Abstract
P2X4 and P2X7 are the predominant P2X receptor subtypes expressed in immune cells. Having previously shown a structural and functional interaction between the two recombinant receptors, our aims here were to identify the preferred assembly pathway of the endogenous receptors in macrophage-like cells and to investigate the trafficking of these receptors between the plasma membrane and intracellular sites. We exploited the difference in size between the two subunits, and we used a combination of cross-linkers and blue native-PAGE analysis to investigate the subunit composition of complexes present in primary cultures of rat microglia and macrophages from wild type and P2X7–/– mice. Our results indicate that the preferred assembly pathway for both receptors is the formation of homotrimers. Homotrimers of P2X7 were able to co-immunoprecipitate with P2X4, suggesting that an interaction occurs between rather than within receptor complexes. In both macrophages and microglia, P2X7 receptors were predominantly at the cell surface, whereas P2X4 receptors were predominantly intracellular. There were clear cell type-dependent differences in the extent to which P2X4 receptors trafficked to and from the surface; trafficking was much more dynamic in microglia than in the macrophages, and further activation of cultured microglia with relatively short (3-h) incubations with lipopolysaccharide caused an ∼4-fold increase in the fraction of receptors at the surface with only a 1.2-fold increase in total expression. The redistribution of intracellular receptors is thus an efficient means of enhancing the functional expression of P2X4 at the plasma membrane of microglia.
Acting via P2X receptors, ATP has several effects on immune cells, including triggering the release of pro-inflammatory cytokines, programmed cell death, and mycobacterial killing (1, 2). Until recently, attention has focused on the P2X7 receptor as the relevant sensor at the cell surface, and there is evidence for its involvement in neuropathic and inflammatory pain (3), arthritis (4), and the control of mycobacterial infection (5). P2X7 has a low affinity for ATP compared with the other predominant subtype expressed by immune cells, P2X4. The role of P2X4 receptors is less well understood, although in microglia they were shown to be up-regulated following peripheral nerve injury and to play an important role in the development of neuropathic pain (6). Regulation of the plasma membrane expression of these two receptors is not understood. Also, despite the considerable interest in both these receptor subtypes as potential targets in development of novel pain therapies, the subunit composition of the native receptors has not been determined.
P2X receptor subunits associate to form trimeric complexes around a central conduction pore (7, 8). With the exception of P2X6, they form functional homomeric receptors, and several subtypes also form functional heterotrimers (9). There is also evidence that P2X receptors form larger signaling complexes, interacting with other neighboring P2X receptors and also with ion channels that belong to different structural classes, including members of the Cys loop receptor family (10) and the gap junction family (11). Higher order structures, with molecular mass corresponding to hexamers and nonamers, have been identified for heterologously expressed P2X2 and P2X4 receptors, suggesting that two or three receptors can form a stable association (12). Consistent with this idea, P2X receptor channels within a patch of membrane have been shown to open in a non-independent manner and to display positive cooperativity (13, 14).
P2X4 and P2X7 are frequently co-expressed, not only in immune cells but also in epithelia and endothelia (15). The P2X7 subtype differs from other members of the family in that it has a very long cytoplasmic C-terminal tail, a low affinity toward ATP, is preferentially activated by BzATP, and couples to the opening of a pore permeable to large molecules up to 900 Da (2). P2X4 receptors have considerably higher affinity for ATP; they are up-regulated by the allosteric modulator ivermectin and are sensitive to the antagonist TNP-ATP3 at micromolar concentrations. It has been widely assumed that P2X7 does not form heteromeric assemblies with other members of the P2X family; however, recent evidence has indicated a structural and functional interaction between P2X4 and P2X7 receptors. First, P2X receptor currents recorded from airway-ciliated cells were reported to show a combination of P2X4-like and P2X7-like properties (16). Second, we showed that P2X4 and P2X7 could be co-immunoprecipitated both from mouse bone marrow-derived macrophages (BMDMs) and also when heterologously co-expressed (17). In addition, we used two nonfunctional point mutants of P2X4 to demonstrate a functional interaction between the two receptors; one mutant exerted a dominant negative effect on P2X7 receptor currents, whereas another conferred P2X4-like properties on the whole cell currents, namely sensitivity to ivermectin and TNP-ATP. Our conclusion was that P2X4 and P2X7 form a heteromeric association but that it remained to be established whether or not they assemble as heterotrimers around the same conduction pore.
In this study, we set out first to address the question of the preferred assembly pathway of native P2X4 and P2X7 receptors in immune cells, and second to compare how these receptors traffic within immune cells. We took advantage of the size difference between P2X4 and P2X7 subunits to analyze the composition of the predominant dimeric and trimeric forms. We found that in rodent macrophages and microglia, P2X4 and P2X7 receptors preferentially assemble as homotrimers. Comparison of the trafficking of these receptors indicated cell type-dependent differences in the extent to which the predominantly intracellular P2X4 receptor traffics to and from the surface.
