Abstract
Peroxiredoxins (Prxs) are a group of peroxidases containing a cysteine thiol at their catalytic site. During peroxidase catalysis, the catalytic cysteine, referred to as the peroxidatic cysteine (CP), cycles between thiol (CP-SH) and disulfide (–S–S–) states via a sulfenic (CP-SOH) intermediate. Hyperoxidation of the CP thiol to its sulfinic (CP-SO2H) derivative has been shown to be reversible, but its sulfonic (CP-SO3H) derivative is irreversible. Our comparative study of hyperoxidation and regeneration of Prx I and Prx II in HeLa cells revealed that Prx II is more susceptible than Prx I to hyperoxidation and that the majority of the hyperoxidized Prx II formation is reversible. However, the hyperoxidized Prx I showed much less reversibility because of the formation of its irreversible sulfonic derivative, as verified with CP-SO3H-specific antiserum. In an attempt to identify the multiple hyperoxidized spots of the Prx I on two-dimensional PAGE analysis, an N-acetylated Prx I was identified as part of the total Prx I using anti-acetylated Lys antibody. Using peptidyl-Asp metalloendopeptidase (EC 3.4.24.33) peptide fingerprints, we found that Nα-terminal acetylation (Nα-Ac) occurred exclusively on Prx II after demethionylation. Nα-Ac of Prx II blocks Prx II from irreversible hyperoxidation without altering its affinity for hydrogen peroxide. A comparative study of non-Nα-acetylated and Nα-terminal acetylated Prx II revealed that Nα-Ac of Prx II induces a significant shift in the circular dichroism spectrum and elevation of Tm from 59.6 to 70.9 °C. These findings suggest that the structural maintenance of Prx II by Nα-Ac may be responsible for preventing its hyperoxidation to form CP-SO3H.
Peroxiredoxins (Prxs)4 are a family of peroxidases that possess a conserved cysteine residue at the catalytic site for the reduction of peroxide/peroxynitrite. Using thiol-based reducing equivalents, like thioredoxin, Prxs catalyze the reduction of hydrogen peroxide, alkylhydroperoxides, and peroxynitrite to water, corresponding alcohols, and nitrite, respectively (1–8). Based on the number and location of conserved cysteine residue(s) directly involved in peroxide reduction, the six isotypes of mammalian Prx can be grouped into three distinct subgroups as follows: 2-Cys Prx, atypical 2-Cys Prx, and 1-Cys Prx, (1–2, 5). Human Prx I (hPrx I) and Prx II (hPrx II) are members of the 2-Cys Prx subgroup and thus contain two conserved cysteine residues that are directly involved in peroxidase activity. Cys52 for hPrx I and Cys51 for hPrx II are designated the peroxidatic cysteines (CP). These residues attack the O–O bond of the peroxide (ROOH) substrate to form the product (ROH) and the sulfenic derivative CP-SOH. This sulfenic derivative then forms a disulfide bond with the other conserved cysteine residue, which is referred to as the resolving cysteine (CR; Cys173 in hPrx I and Cys172 in hPrx II). In the case of 2-Cys Prxs, the disulfide partners, CP and CR, reside within different subunits; therefore, the disulfide bond established between CP and CR (CP-S–S-CR) is intermolecular. The reduced thioredoxin molecule is responsible for reducing the CP-S–S-CR disulfide bond to generate sulfhydryls (1–3, 5, 9).
The CP of eukaryotic 2-Cys Prxs is vulnerable to hyperoxidation, which results in the loss of its peroxidase activity. This feature is referred to as the “floodgate” mechanism, by which Prxs function as a redox sensor for the regulation of cell signaling (10–11). Hyperoxidation of CP does not occur when the disulfide bond (CP-S–S-CR) is formed. However, the thiol (CP-SH) can be hyperoxidized via the sulfenic (CP-SOH) derivative intermediate in the absence of CP-S-S-CR formation during catalysis (12). Two different hyperoxidation products of CP, the reversible sulfinic (CP-SO2H) derivative and the irreversible sulfonic (CP-SO3H) derivative, have been identified. The irreversible CP-SO3H was reported in Tsa1p, a yeast 2-Cys Prx, based on in vivo and in vitro regeneration assay results, and a stronger reactivity to an anti-Tsa1p-SO3H antibody, which exhibits high specificity toward Tsa1p-CP-SO3H relative to Tsa1p-CP-SO2H (13). Both forms of hyperoxidized Prxs, CP-SO2H and CP-SO3H, are superimposed on the acidic migrated spot instead of the Prx-SH spot on a two-dimensional polyacrylamide gel because of the introduction of one negative charge by hyperoxidation (12–16). The protein sulfinic acid reductase, sulfiredoxin, is responsible for reversing 2-Cys Prx-SO2H to Prx-SH in the presence of ATP and thiol-reducing equivalents like thioredoxin or glutathione (17–24). Until now, an intracellular enzymatic regeneration system for Prx-SO3H has not been reported.
Because mammalian Prx I and Prx II have been studied independently in a number of different organisms and cultured cells, the comparative biochemical data supporting their distinctive functional identities is still very limited. Recombinant Prx I (rPrx I) showed a 2.6-fold higher specific activity as a peroxidase than the recombinant Prx II (rPrx II) without any obvious catalytic or mechanistic differences (25, 26). Recent competition kinetics studies of hPrx II revealed a rate constant of 1.3 × 107 m–1 s–1, which is fast enough to favor an intracellular hydrogen peroxide target even in competition with catalase or glutathione peroxidase (27, 28). The kinetic parameters of the competition assay for hPrx I are still not available. Mammalian Prx I interacts with and regulates a broad spectrum of proteins, such as the Src homology domain 3 of c-Abl (29), the Myc box II (MBII) domain of c-Myc (30), the macrophage migration inhibitory factor (MIF, 31), the androgen receptor (32), and the apoptosis signal-regulating kinase-1 (ASK-1) (33). The suggested roles of Prx I in interactions with these molecules are those of a tumor repressor, a survival enhancer, and a growth regulator. Although these suggested functions are controversial (34), all of them can be attributed to the peroxide-scavenging capacity of Prx I (at least in part), except for the enhancement of androgen receptor transactivation (32). Prx II interacts with platelet-derived growth factor receptor and functions as a negative regulator for platelet-derived growth factor signaling (35). Prx II also binds to phospholipase D1 (PLD1) and functions as a hydrogen peroxide-stimulated PLD1 signal terminator (36). Both of these suggested Prx II roles are attributable to the peroxidase activity of Prx II. The major phenotypes of Prx I knock-out mice involve the development of a variety of age-related cancers, hemolytic anemia (37), and dramatic shifts in subcellular reactive oxygen species localization (38). Prx II knock-out mice exhibit splenomegaly and a lack of tumor development in any cell type or tissue (39). Until now, the protein molecule that interacts with Prx I and Prx II has not been characterized, and there is no indication of a heterodimer between Prx I and Prx II. Despite their similar peroxide-scavenging capacities, it is reasonable to conclude that the Prx I and Prx II are unable to compensate for each other in terms of physiological roles. There are several examples of tissue- or cell type-specific expression patterns, such as exclusive Prx I expression in astrocytes and Leydig cells and Prx II expression in neurons and Sertoli cells (40, 41); however, Prx I and Prx II are coexpressed in the majority of mammalian cells and tissues, suggesting distinguished biochemical characteristics of their cellular regulatory mechanisms. Recently, the unique presence of Cys83 in hPrx I, which contributes to the stability of the dimer-dimer interface and suppresses local unfolding, has been claimed to be prone to overoxidation of Prx I (42). The contribution of the dimer-decamer interconversion to the regulation of Prx I activity has also been reported (43).
