif expression (Figs. 6 and 7). Furthermore, we show that Nef7.A3G-containing VLPs potently inhibit HIV-1 replication (Fig. 8). Nef7 has been previously shown to incorporate into viruses at an estimated 1100 molecules per virion, as compared with the estimated 10 molecules of WT Nef per virion, although this difference may not translate linearly to detection on Western blots, and the study did not determine fold incorporation by densitometry analysis of the Western blots (32). Using densitometry analysis, our results showed higher virus incorporation of Nef7 than WT Nef by about 9-fold. Our results also show that Nef7.A3G maintains the higher virion incorporation property of Nef7. A3G by itself was used as a control and, as expected, failed to encapsidate efficiently. A very recent study has shown that fusion of A3A, a Vif-resistant member of the APOBEC3 family that does not restrict HIV-1 infectivity, to the HIV-1 protein Vpr resulted in the alteration of its virion localization from the matrix to the viral core, which granted Vpr.A3A anti-HIV activity (44). A second very recent study showed that fusion of A3G to a virion-targeting peptide derived from Vpr (R88-A3G) resulted in restriction of HIV-1 infectivity (45). This fusion protein is susceptible to Vif-mediated degradation, and the higher virion incorporation is due to increased targeting to the virion by the remaining intracellular R88-A3G. Compared with Vpr, Nef7 is incorporated into virion at a higher level (32, 46). In addition, the higher level of virion incorporation of Nef7 as compared with WT Nef offers Nef7 an advantage over the native HIV-1 Nef protein for virion incorporation. In contrast, Vpr.A3A and R88-A3G are likely to be equally efficient as native HIV-1 Vpr protein in virion incorporation. This is very important when the anti-viral activity of these chimeras is evaluated in the context of HIV-1 infected cells. Furthermore, our results showed that Nef7.A3G had anti-HIV activity in both replication-defective single round HIV infection and the HIV-1 replication assay. However, whether Vpr-A3A would be an effective anti-HIV agent in the context of HIV-1 replication is not established.
Aside from its virion incorporation, a key property of Nef7 that allows it to be used as a carrier for therapeutic proteins is its relative non-toxicity. Although Nef is the most important pathogenic factor of HIV-1, Nef7 has been shown to be defective for a number of key Nef properties. One of the best known characteristics of Nef is its ability to activate the cellular kinase PAK-2, and disease progression in macaques has been associated with reversion of Nef mutants to a PAK-2-activating phenotype (47). Previous work has shown that Nef7 does not activate PAK-2 (31), and our data support that conclusion. In an in vitro kinase assay, both Nef7 and the Nef7.A3G fusion protein failed to activate PAK-2, confirming that Nef7 is less pathogenic than WT Nef and that fusion of A3G to Nef7 does not alter this phenotype. This loss of function does not appear to correlate with a loss of Nef7 binding to PAK-2, as PAK-2 is detected in immunoprecipitates of both Nef7 and Nef7.A3G. Likewise, the expression of Nef7 does not affect the intracellular levels of PAK-2. Taken together, these data suggest that the loss of PAK-2 activation in Nef7 is due to an inability of bound Nef7 to activate PAK-2. Our conclusion is in agreement with the study in which the Nef residues 85, 89, 187, 188, and 191 are found to be critical for Nef binding to PAK-2 (37), as none of these residues are affected in Nef7 as compared with WT Nef.
A second well known property of Nef, the down-regulation of surface molecules such as CD4 and MHC I, has also been shown to be defective in Nef7 (31). Our results show that in HeLa cells WT Nef down-regulates both CD4 and MHC I, with MHC I down-regulation being much more efficient. Both Nef7 and Nef7.A3G show no significant decrease in the surface expression of either CD4 or MHC I. Nef7 had previously been shown to down-regulate CD4 slightly (31) although much less efficiently than WT Nef. In the system that we used, Nef7 does show a slight, although statistically insignificant, decrease in CD4 expression. This difference is likely due to the different cell types used.