EXPERIMENTAL PROCEDURES
Materials—P2X4 and P2X7 antibodies were purchased from Alomone (Israel). Horseradish peroxidase-conjugated anti-rabbit IgG whole antibody was from Health Care Limited (United Kingdom). Culture media were obtained from Invitrogen. Cross-linkers, disuccinimidyl suberate (DSS) and bis[sulfosuccinimidyl] suberate (BS3), were supplied by Pierce. SDS and native gradient gels were purchased from Invitrogen. All other reagents were from Sigma, unless otherwise stated below.
Cell Cultures—Primary cultures of microglial cells were prepared from cerebral cortices of newborn rats, as described previously (18). Glial cultures were maintained for 9–14 days in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 50 units/ml streptomycin and then shaken to dislodge microglia. Microglial cells were seeded in 6-well plates at a density of 9 × 105 cells/well and used for experiments 16–24 h after plating.
BMDMs were prepared from the femora of 5–6-week-old mice, as described previously (18). Cells were cultured for 7–10 days in medium containing RPMI 1640 supplemented with 20% fetal bovine serum, 100 units/ml penicillin, 50 units/ml streptomycin, and 30% L-conditioned medium. L-conditioned medium was collected from cultures of confluent monolayers of L-cells (L929) grown in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Cross-linking Receptors—To cross-link the total receptor population, cells were washed with ice-cold phosphate-buffered saline (PBS) and harvested in 1% Triton X-100 in PBS plus protease inhibitor mixture (Roche Applied Science). Samples were sonicated and incubated on ice for 30 min. Lysates were cleared by centrifugation at 14,000 rpm for 15 min at 4 °C. DSS was freshly prepared in DMSO, diluted to a final concentration of 4 mm, and incubated with the protein samples at room temperature for 30 min. The reaction was quenched by the addition of Tris-HCl, pH 7.5, to a final concentration of 50 mm, followed by incubation for 15 min at room temperature. To cross-link only surface receptors, cells were washed with ice-cold PBS and incubated with BS3 (1 mg/ml) in PBS for 1 h at 4 °C. 50 mm glycine was added to quench unreacted BS3. Cells were washed three times, lysed in Triple Extraction Buffer (TEB; 50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS (Bio-Rad), pH 8), sonicated, and cleared by centrifugation at 14,000 rpm for 15 min at 4 °C. All samples were diluted in Laemmli buffer and analyzed by 4–12% SDS-PAGE and immunoblotting.
Biotinylation of Surface Receptors—Confluent cells were rinsed five times with ice-cold DPBS (PBS, Invitrogen) (–CaCl2, –MgCl2) and incubated with freshly prepared Sulfo-NHS-SS-biotin (Pierce), diluted in DPBS (1 mg/ml), for 30 min at 12 °C. Nonreactive biotin was quenched with 50 mm glycine, followed by three washes with cold DPBS to ensure removal of unbound biotin. The cells were subsequently lysed in 150 μl of TEB and were sonicated to further disrupt and homogenize the cells. The resultant lysate was cleared by centrifugation at 13,000 rpm for 15 min at 4 °C. Of the recovered supernatant, 50 μl was diluted in Laemmli buffer and used to determine total protein levels. The biotinylated proteins in the remaining 100 μl were precipitated by incubation with 30 μl of streptavidin beads (Pierce) on a rotating wheel at 4 °C for 2 h. After three washes in TEB, proteins bound to streptavidin beads were eluted in sample buffer (20 μm dithiothreitol in Laemmli buffer) and subjected to SDS-PAGE and immunoblot analysis. Restriction of biotin labeling to surface receptors was confirmed by lack of reactivity to β-actin.
Membrane Protein Fraction Isolation and Co-immunoprecipitation—To obtain total membrane protein fractions for immunoprecipitation, BMDMs were washed three times with HEPES-buffered saline (HBS)/EDTA buffer (50 mm HEPES, pH 7.4, 100 mm NaCl, 2 mm EDTA), scraped off, and then collected by gentle centrifugation. The cell pellet was resuspended in ice-cold hypotonic buffer (10 mm Tris-HCl, pH 7.0, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture) and incubated for 20 min on ice. Cells were mechanically disrupted by passing the suspension through a needle, and the extract was centrifuged at 14,000 rpm for 15 min. The pellet containing the membrane-derived protein fraction was solubilized by incubation in 1% dodecyl maltoside in HBS buffer for 1 h on ice, and the solution was ultracentrifuged at 30,000 rpm for 1 h.
Before immunoprecipitation, total membrane protein extracts were subjected to DSS cross-linking for 30 min at room temperature, and the reaction was quenched by the addition of Tris-HCl, pH 7.5, to a final concentration of 50 mm. Cross-linked membrane protein extracts were pre-absorbed with anti-rabbit IgG beads (eBioscience) for 30 min. The pre-cleared membrane protein extracts were then incubated with 5 μg of anti-P2X4 antibody in HBS buffer containing 1% dodecyl maltoside for 3 h at 4 °C on a rotating wheel. Anti-rabbit IgG beads were subsequently added to the samples, and the mixture was further incubated for 1 h at 4 °C. The bead-bound protein complexes were washed four times with HBS buffer containing 1% dodecyl maltoside, and proteins were eluted by boiling for 5 min in 40 μl of Laemmli buffer. Samples were analyzed by 4–12% gradient SDS-PAGE and probed by immunoblot using the corresponding primary antibody and rabbit TrueBlot™-horseradish peroxidase-conjugated anti-rabbit IgG (eBioscience) as secondary antibody.