In this study, we confirmed that hPrx II was more susceptible to hyperoxidation as well as more prone to regeneration than hPrx I in HeLa cells. We also found that the difficulty in regenerating hPrx I was caused by irreversible sulfonic (CP-SO3H) hyperoxidation. Using AspN (EC 3.4.24.33) peptide fingerprints, we identified the Nα-terminal acetylation exclusively on hPrx II. In addition, we provide evidence for the structural maintenance offered by Nα-terminal acetylation of hPrx II, which possibly contributes to preventing irreversible overoxidation of CP-SO3H.
EXPERIMENTAL PROCEDURES
Preparation of Proteins—hPrx II was purified from human red blood cells (Red Cross Blood Center, Chonnam National University, Gwangju, Korea) via combined HPLC column chromatography methods as described previously (44). rPrx I and rPrx II were purified from Escherichia coli cells overexpressing each protein by previously described methods (25). Thioredoxin reductase was purified from rat liver as described previously (45) or purchased from Sigma. Recombinant rat thioredoxin (rTrx) was purified from the E. coli overexpression system by a previously described method (44).
Cell Culture and Regeneration Assay—HeLa cells were obtained from the American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's media (WelGENE, Daegu, Korea) supplemented with 10% fetal bovine serum (FBS, WelGENE, Daegu, Korea). For the hyperoxidation of Prxs, designated concentrations of hydrogen peroxide in phosphate-buffered saline (PBS) were applied to the cells for 10 min after removal of the serum-containing media and two washes with PBS. For the regeneration of Prxs, after removal of hydrogen peroxide with two additional PBS washes, cells were incubated in fresh 10% FBS-containing media in the presence of 10 μg/ml cycloheximide (CHX) for the designated time periods. Cells were harvested via the addition of 10% trichloroacetic acid immediately after two washings with PBS. Harvested cells were then ultrasonicated for 3 min, and the protein precipitates were washed with 100% acetone, followed by air drying. Dried cell lysates were then solubilized in lysis buffer (40 mm Tris, 9 m urea, and 4% CHAPS).
Two-dimensional SDS-PAGE—Cell lysates or purified proteins were mixed with rehydration buffer (8 m urea, 2% CHAPS, 0.5% immobilized pH gradient buffer, 20 mm DTT, and 0.005% bromphenol blue) and loaded onto immobilized pH gradient strips (pH 3–10 or pH 4–7, 7 cm; Amersham Biosciences). Isoelectric focusing was carried out in four steps as follows: 50 V, 12 h; 500 V, 0.5 h; 1000 V, 0.5 h; and 8000 V for a total of 20,000 V-h. After reduction and alkylation, second dimensional electrophoresis was conducted on a 12% SDS-polyacrylamide gel using an SE-260 vertical unit (Amersham Biosciences).
In-gel Digestion of Protein Spots on Two-dimensional Polyacrylamide Gel with AspN Endopeptidase—Excised spots from a two-dimensional polyacrylamide gel were washed with distilled water, 50% acetonitrile (ACN), and 100 mm ammonium bicarbonate (ABC, pH 8.5) in microcentrifuge tubes. After 45 min of reduction with a reducing agent (10 mm DTT in a 100 mm ABC, pH 8.5), each piece of the gel was alkylated with 55 mm iodoacetamide in a 100 mm ABC (pH 8.5) for 30 min and then fully dried in a SpeedVac. In-gel digestion of the protein was performed using 12.5 μl of AspN endoproteinase (10 ng/μl, Sigma) in a 100 mm ABC (pH 8.5) for 12 h at 37 °C.
MALDI-TOF-MS Analysis—Mass analysis of AspN digests was performed on a Voyager-DE STR MALDI-TOF mass spectrometer (Perceptive Biosystems). In-gel digested samples were eluted from gels by 50% ACN and 5% formic acid. Each sample was then dissolved with 50% ACN and 0.1% trifluoroacetic acid and mixed with 10 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma), dissolved in 0.1% trifluoroacetic acid and 50% ACN. Samples were spotted on a sample plate, and mass spectra were obtained in the positive-ion reflector mode with delayed extraction. Calibration was performed using angiotensin I (1296.79 Da), adrenocorticotropic hormone (ACTH clip 1–17, 2093.01 Da), and ACTH clip 18–39 (2465.20 Da) as external standards.
Nanoflow Reversed Phase Liquid Chromatography-Tandem Mass Spectrometry—The samples were prepared as described for MALDI-TOF-MS analysis. The extracted peptides were separated and analyzed via nanoflow reverse phase liquid chromatography/electrospray ionization tandem mass spectrometry (RPLC/ESI-MS/MS) to determine their amino acid sequences. The Ultimate nano-RPLC system was used in combination with an autosampler (Famos) and a precolumn switching device (Switchos). The samples were loaded onto a Zorbax 300SB-C18 trap column (5 μm, 10 × 0.2 mm; Agilent) and washed with the loading solvent (0.1% formic acid, flow rate, 4 μl/min) for 10 min to remove salts. Subsequently, flow paths were transferred to the analytical Zorbax 300SB-C18 column (5 μm, 150 × 0.075 mm; Agilent) using a Switchos II column switching device. The nano-flow eluted at a flow rate of 200 nl/min using a 90-min gradient elution from 0% solvent A to 32% solvent B (solvent A, 0.1% formic acid with 5% ACN; solvent B, 0.1% formic acid with 90% ACN). The column outlet was coupled directly to the nano-ESI source, which was interfaced with the QSTAR Mass spectrometer (Applied Biosystems). The nano RPLC/ESI-MS/MS, running on the QSTAR instrument, was acquired in “Information-dependent Acquisition” mode. The data acquisition time was set to 3 s per spectrum over an m/z range of 400–1500 Da for nano-LC-MS/MS analyses. Data base searches were performed on the National Center for Biotechnology Information (NCBI) nonredundant data base using the MASCOT software package (Matrix Sciences) and were confirmed manually.
