Abstract
The low density lipoprotein receptor (LDLR) plays a key role in plasma cholesterol homeostasis by binding and internalizing lipoprotein ligands. Studies have revealed that one or more of the seven LDL type A repeats (LA1–LA7) in the receptor are responsible for apolipoprotein binding. In the present study, protein engineering was performed to swap or replace key LA repeats in a recombinant soluble LDLR (sLDLR). Although wild type sLDLR showed strong ligand binding activity, an sLDLR variant in which LA repeat 5 was replaced by a second copy of LA repeat 2 showed low binding activity. Likewise, a variant wherein LA repeats 2 and 5 were swapped displayed low binding activity. At the same time, substitution of LA repeat 2 with a second a copy of repeat 5 resulted in a receptor with ligand binding activity similar to wild type LDLR. When binding assays were conducted with human low density lipoprotein as ligand, LA repeat order was a less important determinant of binding activity. Taken together, the data indicate that the sequential order of LA repeats plays a key role in ligand binding properties of LDLR.
The low density lipoprotein receptor (LDLR)3 plays an important role in plasma cholesterol homeostasis (1). A fundamental function of LDLR is transport of cholesterol-rich lipoproteins into cells via receptor-mediated endocytosis (2). Human LDLR is 839 amino acids in length and is comprised of five distinct modules that arose from gene duplication. At the N terminus of LDLR, there exists a series of seven imperfect, disulfide bond-rich, LDL type A (LA) repeats, each ∼40 amino acids in length. Calcium binding induces LA repeats to fold into a ligand binding-competent conformation (3). Adjacent to the ligand binding module is a ∼400-residue module that bears homology to epidermal growth factor (EGF) precursor. This module consists of two disulfide bond-rich EGF-like repeats (A and B) and a YWTD β-propeller motif followed by a third EGF-like repeat C (4). The third module of LDLR is distinguished by an abundance of O-linked sugars, whereas the fourth module is comprised of a single membrane-spanning sequence. Finally, a short intracellular C-terminal cytoplasmic domain, required for receptor internalization, is present (5).
LDLR binds two apolipoprotein ligands, apolipoprotein (apo) E and apoB (6). Although these proteins do not share structural similarity, sequence elements rich in positively charged amino acid side chains are present in each that are required for binding. Deletion studies have demonstrated that specific LA repeats are required for apolipoprotein binding to LDLR (7, 8).
Recently, another LDLR ligand, termed proprotein convertase subtilisin-like kexin type 9 (PCSK9), has emerged (9): PCSK9 serves to regulate cholesterol homeostasis by modulating LDLR processing. Unlike lipoprotein ligands, PCSK9 binds EGF repeat A and, apparently, is not released from the receptor at endosomal pH.
LA1–LA7 are ∼40–50% identical in primary sequence. Each repeat contains a Ca2+ binding site and three disulfide bonds. The importance of these structural features for ligand binding is widely recognized. For example, it is known that LA5 is essential for optimal binding of apoB- and apoE-containing ligands (7, 8). On the other hand, deletion of LA2 had no effect on binding of apoE-containing lipoproteins. X-ray crystal structure information is available for isolated LA5 at pH 5.0 (10) as well as the entire ectodomain of LDLR (residues 1–699) at endosomal pH (11). Based on this structural information and complementary data on apolipoprotein ligands, it has been postulated that electrostatic interactions modulate LDLR conformation and ligand binding. Given this, it remains unclear whether the precise order of LA repeats within the ligand binding module may impact ligand binding.
In the present study, protein engineering of a soluble LDLR (sLDLR) was performed to swap or replace specific LA repeats within the ligand binding module of sLDLR. Ligand binding to wild type (WT) and engineered sLDLR was then determined. The results show that LA repeat 5 must not only be present, it must exist in the correct context with respect to other LA repeats within the ligand binding module.
EXPERIMENTAL PROCEDURES
Recombinant sLDLR and ApoE3-NT Isolation and Characterization—Recombinant Trp-null apoE-N-terminal (NT) domain was produced and isolated from Escherichia coli as described earlier (12). WT and variant sLDLR (N-terminal residues 1–699) were isolated from conditioned medium of stably transfected HEK 293 cells as described (13). Site-directed mutagenesis was performed using the QuikChange XL kit (Stratagene) according to the manufacturer's instructions. LA repeat swapping/replacement design and execution were conducted using an approach similar to that described earlier (14). All mutations were verified by dideoxy automated DNA sequencing. Variant sLDLRs generated were analyzed by SDS-PAGE under reducing and non-reducing conditions as a measure of native protein folding and disulfide bond formation (15). Human LDL was obtained from Intracel.