Immunoblotting—Immunoblotting was carried out essentially as described previously (18) except that protein samples were resolved on 4–12% gradient SDS-BisTris gels.
Blue Native (BN)-PAGE—Cells were washed three times with ice-cold PBS containing protease inhibitors, harvested, and lysed in hypotonic buffer (10 mm Tris-HCl, pH 8, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture (Roche Applied Science)). Cells were further disrupted by repeated passes through 23- and 27-gauge needles. The homogenate was centrifuged at 2000 rpm for 10 min at 4 °C to remove nuclei and then spun at 35,000 rpm for 35 min at 4 °C. The pellet was resuspended in BN sample buffer (Invitrogen) and solubilized in 1% digitonin for 1 h on ice. For partial denaturation, samples were subsequently incubated with 0.1% SDS at 37 °C for 1 h. Solubilized membrane fractions were supplemented with 5% G250 additive buffer (Invitrogen) and centrifuged at 13,000 rpm for 10 min at 4 °C. Samples were run on 4–16% native BisTris gels, transferred onto polyvinylidene difluoride membranes, and immunoblotted with appropriate antibodies.
Data Analysis—Densitometric analysis was carried out using GelEval 1.21b software. Statistical analyses were performed with Student's unpaired t test using GraphPad software.
RESULTS
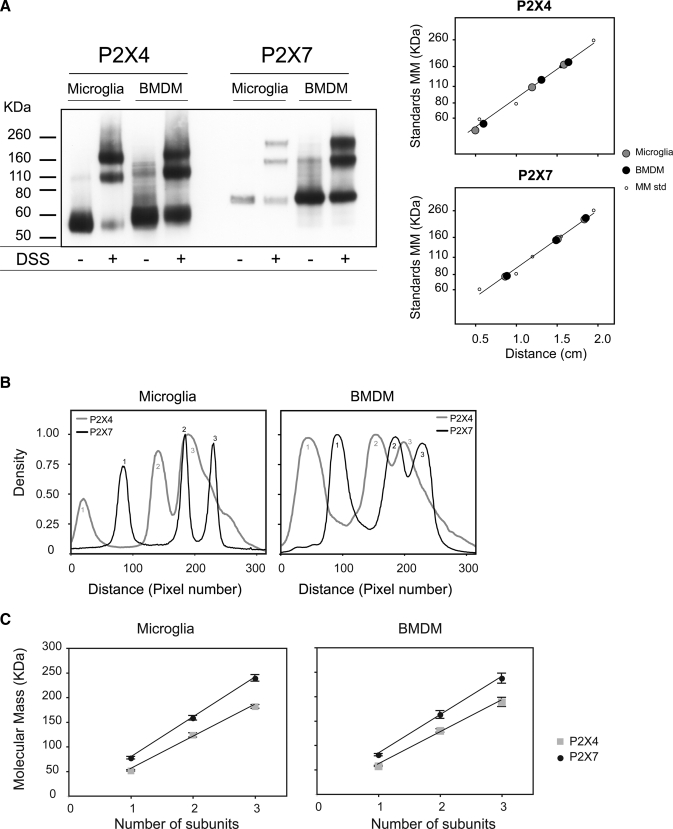
The molecular masses of P2X4 and P2X7 receptor complexes in primary cultures of rat microglia and mouse BMDMs were examined by cross-linking solubilized proteins with DSS and resolving by SDS-PAGE followed by immunoblotting. In the absence of cross-linker, there was a predominant band for P2X4 at 60 kDa and for P2X7 at 76 kDa, which are the expected molecular masses for the glycosylated forms of the monomers (Fig. 1A). In the presence of DSS, there were three prominent bands for P2X4 and P2X7 in both microglia and BMDMs. The relative positions of the bands are shown in the density versus distance plots (Fig. 1B); notably, the middle and upper bands for P2X4 were of different sizes from those for P2X7. The molecular masses were proportional to the predicted subunit number, consistent with the presence of monomers, homodimers, and homotrimers (Fig. 1C).
FIGURE 1.
Cross-linking analysis of P2X4 and P2X7 receptors in microglia and BMDMs. A, Triton X-100-solubilized cell extracts were chemically cross-linked with 4 mm DSS. Proteins were resolved by SDS-PAGE on a 4–12% gradient gel and immunoblotted with anti-P2X4 or anti-P2X7 antibodies. Calibration curves generated using molecular mass (MM) markers are shown at left. B, densitometric analysis of the blot measured from the bottom of the gel showing the relative positions of the three peaks for P2X4 and P2X7 complexes. C, for both receptors, the molecular masses of the three major bands plotted against subunit number lie on a straight line, consistent with the presence of monomers, homodimers, and homotrimers. Each point represents the mean ± S.E. from at least three different experiments.