Peroxidase Activity Assay—The peroxidase activity of Prxs was measured by monitoring the rate of NADPH oxidation at 340 nm in the presence of the thioredoxin system. Reaction mixtures of 150 μl (final volume) contained 50 mm Hepes-NaOH, pH 7.0, 0.2 mm NADPH, 1.1 μm rTrx, 0.12 μm of thioredoxin reductase, and 0.15 μm of each Prx. After adjusting the reaction mixture temperature to 30 °C, reactions were initiated immediately via the addition of hydrogen peroxide. The rate of NADPH oxidation was monitored via the loss of absorbance at 340 nm using a Jasco V-530 UV-visible spectrophotometer equipped with a thermostatic cell holder. An initial linear portion of the absorbance change (10 s) was used for the calculation of peroxidase activity.
Preparation of hPrx II-SO2H and hPrx II-SO3H in Vitro—Reaction mixtures of 500 μl (final volume) contained 50 mm Hepes-NaOH, pH 7.0, 2 mm DTT, 1 mm EDTA, 0.4 μm rTrx, and 2 μm hPrx II. After preincubation at 37 °C, reactions were initiated via the addition of hydrogen peroxide. At 60 s after the start of each reaction, 0.1% trifluoroacetic acid (final concentration) was added to stop the reaction. For the preparation of hPrx II-SO2H or hPrx II-SO3H, 100 μm or 3 mm of hydrogen peroxide concentration was used, respectively. The hyperoxidation status was validated by mass spectrometric analysis. The reaction mixtures were separated by reverse phase HPLC with both spectrophotometric and mass spectrometric detection (Agilent Technology, Series 1100 MSD) using a Vydac narrow bore C18 column (Vydac 218TP5205) as described previously (46). Reaction mixtures of 20 μl were routinely analyzed, and the data were collected in a mass unit range of 21,700–22,500 Da.
CD Analysis—hPrx II and rPrx II proteins were dialyzed against 20 mm sodium phosphate (pH 7.0) containing 100 mm NaCl, with the final protein concentration adjusted to 0.15 mg/ml. CD measurements were performed using a J-715 spectropolarimeter (Jasco) equipped with a PTC-348WI Peltier thermostat (Jasco). For the standard scan, the far-UV was scanned from 260 to 190 nm at 25 °C in a cell with a path length of 0.1 cm. To investigate the stability of the protein, thermal denaturation was monitored from 25 to 90 °C at 222 nm. The constant heating rate was 1 °C/min and the CD signal was recorded at 0.5 °C increments. The denatured fraction (FDX) at each temperature was defined as Fd = (Yn – Y)/(Yn – Yd), where Yn and Yd are the base lines of native protein and denatured proteins, respectively. These values were obtained from the extrapolation of the linear-regression of data points from 25 to 45 °C and from 75 to 90 °C, respectively. Y is the experimental CD signal at each temperature, and the apparent melting temperature (Tm) values were determined as the temperature at Fd = 0.5.
RESULTS
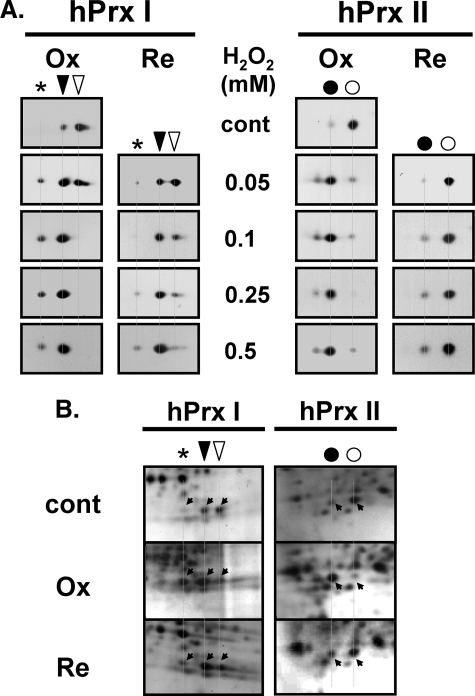
hPrx II Is More Sensitive than hPrx I to Hyperoxidation and Regeneration in HeLa Cells—The sensitivities to hyperoxidation and regeneration of hPrx I and hPrx II were compared in oxidatively stressed HeLa cells based on their migration shifts on two-dimensional polyacrylamide gels. hPrx II hyperoxidized more easily than hPrx I in hydrogen peroxide-treated HeLa cells. More than 80% of hPrx II was hyperoxidized by 10 min of exposure to 50 μm hydrogen peroxide in HeLa cells, whereas less than 40% of hPrx I was hyperoxidized under the same conditions (Fig. 1A). Most of the hPrx I that appeared at the acidic migrated location under normal culture conditions (Fig. 1A) was supposed to be modified; however, we found that it also contained a reduced form (CP-SH) as well as a covalently modified hPrx I. The hyperoxidized form of this modified hPrx I migrated at a position indicated by an asterisk (see Fig. 1A). This observation is in agreement with the previous report that some reduced hPrx I from HeLa cells co-migrates with hyperoxidized hPrx I (47). Therefore, hyperoxidized hPrx I was calculated as the decrease in intensity of the reduced spot relative to control, plus the relative increase in the spot as designated by an asterisk (see Fig. 1A). To hyperoxidize greater than 80% of hPrx I in HeLa cells, treatment with more than 100 μm hydrogen peroxide was needed. Hyperoxidation of hPrx II was mostly reversible with up to 0.5 mm hydrogen peroxide, as seen by the migration back to the reduced spots in the presence of CHX during regeneration (Fig. 1A). At the acidic migration spots after regeneration, the irreversible portion of the hyperoxidized hPrx I was elevated in a hydrogen peroxide concentration-dependent manner (Fig. 1A). Irreversibly hyperoxidized hPrx I appeared in the 50 μm hydrogen peroxide-treated sample. The majority of hPrx I oxidized by 0.5 mm hydrogen peroxide was irreversible for up to 12 h of regeneration (Fig. 1A). Evaluation through direct visualization of protein spots on two-dimensional polyacrylamide gels with silver staining resulted in a pattern comparable with those of the immunoblots (Fig. 1B). Cell death or detachment of cells was not observed in our experimental condition.