ApoE3-NT·Phospholipid Complexes—1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) was purchased from Avanti Polar Lipids. The phospholipid was dissolved in chloroform:methanol (3:1; v/v) and dried into a thin film in a glass tube. Following dispersion of the lipid in 50 mm sodium phosphate, pH 7.0, 150 mm NaCl, apoE3-NT was added. This mixture was bath-sonicated at 24 °C until clear (16).
LDLR Binding Assay—Four μg of sLDLR (in 50 mm Tris base or Tris succinate, 2 mm CaCl2, 100 mm NaCl; adjusted to specified pH values) was incubated with 1 μg of a Trp-null apoE3-NT previously labeled on Cys-112 with the fluorescent probe, N-(iodoacetyl)-N′-(5-sulfo-1-napthyl) ethylenediamine (AEDANS), and complexed with DMPC. Interaction between AEDANS-Trp-null apoE3-NT·DMPC and sLDLR was detected by fluorescence resonance energy transfer between excited Trp residues in sLDLR and the AEDANS moiety covalently attached to Trp-null apoE3-NT·DMPC (13, 17). Following incubation, samples were excited at 280 nm, and fluorescence emission intensity at 470 nm was determined (slit width 5.0 nm) on a PerkinElmer Life Sciences model LS 50B luminescence spectrometer.
Analytical Procedures—Protein concentrations were determined by the bicinchoninic acid assay using bovine serum albumin as standard. SDS-PAGE was performed on 4–20% acrylamide gradient slabs at 35 mA constant current under reducing or non-reducing conditions and stained with Coomassie Blue.
RESULTS
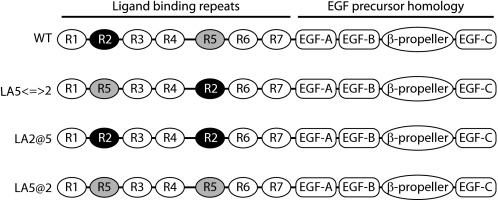
The hypothesis that LA repeat order is critical for ligand binding/release was tested. A known LDLR ligand, apoE3 NT·DMPC, was employed in fluorescence-based ligand binding assays. When AEDANS-labeled Trp-null apoE3 NT·DMPC binds the receptor, a stable fluorescence signal is established via intermolecular fluorescence resonance energy transfer from excited Trp residues in the receptor to the AEDANS moiety covalently bound to apoE. A corresponding decrease in AEDANS fluorescence intensity upon ligand dissociation can be monitored as a real-time measure of ligand release (13). This concept provides a general experimental design for investigating structural requirements for ligand binding and may be adapted to other LDLR family members. In the present study, three variants were generated for studies of the effect of LA repeat order on ligand binding. The overall design strategy was based on structural information gained from previous mutagenesis experiments that revealed LA5 is crucial for apoE-containing lipoprotein and LDL binding, whereas LA2 is less important (8). In the present approach, LA5 was replaced by a second LA2, creating an sLDLR that lacks LA5 while possessing two LA2 repeats (LA2@5 sLDLR). Second, LA5 and LA2 were swapped (LA5⇔2 sLDLR) and, third, LA2 was replaced by LA5, generating an sLDLR variant that contains two LA5 repeats but no LA2 repeat (LA5@2 sLDLR). The organization of LA repeat segments in the variant sLDLR generated are depicted schematically in Fig. 1.
FIGURE 1.
Schematic diagram of wild type and variant sLDLR ligand binding module organization.