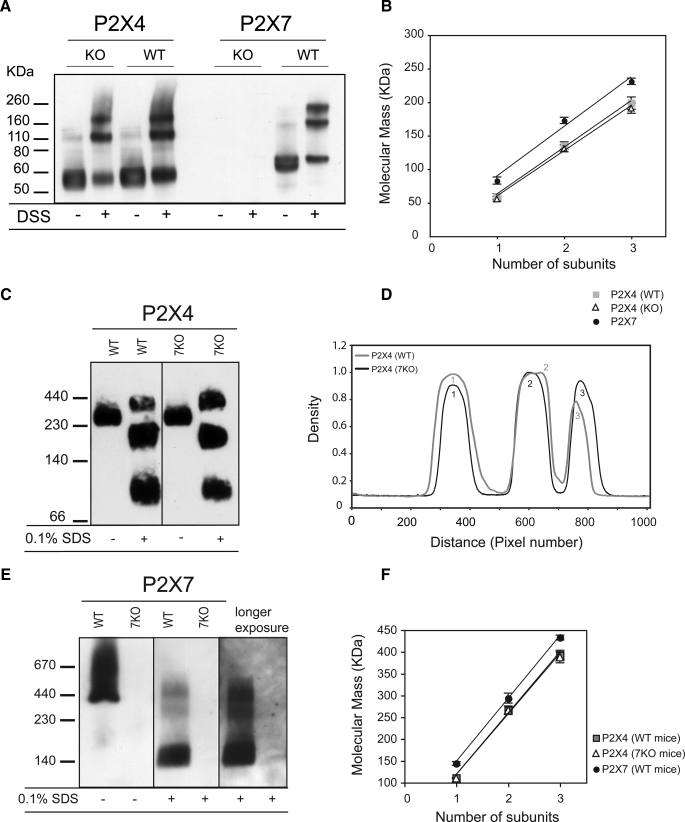
Although these results suggest that the preferred assembly pathway is formation of homotrimers, it is possible that a minor population of heterotrimers, co-existing with the homotrimeric form, would not be resolved as a distinct band. For example, in the P2X4 immunoblots, complexes with three P2X4 subunits would be difficult to resolve from those containing two P2X4 subunits and one P2X7 subunit. To circumvent this problem, we compared lysates from BMDMs obtained from wild type mice with those obtained from P2X7–/– mice. We hypothesized that if there was a significant population of P2X4/7 heterotrimers in wild type cells, we would see a shift in the molecular mass of the trimer detected by the anti-P2X4 antibody, as estimated from the peak of the density versus distance plot. Following incubation with DSS, however, the P2X4 bands appeared at the same positions for both wild type and P2X7–/– cells (Fig. 2, A and B). We also blotted for P2X7, expecting immunoreativity to be completely abolished in the P2X7–/– cells. There was, however, a very faint band running at the same molecular weight as P2X7 suggesting that there might be some residual expression of an alternatively spliced variant, although this was not apparent when we blotted for P2X7 following BN-PAGE (Fig. 2E). If there is some residual expression of a P2X7 variant in the knock-out mouse BMDMs, it is sufficiently low so as to not affect the interpretation of the results.
FIGURE 2.
Comparison of the size of P2X4 receptors in BMDMs from wild type and P2X7–/– mice. A, immunoblot analysis of DSS-treated cell extracts from WT and P2X7–/– (7KO) BMDMs. B, plot of molecular mass versus subunit number shows no difference between WT and P2X7–/– cells in the sizes of the three bands detected for P2X4. C–F, membrane preparations from WT and P2X7–/– cells were solubilized in 1% digitonin, incubated with or without 0.1% SDS, analyzed by BN-PAGE, and immunoblotted with either anti-P2X4 (C) or anti-P2X7 (E) antibodies. Densitometric analysis (D) confirms that the three bands detected with an anti-P2X4 antibody are the same sizes for wild type and P2X7–/– cells. Plot of the calculated molecular mass versus number of subunits (F) shows a linear correlation. Each point represents the mean ± S.E. from three different experiments.
Native P2X receptor complexes from wild type and P2X7–/– mice were solubilized in 1% digitonin and resolved by BN-PAGE. The complexes ran at a higher molecular mass following BN-PAGE than on SDS-PAGE, as has been reported previously (12) (Fig. 2, C and D). Consistent with the results obtained using cross-linkers, the sizes of native P2X4 complexes were the same in wild type and P2X7–/– cells; there was a dominant band running at ∼230 kDa, and after partial denaturation in 0.1% SDS, three bands were detected with sizes consistent with monomers, homodimers, and homotrimers. The bands detected with the anti-P2X7 antibody were not as clearly resolved (Fig. 2, E and F). In the absence of SDS there was a band at 440 kDa with a large smear above it. The smear probably represents mis-assembled complexes running as an aggregate; after treatment with 0.1% SDS, bands corresponding to P2X7 homotrimers, dimers, and monomers were apparent.