FIGURE 1.
Hyperoxidation and regeneration of hPrx I and hPrx II in HeLa cells. A, HeLa cells were treated with the indicated concentrations of hydrogen peroxide for 10 min in PBS for hyperoxidation (Ox), followed by incubation for 12 h for regeneration (Re) in fresh media containing 10% FBS and 10 μg/ml CHX. Crude lysates were separated by two-dimensional PAGE, and the blots were probed with anti-Prx I or anti-Prx II antibodies. B, crude lysates from 250 μm hydrogen peroxide-treated (Ox) and -regenerated for 12 h (Re) were separated on two-dimensional polyacrylamide gels, and the proteins were visualized by silver staining. The spot positions of hyperoxidized (acidic migrated, ▾) and reduced (▿) forms of hPrx I and hyperoxidized (acidic migrated, •) and reduced (○) forms of hPrx II are indicated. An asterisk indicates the further acidic migrated positions of hPrx I. cont, control.
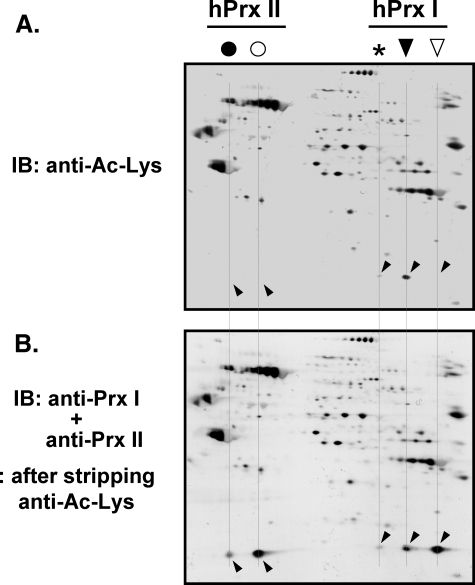
N-Acetylation Was Found Exclusively on Acidic Migrated hPrx I—Oxidation induced further acidic migration of acidic migrated hPrx I (Fig. 1A), suggesting the possibility of a post-translational modification other than hyperoxidation. The criteria for modifications were the gain of one negative charge or the loss of one positive charge without affecting mobility on the SDS-PAGE. N-Acetylation was evaluated because hPrxs I and II were recently identified as targets of histone deacetylase 6 (48), and this modification met the criteria for modification. Acidic migrated hPrx I had a strong immunoblot signal with an anti-acetylated Lys (anti-Ac-Lys) antibody (Fig. 2A). Neither the hPrx II spots nor the reduced hPrx I spot cross-reacted with anti-Ac-Lys antibody (Fig. 2, A and B), suggesting N-acetylation on one of the Lys residues of hPrx I located near the C terminus (48).
FIGURE 2.
N-Acetylation of the acidic migrated hPrx I in HeLa cells. HeLa cell lysate (50 μg) was separated on a two-dimensional polyacrylamide gel, followed by immunoblotting with anti-acetylated lysine antibody (anti-Ac-Lys; Cell Signaling Technology). For accurate control of relative sensitivity, alkaline phosphatase-conjugated anti-rabbit IgG with nitro blue tetrazolium/5-bromo-4-chloro-3′-indolyl phosphate was used for the visualization of immune complexes (A). After mild stripping of the antibodies, as described in the manufacturer's protocol (Abcam), the membranes were reprobed with a mixture of anti-Prx I and anti-Prx II antibodies (B). The spot positions of acidic migrated (▾) and reduced (▿) forms of hPrx I and acidic migrated (•) and reduced (○) forms of hPrx II are indicated. An asterisk indicates the further acidic migrated positions of hPrx I. Spots that cross-reacted with anti-Prx I and anti-Prx II are indicated with slanted downward and slanted upward arrowheads, respectively. IB, immunoblot.
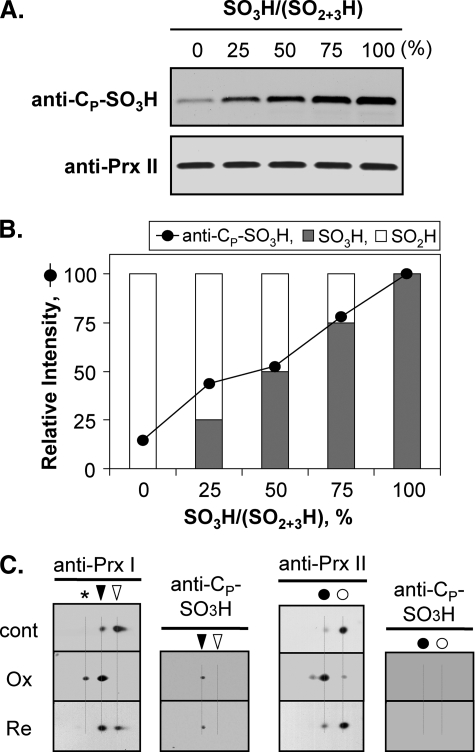
Irreversible Hyperoxidation Was Because of Further Oxidation of CP-SO3H—Oxidation of CP-SH to CP-SO3H and its irreversibility have been reported in Tsa1p, a yeast 2-Cys Prx (13). Although the regeneration of CP-SO2H to CP-SH has been reported in yeast and mammalian systems (17–24), detailed information about irreversibly hyperoxidized Prxs in mammalian systems is still limited. On two-dimensional polyacrylamide gels, certain portions of the hyperoxidized Prxs remained at the acidic location (hyperoxidized state) despite a prolonged regeneration time. Moreover, the intensity varied mainly with cell type and Prx isotype (18, 47). To explore the nature of these irreversible species, hyperoxidized spots of hPrx I and hPrx II were probed with an anti-CP-SO3H antibody. Anti-CP-SO3H antibody was initially prepared in response to a sulfonylated peptide of the active site sequence of the yeast 2-Cys Prx, Tsa1p. This antibody exhibits high specificity toward Tsa1p-SO3H relative to Tsa1p-SO2H (13). The use of an anti-Tsa1p-SO3H antibody to probe CP-SO3H of mammalian 2-Cys Prxs has been suggested because the active site sequence between Tsa1p (-AFTVCPPTEI-) and the mammalian 2-Cys Prxs (-DFTVCPPTEI-) is conserved (13). To analyze the specificity of the anti-Cp-SO3H antibody to hyperoxidized mammalian 2-Cys Prxs, hPrx II-CP-SO2H and hPrx II-CP-SO3H were prepared from purified hPrx II in the presence of rTrx and DTT. Careful preincubation and quenching of reaction mixtures 60 s after the addition of 100 μm or 3 mm of hydrogen peroxide produced a near 100% relative abundance of hPrx II-SO2H or hPrx II-SO3H. Molecular masses were verified using our LC/MS system, and the masses were 21,834.7 ± 1.1 Da for hPrx II-SO2H and 21,850.7 ± 1.1 Da for hPrx II-SO3H. Other oxidation adducts or mixtures of CP-SO2H and CP-SO3H were not detected under our experimental conditions. As shown in Fig. 3, A and B, anti-CP-SO3H immunoblot signals increased in proportion to the hPrx II-SO3H concentration. Hyperoxidized hPrx II in HeLa cells treated with 250 μm hydrogen peroxide did not cross-react with the anti-CP-SO3H antibody, suggesting the CP of hPrx II was hyperoxidized to the reversible sulfinic form (CP-SO2H) (Fig. 3C). A portion of the acidic migrated hPrx I was recognized by the anti-CP-SO3H antibody. The immunoblot signal intensity did not change, even after 12 h of regeneration (Fig. 3C). These results suggest the CP of hPrx I was, in part, hyperoxidized to the irreversible sulfonic form (hPrx I-SO3H).