Because Nef7.A3G is incorporated into progeny viruses we expected that it would retain the A3G ability to restrict the infectivity of HIV-1, and this was indeed the case. In a single-cycle HIV infectivity assay Nef7.A3G-containing HIV virions showed a dramatic reduction in infectivity. This reduction was consistent across all cell types used and with both VSV-G pseudotyped viruses and viruses expressing X4- and R5-tropic HIV receptors, confirming that fusion to Nef7 restores the anti-HIV phenotype of A3G. To determine whether the Nef7.A3G fusion protein was Vif-resistant, we expressed Nef7.A3G, Nef7.A3G(D128K), and A3G in the presence or absence of Vif and analyzed both cellular expression levels and virion incorporation. Nef7.A3G(D128K) had the highest level of intracellular expression followed by Nef7.A3G and Nef7.A3G(E259Q). The A3G protein by itself had lower expression than any of the Nef7 fusion proteins, indicating that Nef7.A3G fusion proteins are partially Vif-resistant, possibly due to structural constraints on Vif binding. In the virus blots the difference between the Nef7.A3G and Nef7.A3G(D128K) is not as great as in the cell lysate blot. This suggests that the slight Vif resistance of Nef7.A3G is not solely responsible for its increased virion incorporation.
We theorized that specific targeting of Nef7.A3G fusion proteins to HIV-infected cells could inhibit the spread of HIV infection by allowing it to be incorporated into the progeny viruses produced from those cells and, thus, restrict the further spread of HIV from all targeted cells. To test this we created VLPs that were pseudotyped with CD4 and CXCR4, allowing their targeting to HIV-infected cells through an “inverse fusion” process where the CD4 and CXCR4 on the VLPs bind to the gp120 expressed on the surface of HIV-infected cells. We infected Jurkat T cells with HIV and then treated them with pseudotyped VLPs containing Nef7.A3G and monitored virus production over multiple rounds of infection. In this study we used a ratio of input virus to VLP of 1:10. This ratio is very important, as a higher ratio would result in a higher magnitude of antiviral effect. The results confirm that Nef7.A3G-containing VLPs inhibit the spread of HIV in infected Jurkat cells. Furthermore, the specific targeting of infected cells by inverse fusion should minimize bystander toxicity, increasing the usefulness of this strategy as a therapeutic platform to inactivate HIV-1 and suppress HIV-1 replication.
This strategy has many potential uses aside from the Nef7.A3G fusion protein. Because of its high virion incorporation, Nef7 fusion proteins can be used to target several components of the HIV virion, such as the tRNA primer for reverse transcription. Nef7 fusion proteins could also be used for novel experimental procedures in the laboratory. For example, a Nef7.GFP fusion protein could be used to track viral cores at early stages of infection, and a Nef7.luciferase fusion protein could be used as a sensitive assay for measuring virus production. Moreover, Nef fusion proteins can be adapted to deliver anti-HIV therapeutic proteins or components directly into HIV or HIV-infected cells.
Further characterization of the molecular mechanism responsible for Nef7's higher virion incorporation and the structure-function relationship of Nef7 shall surely help fine-tune this strategy to minimize any residual pathogenic activity of Nef7 and to maximize the benefits of this novel strategic anti-HIV platform for its potential clinical translation.
Acknowledgments
We thank Dr. W. C. Green for pcDNA3.1-APOBEC3G-HA, Dr. D. Trono for psPAX2, Dr. C. Touloukian for pHLA-A2, Dr. M. Emmerman for HIV.env–.nef–, Dr. J. Blum for anti-MHC I antibody, and Dr. D. Littman for U87.CD4.CXCR4 and U87.CD4.CCR5 cells.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS039804, R21DA022986, and R21DA025487 (to J. J. H.).
Footnotes
The abbreviations used are: HIV-1, human immunodeficiency virus type 1; PAK-2, p21-activated kinase-2; PBS, phosphate-buffered saline; MHC, major histocompatibility complex; WT, wild type; HA, hemagglutinin; RT, reverse transcription; GFP, green fluorescent protein; VLP, virus-like particles; Vif, virion infectivity protein; Luc, luciferase.