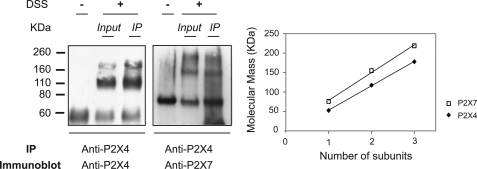
Having previously shown that P2X7 co-immunoprecipitates with P2X4 in mouse BMDMs and when heterologously co-expressed, we wished to determine the stoichiometry of the P2X7 receptor complexes that co-precipitate with P2X4. We therefore incubated with DSS prior to immunoprecipitating with the anti-P2X4 antibody. After blotting with anti-P2X7 antibody, bands in the precipitate corresponded in size to those in the total lysate, namely monomers, homodimers, and homotrimers (Fig. 3), thus indicating that an interaction occurs between P2X4 and P2X7 homotrimers.
FIGURE 3.
Co-immunoprecipitation of P2X7 homotrimers with P2X4. Membrane protein fractions from BMDMs were subjected to DSS cross-linking followed by immunoprecipitation (IP) using a polyclonal anti-P2X4 antibody. Isolated complexes were resolved on 4–12% SDS-PAGE and immunoblotted with anti-P2X4 (left) or anti-P2X7 (right) antibodies. Bands in the precipitate were of the same size as those in the total cell lysate; they correspond to monomers, homodimers, and homotrimers.
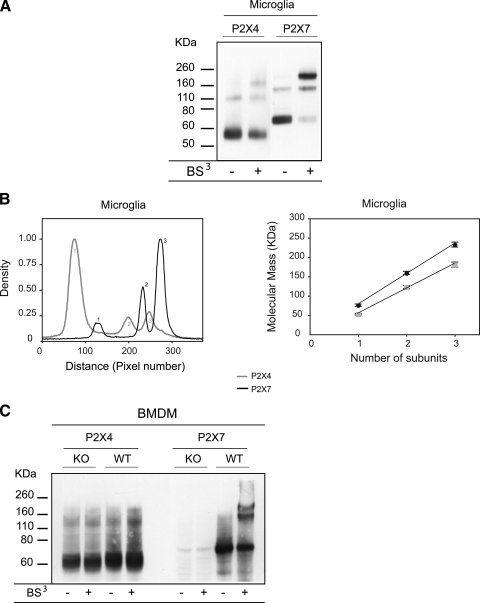
The membrane-impermeant reagent BS3 has the same arm length as DSS and can be used to selectively cross-link receptors at the plasma membrane. As well as enabling us to determine their size, it provides a measure of the relative proportions of receptors that are exposed at the surface versus located intracellularly. The blot and corresponding densitometry plot (Fig. 4, A and B) show that in microglia the targeting of P2X4 and P2X7 receptors is very different; P2X4 receptors were predominantly inaccessible to the cross-linking reagent and ran as monomers, whereas the majority of P2X7 receptors were cross-linked at the plasma membrane and ran as homotrimers. The small fraction of P2X4 that was accessible to BS3 was also in the form of homotrimers. In three experiments the percentage in the form of trimers was 20.1 ± 3.5% (n = 3) for P2X4 and 80.7 ± 3.9% (n = 3) for P2X7. The intracellular localization of P2X4 was consistent with our previous finding that these receptors co-localize with the marker for lysosomes, Lamp-1 (18). Accessibility of receptors to BS3 was tested in BMDMs from wild type and P2X7–/– mice (Fig. 4C), and the results were similar, although surface expression of P2X4 was lower than in microglia; it was below what we could reliably detect with this assay. For P2X7 the proportion cross-linked at the surface was similar to that in microglia (71.2 ± 1.9%, n = 3).
FIGURE 4.
Use of a surface cross-linking reagent (BS3) shows high levels of P2X7 at the surface of microglia and BMDMs but very low levels of P2X4. A, microglia were incubated with the membrane-impermeant cross-linker, BS3 (1 mg/ml), lysed. and solubilized. Proteins were subjected to SDS-PAGE and immunoblot analysis. B, densitometric analysis indicates that P2X4 receptors were mostly inaccessible to BS3 and ran as monomers, giving rise to a dominant first peak. In contrast, P2X7 receptors were mostly accessible to BS3 and stabilized in a trimeric form. The percentage of trimers was 20.1 ± 3.5% (n = 3) for P2X4 and 80.7 ± 3.9% (n = 3) for P2X7. Analysis of molecular mass indicates that trimers were of a size consistent with homomeric complexes. C, BMDMs from WT and P2X7–/– cells were subjected to cross-linking by BS3, followed by SDS-PAGE and immunoblotting. In WT cells the proportion of cross-linked P2X7 was 71.2 ± 1.9% (n = 3), whereas we were unable to reliably detect any cross-linked P2X4 receptors. KO, knock-out.