FIGURE 3.
Specificity of anti-CP-SO3H antiserum and the hyperoxidation status of hPrx I and hPrx II in HeLa cells. Reaction mixtures (500 μl) contained 50 mm Hepes-NaOH, pH 7.0, 2 mm DTT, 1 mm EDTA, 0.4 μm rTrx, and 2 μm hPrx II. Reactions were initiated by adding hydrogen peroxide (100 μm for CP-SO2H or 3 mm for CP-SO3H) and terminated by adding trifluoroacetic acid (0.1% final concentration) after 60 s of incubation. Hyperoxidation status (CP-SO2H or CP-SO3H) was verified by LC/MS (see “Experimental Procedures”). A total of 80 ng of hPrx II proteins, with the indicated ratios of hPrx II-SO2H and hPrx II-SO3H, were separated by SDS-PAGE and visualized with anti-CP-SO3H or anti-Prx II antiserum (A). Anti-CP-SO3H immunoblot signals (A, upper panel) were quantified using Fuji Film MultiGauge software (B). C, HeLa cell crude extracts, treated with 250 μm hydrogen peroxide for 10 min (Ox) followed by 12 h of regeneration (Re), were separated on two-dimensional polyacrylamide gels. Prx proteins with CP-SO3H was visualized with anti-CP-SO3H antibody. hPrx I and hPrx II were visualized with anti-Prx I and anti-Prx II antisera, respectively. The spot positions of hyperoxidized (▾) and reduced (▿) forms of hPrx I and hyperoxidized (•) and reduced (○) forms of hPrx II are indicated. An asterisk indicates the further acidic migrated position of hPrx I. cont, control; Ox, hyperoxidized; Re, regenerated.
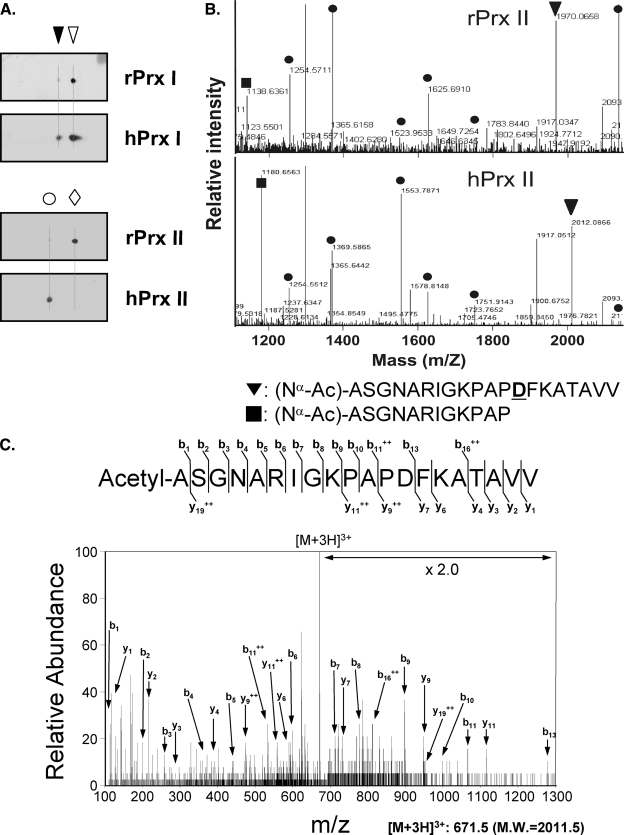
Nα-terminal Acetylation of hPrx II—hPrx I and hPrx II cDNAs were overexpressed in E. coli, and the crude extracts were analyzed by two-dimensional PAGE. rPrx I in E. coli and hPrx I in HeLa cells had the same two-dimensional PAGE migration patterns (theoretical pI = 8.27 and determined pI = 8.3 for the reduced form; Fig. 4A). hPrx II in HeLa cells (theoretical pI = 5.67) migrated to a more acidic location (determined pI = 5.45) relative to E. coli-expressed rPrx II (determined pI = 5.7, Fig. 4A). To identify the modification responsible for the change in pI, we performed AspN peptide fingerprinting on the protein spots of rPrx II and hPrx II. The masses of AspN peptides from rPrx II and hPrx II were matched with a coverage of 40%, except for two peptide pairs. One of the mismatched peptide pairs was 1138.64 Da from rPrx II and 1180.66 Da from hPrx II (Fig. 4B), which corresponded to the P2–13 peptide from the N terminus with an additional 42.02 Da ((Nα-Ac)-ASGNARIGKPAP). The masses of the other mismatched peptide pairs were 1970.07 Da from rPrx II and 2012.09 Da from hPrx II (Fig. 4B), which corresponded to the P2–21 peptide from the N terminus with an extra 42.02 Da ((Nα-Ac)-ASGNARIGKPAPDFKATAVV; the site of mis-cleavage is underlined). Therefore, the site of modification was limited to P2–13 from the N terminus. To locate the site of the 42-Da modification, the sequence of the 2012.09-Da peptide from hPrx II was analyzed by ESI MS/MS. Although the peptide included two Lys residues, the prominent b-ion series in the low mass range indicates the acetyl modification of an Ala residue at the N terminus. The b1 ion of the 114.05 Da peptide indicates an Ala residue with an acetyl group (Fig. 4C). These results support demethionylation at the N terminus and Nα-terminal acetylation of the penultimate Ala of hPrx II.