References
- 1.Simon, J. H., Gaddis, N. C., Fouchier, R. A., and Malim, M. H. (1998) Nat. Med. 4 1397–1400 [DOI] [PubMed] [Google Scholar]
- 2.Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002) Nature 418 646–650 [DOI] [PubMed] [Google Scholar]
- 3.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S., and Malim, M. H. (2003) Cell 113 803–809 [DOI] [PubMed] [Google Scholar]
- 4.Miyagi, E., Opi, S., Takeuchi, H., Khan, M., Goila-Gaur, R., Kao, S., and Strebel, K. (2007) J. Virol. 81 13346–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M., and Gabuzda, D. (2004) Genes Dev. 18 2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huthoff, H., and Malim, M. H. (2007) J. Virol. 81 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marin, M., Rose, K. M., Kozak, S. L., and Kabat, D. (2003) Nat. Med. 9 1398–1403 [DOI] [PubMed] [Google Scholar]
- 8.Sheehy, A. M., Gaddis, N. C., and Malim, M. H. (2003) Nat. Med. 9 1404–1407 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, L., Saadatmand, J., Li, X., Guo, F., Niu, M., Jiang, J., Kleiman, L., and Cen, S. (2008) Virology 370 113–121 [DOI] [PubMed] [Google Scholar]
- 10.Wolf, D., Giese, S. I., Witte, V., Krautkramer, E., Trapp, S., Sass, G., Haller, C., Blume, K., Fackler, O. T., and Baur, A. S. (2008) Virology 370 45–54 [DOI] [PubMed] [Google Scholar]
- 11.Giese, S. I., Woerz, I., Homann, S., Tibroni, N., Geyer, M., and Fackler, O. T. (2006) Virology 355 175–191 [DOI] [PubMed] [Google Scholar]
- 12.Cullen, B. R. (1998) Cell 93 685–692 [DOI] [PubMed] [Google Scholar]
- 13.Mariani, R., Kirchhoff, F., Greenough, T. C., Sullivan, J. L., Desrosiers, R. C., and Skowronski, J. (1996) J. Virol. 70 7752–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamieson, B. D., Aldrovandi, G. M., Planelles, V., Jowett, J. B., Gao, L., Bloch, L. M., Chen, I. S., and Zack, J. A. (1994) J. Virol. 68 3478–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kestler, H. W., III, Ringler, D. J., Mori, K., Panicali, D. L., Sehgal, P. K., Daniel, M. D., and Desrosiers, R. C. (1991) Cell 65 651–662 [DOI] [PubMed] [Google Scholar]
- 16.Crotti, A., Neri, F., Corti, D., Ghezzi, S., Heltai, S., Baur, A., Poli, G., Santagostino, E., and Vicenzi, E. (2006) J. Virol. 80 10663–10674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swigut, T., Shohdy, N., and Skowronski, J. (2001) EMBO J. 20 1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venzke, S., Michel, N., Allespach, I., Fackler, O. T., and Keppler, O. T. (2006) J. Virol. 80 11141–11152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua, J., and Cullen, B. R. (1997) J. Virol. 71 6742–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua, J., Blair, W., Truant, R., and Cullen, B. R. (1997) Virology 231 231–238 [DOI] [PubMed] [Google Scholar]
- 21.Williams, M., Roeth, J. F., Kasper, M. R., Filzen, T. M., and Collins, K. L. (2005) J. Virol. 79 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mangino, G., Percario, Z. A., Fiorucci, G., Vaccari, G., Manrique, S., Romeo, G., Federico, M., Geyer, M., and Affabris, E. (2007) J. Virol. 81 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenard, D., Yonemoto, W., de Noronha, C., Cavrois, M., Williams, S. A., and Greene, W. C. (2005) J. Immunol. 175 6050–6057 [DOI] [PubMed] [Google Scholar]