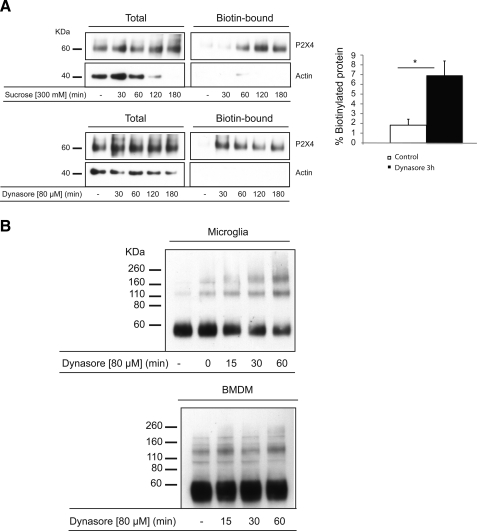
To determine whether or not intracellular P2X4 receptors are trafficking to and from the plasma membrane, we tested the effects of two inhibitors of endocytosis, hypertonic sucrose and Dynasore (19), on the proportion of P2X4 receptors biotinylated at the surface of microglia (Fig. 5A). Blotting for β-actin was carried out as a control to ensure that intracellular proteins were not also biotinylated. Both inhibitors caused an increase in surface biotinylation of P2X4 within 30–60 min. The increase with Dynasore was ∼4-fold, and incubations with Dynasore longer than 60 min did not appear to further increase surface P2X4 suggesting that the rapidly recycling intracellular pool was emptied by this time point. Similar experiments were carried out comparing the rate of accumulation of P2X4 at the surface of BMDMs and microglia in the presence of Dynasore but this time using the surface cross-linker BS3 (Fig. 5B). The accumulation of P2X4 at the surface of microglia was again evident within 30–60 min, whereas in BMDMs we were not able to detect surface P2X4 receptors, even after a 60-min incubation with Dynasore. We also tested the effects of Dynasore on the surface cross-linking of P2X7 but saw no effect (results not shown), suggesting that this receptor is stably expressed at the plasma membrane and not rapidly endocytosed. We conclude that in cultured microglia, P2X4 receptors are trafficking to and from the surface while being predominantly located intracellularly, whereas in cultured macrophages we have no evidence for the dynamic trafficking of this receptor.
FIGURE 5.
Inhibition of endocytosis rapidly enhances surface expression of P2X4 receptors in microglia but not BMDMs. A, microglia incubated with or without sucrose (300 mm) or Dynasore (80 μm) for various times were subjected to surface biotinylation; the biotinylated protein was captured using streptavidin beads, released under reducing conditions, and analyzed by SDS-PAGE and immunoblotting. Samples were loaded to obtain approximately the same levels of total P2X4 protein for the different treatments (prior to incubation with streptavidin beads) along with the corresponding fractions of streptavidin-purified (i.e. biotinylated) protein. The band intensities for the control and 3-h time points with Dynasore were measured using GelEval 1.21b, and the fraction of biotinylated protein was calculated. Dynasore (3 h) significantly increased the fraction of biotinylated protein (*, p < 0.01). Each time point represents the mean ± S.E. from seven independent experiments. B, comparison of the kinetics of Dynasore-evoked accumulation of P2X4 receptors at the cell surface in microglia and BMDMs. Cells were incubated with Dynasore (80 μm) for various times and then incubated with BS3. Dynasore caused a time-dependent increase in cross-linked P2X4 species in microglia as early as 15 min. In the absence of Dynasore, the proportion cross-linked was 24.4 ± 1.8% (n = 3), and after 60 min in Dynasore this was increased to 55.1 ± 9.4% (n = 3), p < 0.05. In BMDMs we could not reliably detect any cross-linked P2X4 receptors even after 60 min in Dynasore.
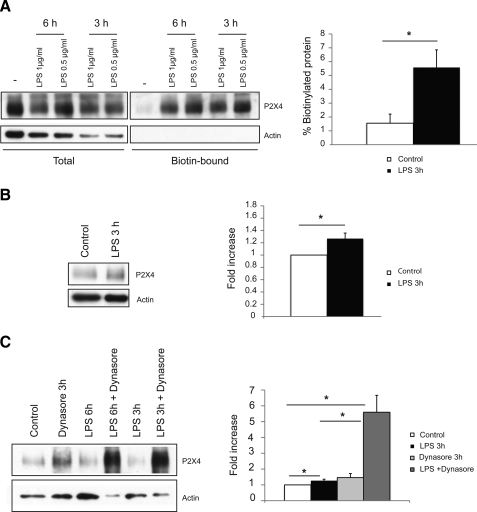
Microglia in culture are known to be in an activated state. They can, however, be further activated by incubation with bacterial lipopolysaccharide (LPS) (20). In a microglial cell line, a 48-h incubation with LPS (0.5 μg/ml) was shown to significantly increase the amplitude of P2X4 receptor currents (21). We tested the effects of much shorter incubations with LPS (3–6 h) on both the surface expression and total expression of P2X4 in our primary cultures of microglia. Within 3 h there was an ∼4-fold increase in the proportion of P2X4 receptors at the surface (Fig. 6A), whereas overall expression of the protein had increased by only 1.2-fold (Fig. 6B). Thus the redistribution of intracellular P2X4 receptors is a more rapid mechanism for up-regulating plasma membrane receptors than an increase in overall expression levels of this protein. When we combined LPS and Dynasore treatment, there was a very dramatic increase in the total expression of P2X4 (Fig. 6C), but we were unable to reliably estimate surface levels because cells became permeabilized as indicated by the biotinylation of β-actin (results not shown). It is unclear why the combination of LPS and Dynasore proved toxic to the cells.