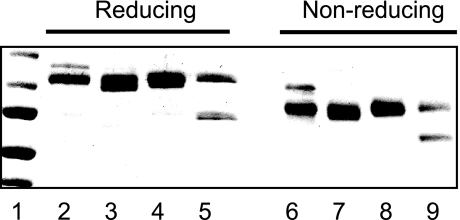
Prior to performing binding studies, isolated WT and variant sLDLRs were subjected to SDS-PAGE under reducing and non-reducing conditions (Fig. 2). It is known that, when correctly folded, LDLR displays a characteristic migration behavior in the presence/absence of β-mercaptoethanol (15). The pattern observed with WT sLDLR was consistent with previous results, manifesting a slower migration under reducing conditions. Each of the variant sLDLRs behaved in a similar manner, indicating that they are correctly folded in solution. One anomaly noted was the presence of two bands in the case of LA5@2 sLDLR. The upper band corresponds to the correct size for this protein, whereas the lower band is identified as an sLDLR on the basis of its detection by anti-LDLR (data not shown). Thus, this band likely corresponds to a partially cleaved byproduct of intact LA5@2 sLDLR or an unglycosylated immature form of the protein. Because the smaller band also migrates faster under non-reducing conditions when compared with reducing conditions (lanes 5 and 9), we conclude that the corresponding protein is folded correctly.
FIGURE 2.
Characterization of wild type and variant sLDLR. Isolated recombinant WT and variant sLDLRs were subjected to SDS-PAGE under reducing (lanes 2–5) and non-reducing (lanes 6–9) conditions. Lanes 2 and 6, WT sLDLR; lanes 3 and 7, LA2@5 sLDLR; lanes 4 and 8, LA5⇔2 sLDLR; and lanes 5 and 9, LA5@2 sLDLR. Samples were separated on a 4–20% acrylamide gradient slab gel electrophoresed at 35 mA constant current and stained with Coomassie Blue. The relative mobility of standard proteins is shown in lane 1 (from top to bottom: 150, 100, 75, 50, and 37 kDa, respectively).
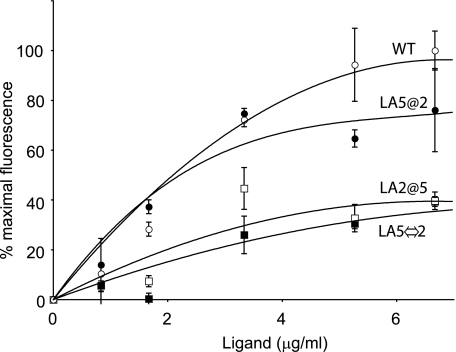
Effect of Ligand Concentration on Receptor Binding—A hallmark feature of ligand interactions with LDLR family members is saturability. When increasing amounts of AEDANS-Trp-null apoE3 NT·DMPC were incubated with a fixed amount of WT sLDLR, background-subtracted AEDANS fluorescence emission intensity increased in a ligand concentration-dependent manner and reached a plateau, after which no further fluorescence emission enhancement was observed (Fig. 3). Contrary to this, AEDANS-Trp-null apoE3 NT·DMPC binding to LA2@5 sLDLR was lower. These data support the concept that LA5 is critical for ligand binding. To assess whether the relative position of LA5 within the context of the ligand binding module is a factor, AEDANS-Trp-null apoE3 NT·DMPC binding to LA5⇔2 sLDLR was examined. In this case, low binding activity was observed. To distinguish whether ligand binding in sLDLR may be affected by a second LA5 in the module, LA5@2 sLDLR was studied. In this case, binding activity roughly equivalent to WT sLDLR was noted.
FIGURE 3.
Ligand concentration-dependent fluorescence emission intensity of wild type and variant sLDLR. Fourμg of a given sLDLR was incubated with increasing amounts of AEDANS-Trp-null apoE3-NT·DMPC. Open circles, WT sLDLR; filled circles, LA5@2 sLDLR; open squares, LA2@5 sLDLR; and filled squares, LA5⇔2 sLDLR. Fluorescence emission intensity values reported were obtained after subtraction of fluorescence emission intensity of ligand alone at each concentration.