- 24.Wei, B. L., Arora, V. K., Raney, A., Kuo, L. S., Xiao, G. H., O'Neill, E., Testa, J. R., Foster, J. L., and Garcia, J. V. (2005) J. Virol. 79 14976–14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trible, R. P., Emert-Sedlak, L., and Smithgall, T. E. (2006) J. Biol. Chem. 281 27029–27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi, M., and Aiken, C. (2008) Virology 373 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Ronde, A., Klaver, B., Keulen, W., Smit, L., and Goudsmit, J. (1992) Virology 188 391–395 [DOI] [PubMed] [Google Scholar]
- 28.Welker, R., Harris, M., Cardel, B., and Krausslich, H. G. (1998) J. Virol. 72 8833–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chazal, N., Singer, G., Aiken, C., Hammarskjold, M. L., and Rekosh, D. (2001) J. Virol. 75 4014–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Aloja, P., Olivetta, E., Bona, R., Nappi, F., Pedacchia, D., Pugliese, K., Ferrari, G., Verani, P., and Federico, M. (1998) J. Virol. 72 4308–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Aloja, P., Santarcangelo, A. C., Arold, S., Baur, A., and Federico, M. (2001) J. Gen. Virol. 82 2735–2745 [DOI] [PubMed] [Google Scholar]
- 32.Peretti, S., Schiavoni, I., Pugliese, K., and Federico, M. (2005) Mol. Ther. 12 1185–1196 [DOI] [PubMed] [Google Scholar]
- 33.Peretti, S., Schiavoni, I., Pugliese, K., and Federico, M. (2006) Virology 345 115–126 [DOI] [PubMed] [Google Scholar]
- 34.Lewis, P., Hensel, M., and Emerman, M. (1992) EMBO J. 11 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He, J., Chen, Y., Farzan, M., Choe, H., Ohagen, A., Gartner, S., Busciglio, J., Yang, X., Hofmann, W., Newman, W., Mackay, C. R., Sodroski, J., and Gabuzda, D. (1997) Nature 385 645–649 [DOI] [PubMed] [Google Scholar]
- 36.Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D., and Naldini, L. (1998) J. Virol. 72 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agopian, K., Wei, B. L., Garcia, J. V., and Gabuzda, D. (2006) J. Virol. 80 3050–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He, J., and Landau, N. R. (1995) J. Virol. 69 4587–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawai, E. T., Khan, I. H., Montbriand, P. M., Peterlin, B. M., Cheng-Mayer, C., and Luciw, P. A. (1996) Curr. Biol. 6 1519–1527 [DOI] [PubMed] [Google Scholar]
- 40.Roeth, J. F., and Collins, K. L. (2006) Microbiol. Mol. Biol. Rev. 70 548–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubben, N. B., Sahlender, D. A., Motley, A. M., Lehner, P. J., Benaroch, P., and Robinson, M. S. (2007) Mol. Biol. Cell 18 3351–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schumacher, A. J., Hache, G., Macduff, D. A., Brown, W. L., and Harris, R. S. (2008) J. Virol. 82 2652–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endres, M. J., Jaffer, S., Haggarty, B., Turner, J. D., Doranz, B. J., O'Brien, P. J., Kolson, D. L., and Hoxie, J. A. (1997) Science 278 1462–1464 [DOI] [PubMed] [Google Scholar]
- 44.Aguiar, R. S., Lovsin, N., Tanuri, A., and Peterlin, B. M. (2008) J. Biol. Chem. 283 2518–2525 [DOI] [PubMed] [Google Scholar]
- 45.Ao, Z., Yu, Z., Wang, L., Zheng, Y., and Yao, X. (2008) PLoS ONE 3 e1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, S. P., Tungaturthi, P., Cartas, M., Tomkowicz, B., Rizvi, T. A., Khan, S. A., Kalyanaraman, V. S., and Srinivasan, A. (2001) Virology 283 78–83 [DOI] [PubMed] [Google Scholar]
- 47.Khan, I. H., Sawai, E. T., Antonio, E., Weber, C. J., Mandell, C. P., Montbriand, P., and Luciw, P. A. (1998) J. Virol. 72 5820–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]