FIGURE 4.
Nα-terminal acetylation of hPrx II. A, lysates from rPrx I or rPrx II overexpressing E. coli cells (rPrx I or rPrx II) and lysates from HeLa cells (hPrx I and hPrx II) were separated by two-dimensional PAGE. Blots were probed with anti-Prx I (upper panel) or anti-Prx II (lower panel) antibodies. B, AspN fingerprinting analysis of the rPrx II (upper panel) and the hPrx II (lower panel) spots using MALDI-TOF/MS. Peptides with compatible (•) and incompatible mass pairs (▪ and ▾) are indicated. Ac-, acetyl-; D, site of mis-cleavage. C, MS/MS spectrum of the [M + 3H]3+ ion peak at m/z 671.5, which corresponds to the M + H+ ion peak at m/z 2012.09 in B, lower panel.
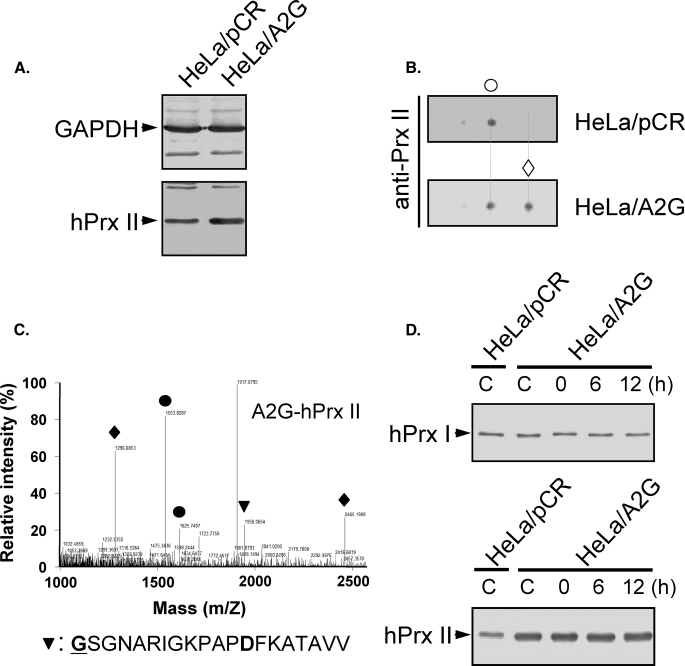
A2G-hPrx II Mutant Lacking Nα-terminal Acetylation—To identify the functional roles of the Nα-terminal acetylation of hPrx II, the penultimate Ala of hPrx II was mutated to a Gly residue (A2G-hPrx II) to prevent Nα-terminal acetylation. A2G-hPrx II was successfully expressed in HeLa cells via transient transfection with pCR plasmids harboring the mutant A2G-hPrx II cDNA (Fig. 5A). A2G-hPrx II showed basic migration on a two-dimensional polyacrylamide gel relative to the intrinsic hPrx II (Fig. 5B), suggesting the absence of Nα-acetylation. Excision of the initial Met residue and the absence of Nα-terminal acetylation were confirmed by mass spectrometric analysis, which revealed a mass of 1956.065 Da for the AspN fragment (theoretical molecular mass, 1956.066 Da, GSGNARIGKPAPDFKATAVV, the mutated Gly is boldface and underlined, and the mis-cleaved Asp is boldface; see Fig. 5C). The possibility of Nα-terminal myristoylation, which occasionally occurs at the N-terminal Gly residue, was negligible based on our two-dimensional PAGE and AspN fingerprinting results. After oxidative stress induced by 500 μm hydrogen peroxide, we did not see any stability differences between the intrinsic hPrx II and the overexpressed A2G-hPrx II in HeLa cells after up to 12 h in the presence of CHX (Fig. 5D).
FIGURE 5.
Expression of non-Nα-acetylated A2G-hPrx II and its stability in oxidatively stressed HeLa cells. A, HeLa cells were transiently transfected with pCR plasmids harboring A2G-hPrx II cDNA (HeLa/A2G) or empty vector (HeLa/pCR). Immunoblots were visualized with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (upper) and anti-Prx II (lower) antibodies. B, two-dimensional PAGE analysis of HeLa/pCR and HeLa/A2G cell lysates. Blots were probed with anti-Prx II antibody. Intrinsic hPrx II spot position (○) and exogenously overexpressed A2G-hPrx II spot position (♦) are indicated. C, MALDI-TOF/MS analysis of AspN digests of the A2G-hPrx II spot. Peptide (P2–P21) containing the penultimate A to G mutation without Nα-terminal acetylation is indicated (▾). Closed circles indicate peptides matched to the theoretical molecular mass of AspN fragments of hPrx II. External mass standards were angiotensin (1296.69) and ACTH (2465.20) (♦). D, cells were treated with 500 μm hydrogen peroxide for 10 min and then incubated for 6 and 12 h in fresh Dulbecco's modified Eagle's media containing 10 μg/ml CHX and 10% FBS. Samples were subjected to SDS-PAGE analysis followed by immunoblotting with anti-Prx I or anti-Prx II antibodies.
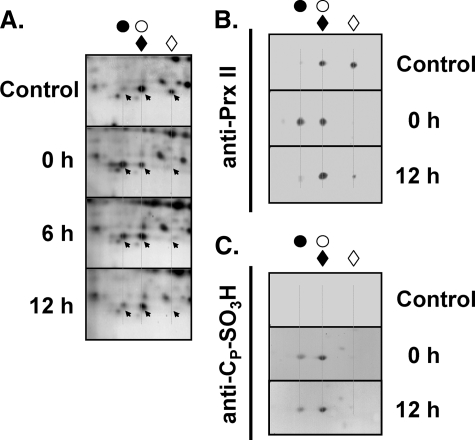
Nα-terminal Acetylation Protects CP of hPrx II against Irreversible Hyperoxidation—Because the reduced hPrx II spot and the hyperoxidized A2G-hPrx II spot were superimposed on the two-dimensional polyacrylamide gel, a precise evaluation of the ratio of hyperoxidized/reduced proteins seemed infeasible (Fig. 6). The extent of hyperoxidation and regeneration of A2G-hPrx II was thus evaluated by the change in the reduced spot signal, whereas that of intrinsic hPrx II was evaluated by the change in the hyperoxidated spot signal (Fig. 6, A and B). Virtually all of the reduced A2G-hPrx II spot signal was lost after 10 min of treatment with 250 μm hydrogen peroxide, and restoration to the reduced spot was negligible up to 12 h of regeneration, indicating irreversible hyperoxidation. Contrary to A2G-hPrx II, the majority of hyperoxidized hPrx II spots were retro-reduced after 12 h of regeneration, suggesting reversible hyperoxidation (Fig. 6, A and B). The irreversible sulfonic (CP-SO3H) state of hyperoxidized A2G-hPrx II was confirmed by Western blot with an anti-CP-SO3H antibody (Fig. 6C).