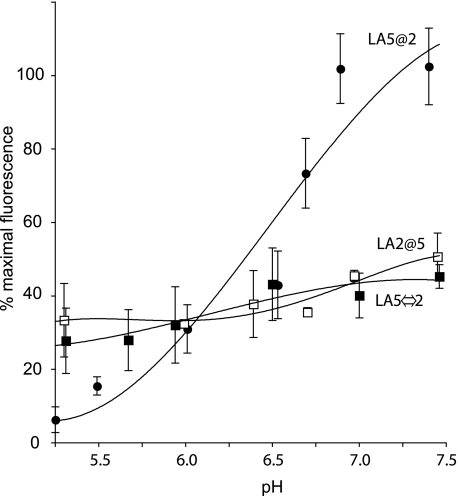
Effect of Solution pH on Ligand Binding—In previous studies with WT sLDLR, as the solution pH was decreased from 7 to 5, a corresponding decrease in AEDANS fluorescence intensity was detected, consistent with ligand release from the receptor (16). Control experiments showed that AEDANS fluorescence intensity via direct excitation was unaffected by pH over this range (data not shown). In the case of LA5@2 sLDLR, fluorescence changes observed upon lowering the solution pH were similar to those seen with WT sLDLR. On the other hand, pH-dependent effects on ligand binding of LA5⇔2 and LA2@5 sLDLR variants were attenuated (Fig. 4). The lesser decline in fluorescence observed as a function of decreasing pH for these variants may be related to the apparent lack of ligand saturation at neutral pH under these conditions.
FIGURE 4.
The effect of solution pH on ligand binding to WT and variant sLDLR. One μg of AEDANS-Trp-null apoE3-NT·DMPC and 4 μg of a given sLDLR were incubated in 50 mm Tris base or Tris succinate buffer, 2 mm CaCl2, 100 mm NaCl, adjusted to the indicated pH values. Samples (300 μl final volume) were excited at 280 nm, and fluorescence emission intensity at 470 nm was determined. Values reported are the mean ± S.D. (n = 3); filled circles, LA5@2 sLDLR; open squares, LA2@5 sLDLR; and filled squares, LA5⇔2 sLDLR.
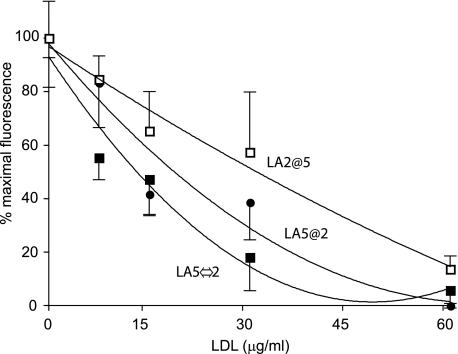
Effect of LA Repeat Order on LDL Binding—To further evaluate the ligand binding specificity of sLDLR variants, competition binding experiments were conducted using LDL isolated from human plasma as ligand. In previous study (13), we showed that LDL can compete with AEDANS-Trp-null apoE3 NT·DMPC for binding to WT sLDLR. In the present study, the ability of LDL to compete with AEDANS-Trp-null apoE3 NT·DMPC for binding to variant sLDLR was monitored by changes in fluorescence intensity at 470 nm (excitation 280 nm). In each case, LDL induced a concentration-dependent decrease in AEDANS fluorescence intensity emission, indicating that each of the three sLDLR variants studied recognize LDL as a ligand (Fig. 5). At the same time, it is difficult to compare sLDLR variants directly in terms of LDL ligand binding activity because the different variants bound different amounts of competitor apoE.
FIGURE 5.
Effect of human LDL on AEDANS-apoE NT binding to sLDLR. One μg of AEDANS-Trp-null apoE3-NT·DMPC and 4 μg of a given variant sLDLR were incubated in the presence of increasing concentrations of LDL. Samples (300 μl final volume) were excited at 280 nm, and fluorescence emission intensity at 470 nm was determined. Open squares, LA2@5 sLDLR; filled circles, LA5@2 sLDLR; and filled squares, LA5⇔2 sLDLR. Values reported are the average ± standard deviation (n = 3).