FIGURE 6.
Short term treatment of microglia with LPS up-regulates P2X4 expression at the cell surface. A, surface biotinylation was carried out on microglia incubated with LPS (0.5 μg/ml) for different time periods. Samples were loaded to obtain approximately the same level of total P2X4 protein for the different treatments, along with the corresponding fractions of streptavidin-purified protein. After 3 h in LPS, there was a significant increase in the fraction of biotinylated protein (*, p < 0.05, n = 3). B, after 3 h in LPS (0.5 μg/ml) there was a small but significant increase in total P2X4 protein, normalized to actin (*, p < 0.05, n = 3). C, microglia were treated with a combination of LPS and Dynasore, and total protein samples were loaded to obtain approximately the same level of actin. Co-treatment with Dynasore and LPS resulted in a dramatic increase in total P2X4 expression (*, p < 0.05). Blots for biotinylated samples are not shown because cells became permeabilized following the combined Dynasore/LPS treatment.
DISCUSSION
Many P2X receptor subtypes are reported to form functional heteromeric assemblies, and yet only for two subunit combinations, P2X2/3 and P2X1/5, is there good evidence that heteromerization is the preferred assembly pathway under physiological conditions (22, 23). Having previously reported a structural and functional interaction between heterologously expressed P2X4 and P2X7 receptors and the co-immunoprecipitation of the native receptors from mouse BMDMs, we wished to establish whether or not P2X4 and P2X7 preferentially form homo- or heterotrimers when endogenously expressed. We conclude from this study that for P2X4 and P2X7 in immune cells, the preferred assembly pathway is the formation of homotrimers.
Our results are in agreement with a recent study by Nicke (24), who reported that the molecular masses of P2X4 and P2X7 receptor complexes when heterologously expressed in Xenopus oocytes and from different tissue extracts are consistent with homotrimeric complexes. We cannot, however, rule out the possibility that a small proportion of heterotrimers exist. Given the large number of P2X7 splice variants that have recently been identified, at least for the human isoform (25), it is possible that the P2X4-containing complexes contain a truncated P2X7 splice variant lacking the C-terminal epitope. It seems more likely, however, that structural and functional interaction between these two receptors involves the close association of neighboring P2X4 and P2X7 homotrimers, perhaps similar in nature to the interactions that underlie the clustering and positive functional cooperativity displayed by neighboring P2X2 receptors (13). Consistent with this idea, we found that homotrimers of P2X7 co-immunoprecipitate with P2X4.
Analysis of the plasma membrane expression of P2X4 and P2X7 indicates substantial differences in the targeting of these two receptors. In microglia, P2X7 was predominantly in the form of homotrimers at the cell surface, whereas P2X4 was intracellular. Inhibitors of endocytosis did, however, show that P2X4 traffics to and from the surface. In addition, treatment with LPS to further activate microglia caused significant stabilization of P2X4 at the surface within 3 h, while producing a much more modest increase in the overall expression of the receptor. Targeting of P2X4 to endolysosomes is therefore likely to be an important mechanism for regulating the plasma membrane expression of this high affinity receptor in microglia, and given the role it plays in the development of neuropathic pain, careful control of its exposure at the surface of microglia is crucially important. In BMDMs, the overall level of P2X4 expression is high, and yet plasma membrane expression was below the levels where we could reliably detect it with the methods used in this study, even when endocytosis was inhibited. We must therefore consider the possibility that the primary site of action of the P2X4 receptor is within intracellular compartments rather than at the cell surface. We previously showed targeting of P2X4 to lysosomes and to the phagosome in freshly isolated peritoneal macrophages (18). A recent study has shown ATP-dependent signaling within the lumen of phagosomes of macrophages, and has proposed that P2X7 receptors are activated within these compartments to regulate actin assembly around the phagosome (26). We propose a role for both P2X4 and P2X7 within the phagosome and suggest that it is here that an interaction between these two subtypes is important for ATP-dependent signaling.
Acknowledgments
We thank GlaxoSmithKline, Harlow, United Kingdom, for providing the P2X7 knock-out mice to D. C. Górecki. We also express our gratitude to Dr. A. Paramavisam for technical support and useful discussions.
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/F001320/1 (to R. M.-L.) and the Interreg IV (AdMiN) (to D. C. G.).
Footnotes
The abbreviations used are: TNP-ATP, 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate; LPS, lipopolysaccharide; DSS, disuccinimidyl suberate; BS3, bis[sulfosuccinimidyl] suberate; BMDM, bone marrow-derived macrophage; PBS, phosphate-buffered saline; BN, blue native; HBS HEPES-buffered saline; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; WT, wild type.