FIGURE 6.
Effects of Nα-terminal acetylation on the hyperoxidation of hPrx II in HeLa cells. HeLa cells expressing A2G-hPrx II were exposed to 250 μm hydrogen peroxide for 10 min and incubated in fresh media containing 10% FBS and 10 μg/ml CHX. Cells were collected at the indicated time and the crude lysates were analyzed by two-dimensional PAGE. Proteins separated on two-dimensional gels were visualized with silver staining (A). Immunoblots were probed with anti-Prx II (B) or anti-CP-SO3H antibodies (C). Filled and open circles represent hyperoxidized and reduced spot positions of hPrx II, respectively. Filled and open diamonds indicate hyperoxidized and reduced spot positions of A2G-hPrx II, respectively.
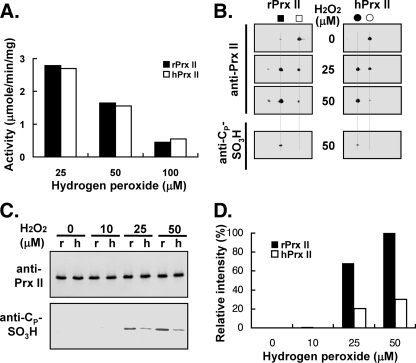
Nα-terminal Acetylation Did Not Have Any Effect on Initial Peroxidase Activity—The initial rates (first 10 s) of peroxide reduction were compared using three different concentrations of the substrate hydrogen peroxide. As shown in Fig. 7A, initial peroxidase activities decreased with increasing concentrations of substrate; however, Nα-acetylation did not affect the initial rate of peroxidase activity. The extent of the acidic migrated portions of rPrx II (non-Nα-acetylated) and hPrx II (Nα-acetylated) exhibited similar initial peroxidase activities. However, the hyperoxidized rPrx II showed a significantly stronger anti-CP-SO3H antibody immunoblot signal than hPrx II, indicating a higher irreversible hyperoxidation (Fig. 7B). The extent of irreversible hyperoxidation was compared with three different substrate concentrations on a single immunoblot (Fig. 7, C and D). When compared with rPrx II, about 30% of hPrx II was irreversibly hyperoxidized by each concentration of hydrogen peroxide used (Fig. 7D). Neither hPrx II nor rPrx II was irreversibly hyperoxidized by 10 μm hydrogen peroxide incubation for up to 150 s.
FIGURE 7.
Nα-terminal acetylation did not affect initial peroxidase activity. A, peroxidase activities of hPrx II and rPrx II were monitored by NADPH oxidation in the presence of thioredoxin system (see “Experimental Procedures”). An initial linear portion of the absorbance change (10 s) was used for the calculation of these activities. The results shown are the means of three independent assays. Samples from the assay mixtures were taken out 150 s after the reaction start and subjected to two-dimensional PAGE (B) and SDS-PAGE (C) analysis to identify and quantitate CP-SO3H, respectively. Immunoblots were visualized by anti-Prx II and anti-CP-SO3H antisera. r, rPrx II; h, hPrx II. D, graphical representation of the Tsa1p-SO3H immunoblot signal of C. The band intensities were quantitated by a densitometer (Gel Doc XR system, Bio-Rad).
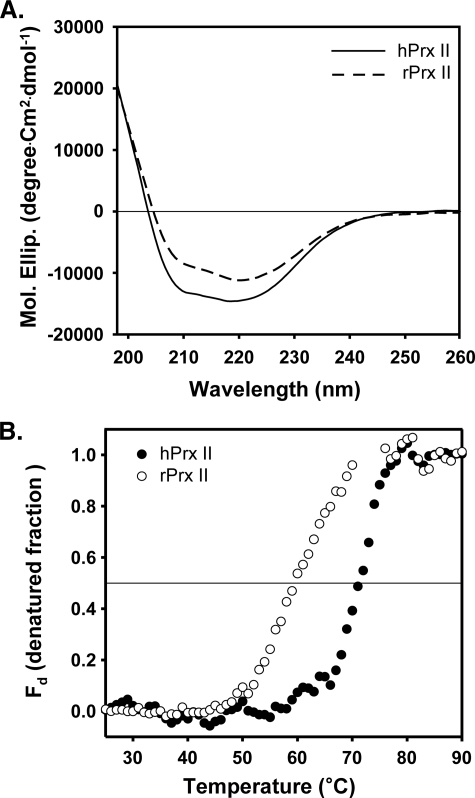
Structural Maintenance of hPrx II by Nα-terminal Acetylation—To investigate the role of Nα-terminal acetylation in the intrinsic protein structure of hPrx II, we compared the far-UV CD spectrogram of hPrx II with that of rPrx II. Although acquisition of the CD signal below 200 nm was difficult because of noise, hPrx II showed up to 27% more negative values in the mean residual molar ellipticity at 222 nm than rPrx II (Fig. 8A). The larger negative CD values near 220 nm represent a more ordered protein structure because of changes in the secondary structure (49, 50). This suggests a more ordered structure in hPrx II than rPrx II, which was conferred by Nα-terminal acetylation of the secondary structure. Next, we investigated the effect of Nα-terminal acetylation on protein thermostability. Our results revealed a Tm of 70.9 and 59.6 °C for hPrx II and rPrx II, respectively (Fig. 8B). These data indicate that Nα-terminal acetylation may contribute to the thermostability of Prx II.
FIGURE 8.
The effect of Nα-terminal acetylation on the secondary structure and thermal stability of hPrx II. A, mean residual molar ellipticity of Nα-terminal acetylated hPrx II (—) and nonacetylated rPrx II (- - -) in a 20 mm sodium phosphate, pH 7.4, buffer containing 100 mm NaCl at 25 °C. Because of the noise of the CD signal below 197 nm, the spectra are shown from 197 to 260 nm. B, thermal denaturation of hPrx II (closed circle) and rPrx II (open circle) in 20 mm sodium phosphate, pH 7.4, with 100 mm NaCl was monitored by the increase in CD signal at 222 nm with a rate of 1.0 °C/min. The denatured fraction (Fd) was calculated from the CD data as described under “Experimental Procedures.”