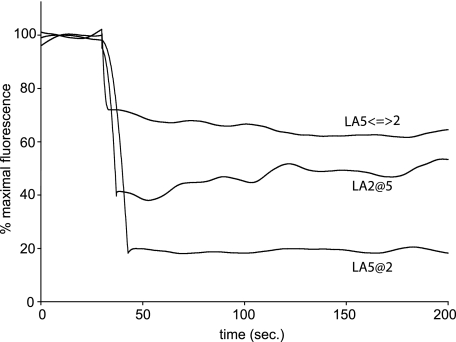
Evaluation of Ligand Release—Although the data presented above illustrate the effect of LA repeat order in sLDLR on ligand binding, we sought to evaluate the potential effects on ligand release. pH-dependent ligand release was monitored by fluorescence resonance energy transfer as a function of time following a pH transition from 7.0 to 6.0. In the case of the LA5@2 variant, a rapid decrease in AEDANS fluorescence intensity accompanied the shift in pH, similar to results reported earlier for WT sLDLR (13, 16). Within seconds of pH lowering, a greater than 80% decline in AEDANS fluorescence was noted (Fig. 6), consistent with discharge of bound ligand from the receptor. In the case of LA5⇔2 and LA2@5 sLDLR variants, pH-induced fluorescence intensity changes were less pronounced, and the end points reached were ∼32 and 46% of maximal AEDANS fluorescence intensity (i.e. pH 7), respectively.
FIGURE 6.
Effect of solution pH on ligand release from variant sLDLR. One μg of AEDANS-Trp-null apoE3-NT·DMPC and 4 μg of a given sLDLR were incubated in 10 mm Tris base, 2 mm CaCl2, 100 mm NaCl, pH 7.0. At time 30 s, the solution pH was changed by adding an aliquot (6 μl) of 1 m Tris succinate, pH = 6.0. Samples (300 μl final volume) were excited at 280 nm, and fluorescence emission intensity at 470 nm was monitored as a function of time.
DISCUSSION
Consistent with studies by Russell et al. (8), the present results reveal that LA5 plays a critical role in ligand binding and release because LA2@5 sLDLR, which lacks an LA5 repeat altogether, displays lower binding activity than WT sLDLR. The present studies also reveal that the relative position of LA5 within the ligand binding module is important because LA5⇔2 sLDLR, in which LA5 is located between LA1 and LA3 rather than between LA4 and LA6, is less active than WT sLDLR. These results suggest that LA position within the module influences ligand recognition/binding activity.
We previously demonstrated pH-dependent ligand binding activity of WT and various point mutant sLDLR (13, 16). In the current study, we investigated pH-dependent ligand binding with three LA repeat swapping/replacement variants. The LA5@2 sLDLR variant showed the same level of susceptibility as WT sLDLR to pH-dependent ligand release. On the other hand, both the LA5@2 sLDLR and the LA2@5 sLDLR were less sensitive to pH-dependent ligand release. These data may be explained by the x-ray crystal structure of sLDLR at pH 5.3 (11). In this study, it was shown that LA5 makes contact with the β-propeller motif. These interactions are thought to be important to maintain a “closed” receptor conformation that has been proposed to drive ligand release. In this manner, the β-propeller motif functions as an intramolecular alternative ligand (17). If the location of LA5 is disrupted, it may be postulated that the β-propeller domain may be unable to induce ligand release at endosomal pH. Indeed, in the present study, we observed that LA5⇔2 sLDLR and LA2@5 sLDLR are less sensitive to pH-dependent binding and release.
We previously demonstrated that three key histidine residues are critical for pH-dependent binding and release of apoE ligands from sLDLR (16). Although current results also support this hypothesis, Zhao and Michaely (18) recently proposed the new model for LDLR ligand release. In their model, at endosomal pH, the receptor adopts a closed conformation and EGF precursor homology repeats engage with LA repeats via the other side of the ligand binding region. This interaction is postulated to drive a conformational change in LA5 that disrupts the binding site for ligands, thereby facilitating release. Our current results cannot exclude this interesting hypothesis, and further studies will be required to establish the precise molecular basis for ligand release from LDLR.
Both WT sLDLR and LA5@2 sLDLR showed a nearly identical pattern in LDL competition binding assays. Although LA5⇔2 sLDLR displays lower affinity for apoE-containing ligands, the pattern observed with LDL was very similar to both WT sLDLR and LA5@2 sLDLR. On the other hand, LA2@5 sLDLR was less susceptible to competition by LDL. These results are consistent with findings that LDL binding is affected by LA deletion (7, 8). It is noteworthy that LDL contains a single apoB ligand such that its binding interaction is monovalent. By contrast, multiple apoEs exist on ligand complexes such that multivalent binding is possible.