References
- 1.Khakh, B. S., and North, R. A. (2006) Nature 442 527–532 [DOI] [PubMed] [Google Scholar]
- 2.North, R. A. (2002) Physiol. Rev. 82 1013–1067 [DOI] [PubMed] [Google Scholar]
- 3.Chessell, I. P., Hatcher, J. P., Bountra, C., Michel, A. D., Hughes, J. P., Green, P., Egerton, J., Murfin, M., Richardson, J., Peck, W. L., Grahames, C. B., Casula, M. A., Yiangou, Y., Birch, R., Anand, P., and Buell, G. N. (2005) Pain 114 386–396 [DOI] [PubMed] [Google Scholar]
- 4.Labasi, J. M., Petrushova, N., Donovan, C., McCurdy, S., Lira, P., Payette, M. M., Brissette, W., Wicks, J. R., Audoly, L., and Gabel, C. A. (2002) J. Immunol. 168 6436–6445 [DOI] [PubMed] [Google Scholar]
- 5.Fairbairn, I. P., Stober, C. B., Kumararatne, D. S., and Lammas, D. A. (2001) J. Immunol. 167 3300–3307 [DOI] [PubMed] [Google Scholar]
- 6.Tsuda, M., Shigemoto-Mogami, Y., Koizumi, S., Mizokoshi, A., Kohsaka, S., Salter, M. W., and Inoue, K. (2003) Nature 424 778–783 [DOI] [PubMed] [Google Scholar]
- 7.Nicke, A., Baumert, H. G., Rettinger, J., Eichele, A., Lambrecht, G., Mutschler, E., and Schmalzing, G. (1998) EMBO J. 17 3016–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrera, N. P., Ormond, S. J., Henderson, R. M., Murrell-Lagnado, R. D., and Edwardson, J. M. (2005) J. Biol. Chem. 280 10759–10765 [DOI] [PubMed] [Google Scholar]
- 9.Murrell-Lagnado, R. D., and Qureshi, O. S. (2008) Mol. Membr. Biol. 25 321–331 [DOI] [PubMed] [Google Scholar]
- 10.Khakh, B. S., Zhou, X., Sydes, J., Galligan, J. J., and Lester, H. A. (2000) Nature 406 405–410 [DOI] [PubMed] [Google Scholar]
- 11.Pelegrin, P., and Surprenant, A. (2006) EMBO J. 25 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aschrafi, A., Sadtler, S., Niculescu, C., Rettinger, J., and Schmalzing, G. (2004) J. Mol. Biol. 342 333–343 [DOI] [PubMed] [Google Scholar]
- 13.Ding, S., and Sachs, F. (2002) BMC Neurosci. 3 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara, Y., and Kubo, Y. (2004) J. Physiol. (Lond.) 558 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surprenant, A., and North, R. A. (2009) Annu. Rev. Physiol. 71 333–359 [DOI] [PubMed] [Google Scholar]
- 16.Ma, W., Korngreen, A., Weil, S., Cohen, E. B., Priel, A., Kuzin, L., and Silberberg, S. D. (2006) J. Physiol. (Lond.) 571 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, C., Masin, M., Qureshi, O. S., and Murrell-Lagnado, R. D. (2007) Mol. Pharmacol. 72 1447–1456 [DOI] [PubMed] [Google Scholar]
- 18.Qureshi, O. S., Paramasivam, A., Yu, J. C., and Murrell-Lagnado, R. D. (2007) J. Cell Sci. 120 3838–3849 [DOI] [PubMed] [Google Scholar]
- 19.Macia, E., Ehrlich, M., Massol, R., Boucrot, E., Brunner, C., and Kirchhausen, T. (2006) Dev. Cell 10 839–850 [DOI] [PubMed] [Google Scholar]
- 20.Lehnardt, S., Massillon, L., Follett, P., Jensen, F. E., Ratan, R., Rosenberg, P. A., Volpe, J. J., and Vartanian, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8514–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raouf, R., Chabot-Dore, A. J., Ase, A. R., Blais, D., and Seguela, P. (2007) Neuropharmacology 53 496–504 [DOI] [PubMed] [Google Scholar]
- 22.Lewis, C., Neidhart, S., Holy, C., North, R. A., Buell, G., and Surprenant, A. (1995) Nature 377 432–435 [DOI] [PubMed] [Google Scholar]
- 23.Lalo, U., Pankratov, Y., Wichert, S. P., Rossner, M. J., North, R. A., Kirchhoff, F., and Verkhratsky, A. (2008) J. Neurosci. 28 5473–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicke, A. (2008) Biochem. Biophys. Res. Commun. 377 803–808 [DOI] [PubMed] [Google Scholar]
- 25.Cheewatrakoolpong, B., Gilchrest, H., Anthes, J. C., and Greenfeder, S. (2005) Biochem. Biophys. Res. Commun. 332 17–27 [DOI] [PubMed] [Google Scholar]
- 26.Kuehnel, M. P., Rybin, V., Anand, P. K., Anes, E., and Griffiths, G. (2009) J. Cell Sci. 122 499–504 [DOI] [PubMed] [Google Scholar]