In 2004, Fisher et al. (19) showed that the LA4-LA5 pair was sufficient to bind apoE-containing ligands. These authors concluded that LA5 alone is not sufficient to recognize apoE-containing ligands. In addition, LA3-LA4 and LA5-LA6, two repeat pairs, were unable to bind ligand with high affinity. When taken together with the results of the present study, it may be concluded that not only must a minimum LA repeat sequence be present, it must also exist in the correct sequence to maximize ligand recognition/binding. Thus, very different experimental approaches support the conclusion that LA5 is important, if not indispensable, for LDLR ligand binding. Furthermore, using different approaches, there is agreement that the context in which LA5 resides in is also critical, whereas LA5 in isolation is not sufficient for binding.
Acknowledgments
We thank Hyung Choi for skilled technical assistance and Dr. Vinita Gupta for assistance in generation of the sLDLR variants.
This work was supported, in whole or in part, by National Institutes of Health Grant HL 64159 (to R. O. R.).
Footnotes
The abbreviations used are: LDL, low density lipoprotein; LDLR, LDL receptor; sLDLR, soluble LDLR; LA, LDL type A repeat; AEDANS, N-(iodoacetyl)-N′-(5-sulfo-1-napthyl) ethylenediamine; apo, apolipoprotein; DMPC, dimyristoyl-phosphatidylcholine; NT, amino terminal; WT, wild type; EGF, epidermal growth factor.
References
- 1.Brown, M. S., and Goldstein, J. L. (1986) Science 232 34–47 [DOI] [PubMed] [Google Scholar]
- 2.Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W., and Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1 1–39 [DOI] [PubMed] [Google Scholar]
- 3.Daly, N. L., Scanlon, M. J., Djordjevic, J. T., Kroon, P. A., and Smith, R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6334–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, C. G., Goldstein, J. L., Sudhof, T. C., Anderson, R. G., Russell, D. W., and Brown, M. S. (1987) Nature 326 760–765 [DOI] [PubMed] [Google Scholar]
- 5.Lehrman, M. A., Goldstein, J. L., Brown, M. S., Russell, D. W., and Schneider, W. J. (1985) Cell 41 735–743 [DOI] [PubMed] [Google Scholar]
- 6.Kong, W. J., Liu, J., and Jiang, J. D. (2006) J. Mol. Med. 84 29–36 [DOI] [PubMed] [Google Scholar]
- 7.Esser, V., Limbird, L. E., Brown, M. S., Goldstein, J. L., and Russell, D. W. (1988) J. Biol. Chem. 263 13282–13290 [PubMed] [Google Scholar]
- 8.Russell, D. W., Brown, M. S., and Goldstein, J. L. (1989) J. Biol. Chem. 264 21682–21688 [PubMed] [Google Scholar]
- 9.Horton, J. D., Cohen, J. C., and Hobbs, H. H. (2009) J. Lipid Res. 50 S172-S177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fass, D., Blacklow, S., Kim, P. S., and Berger, J. M. (1997) Nature 388 691–693 [DOI] [PubMed] [Google Scholar]
- 11.Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., and Deisenhofer, J. (2002) Science 298 2353–2358 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto, T., and Ryan, R. O. (2006) Biochim. Biophys. Acta 1761 1100–1106 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto, T., Lamoureux, J., and Ryan, R. O. (2006) J. Lipid Res. 47 1091–1096 [DOI] [PubMed] [Google Scholar]
- 14.Kiss, R. S., Weers, P. M., Narayanaswami, V., Cohen, J., Kay, C. M., and Ryan, R. O. (2003) J. Biol. Chem. 278 21952–21959 [DOI] [PubMed] [Google Scholar]
- 15.Bajari, T. M., Strasser, V., Nimpf, J., and Schneider, W. J. (2005) Methods (Amst.) 36 109–116 [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto, T., Chen, H. C., Guigard, E., Kay, C. M., and Ryan, R. O. (2008) Biochemistry 47 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudenko, G., and Deisenhofer, J. (2003) Curr. Opin. Struct. Biol. 13 683–689 [DOI] [PubMed] [Google Scholar]
- 18.Zhao, Z., and Michaely, P. (2008) J. Biol. Chem. 283 26528–26537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher, C., Abdul-Aziz, D., and Blacklow, S. C. (2004) Biochemistry 43 1037–1044 [DOI] [PubMed] [Google Scholar]