DISCUSSION
The major 2-Cys Prxs in humans, hPrx I and hPrx II, share 78% identity in their amino acid sequences and share the same catalytic mechanism without a significant substrate or cofactor preference for peroxidase activity. However, based on data from knock-out mouse experiments and binding protein studies, their functional assignments in cells are significantly different (29–39). It is reasonable to speculate that the physiological function of Prx I and Prx II is mainly through their peroxidase activities because evidence of direct modulation of partner molecules by protein-protein interactions was limited until recently. Thus, there were huge numbers of suggested physiological roles for mammalian Prx I and Prx II, which are attributable to peroxidase catalysis, claiming precise and versatile regulation of peroxidase activity in cells. In this study, we demonstrate that Nα-terminal acetylation occurs exclusively in hPrx II, which could provide a protective effect against irreversible hyperoxidation to form a sulfonic (CP-SO3H) derivative without altering its sensitivity toward hydrogen peroxide as a peroxidase substrate (Figs. 1 and 7). We also present evidence of N-acetylation of the acidic migrated portion of hPrx I. These changes might coincide with the acute requirements of the cell for maintaining proper levels of hydrogen peroxide in cells. Based on the observation that rPrx II exhibited a higher activity than rPrx I in purified proteins, Lee et al. (42) proposed that Prx II is physiologically a better-suited peroxidase than Prx I. Contrary to their proposal, we observed with HeLa cells that hPrx II was more susceptible to hydrogen peroxide, and this hyperoxidative inactivation of hPrx II was mostly reversible (Figs. 1 and 3). Discrepancies between studies often arise because of differences in assay procedure, such as in vitro versus cultured cells. As shown here, a portion of hPrx I in HeLa cells is N-acetylated (Fig. 2), and hPrx II is Nα-terminally acetylated (Fig. 4). These post-translational or cotranslational modifications are hard to reproduce in a prokaryotic expression system. In addition, other differences in assay conditions such as the range of hydrogen peroxide and the efficiency of the thioredoxin system used might also affect the distribution of the oxidized CP derivatives, namely CP-SOH, CP-S-S-CR, CP-SO2H, and the irreversible CP-SO3H.
Western immunoblot identification of N-acetylated hPrx I from an acidic migrated hPrx I spot is a reasonable observation, because N-acetylation causes an acidic pI shift of the protein because of a loss of one positive charge (Fig. 2). Careful evaluation was performed on the composition of the N-acetylated Prx I in the spot of acidic migrated hPrx I, because its quantity fluctuated significantly (up to 20% of total hPrx I) from batch to batch of HeLa cell cultures. In addition, N-acetylation of hPrx I has been reported to induce protection against overoxidation (48). Our results appear to be consistent with the notion that hPrx I is less susceptible to hyperoxidation. However, when hyperoxidation proceeded, hPrx I hyperoxidized to form its irreversible sulfonic derivative, then it cannot be regenerated to its reactive sulfhydryl derivative.
The mass spectrometric data revealed that hPrx II in HeLa cells underwent Nα-terminal acetylation, in which the N-terminal methionine was cleaved, and the acetylation occurred at the penultimate alanine (Fig. 4). Nα-terminal acetylation after N-terminal demethionylation is the most common modification in eukaryotic proteins (51, 52). In the crystal structure analysis of human erythrocyte Prx II (Protein Data Bank code 1QMV), the modification of the penultimate Ala in the N terminus was assumed to be an N-carbamylation (53). Carbamylation of proteins is generally believed to take place via the nonenzymatic binding of the urea-derived cyanate to free amino or sulfhydryl groups (54). Carbamyl modifications are possible in vivo (55–57) as well as in vitro (58); however, these reactions are not likely to be highly site-specific or an all-or-none type of modification, because of its nonenzymatic nature. Our results clearly demonstrate a 42-Da (acetylation) mass difference resulting from N-terminal modification instead of a 43-Da (carbamylation) difference. It was difficult to identify the basic migrated (nonacetylated) hPrx II spot with immunoblot unless the protein samples were overloaded. This result suggests virtually all the Prx II proteins in HeLa cells are Nα-terminal acetylated. Contrary to hPrx II, we did not see Nα-terminal acetylated hPrx I. Instead, we detected a fraction of hPrx I that was N-acetylated (Fig. 2). Unlike the irreversible cotranslational Nα-terminal acetylation, N-acetylation is a reversible post-translational modification (51). These results demonstrate that virtually all the hPrx II is Nα-terminal acetylated, and a portion of hPrx I is N-acetylated with variations. Results agree with the reported characteristics of protein acetylation in the mammalian system. Every crude extract of mammalian (human, rat, and mouse) tissue and cell line we analyzed had acidic migrated Prx II spots compared with the recombinant Prx II protein spots on two-dimensional polyacrylamide gels. This result suggests the possible Nα-terminal acetylation of all mammalian Prx IIs (data not shown).
Compared with hPrx II (Nα-acetylated), rPrx II (nonacetylated) showed no obvious differences in initial rates of peroxide reduction (Fig. 7A). However, relative to hPrx II, rPrx II was about three times more susceptible to irreversible (CP-SO3H) hyperoxidation (Fig. 7, C and D). To reveal the role of each modification on the regulation of peroxidase activity, an analysis of the rate constant by competitive kinetics with other peroxidases such as horseradish peroxidase (27, 28) should be completed. There was no obvious difference in the acidic migration (hyperoxidation) sensitivity between hPrx II and A2G-hPrx II when transiently overexpressed in HeLa cells. However, non-Nα-acetylated A2G-hPrx II was more prone to irreversible hyperoxidation to form sulfonic derivative than hPrx II (Fig. 6). These results demonstrate Nα-terminal acetylation of hPrx II mainly affects the irreversible hyperoxidation (CP-SO2H to CP-SO3H), instead of its peroxidase activity (CP-SH to CP-S-S-CR) or reversible hyperoxidation (CP-SOH to CP-SO2H). The crystal structure of sulfinic hPrx II (hPrx II-SO2H) demonstrates the hyperoxidation-induced trapping of the active site cysteine residue (53). Inaccessibility of substrate to the active site of hPrx II-SO2H may provide protection against further oxidation of CP-SO2H to CP-SO3H. Nevertheless, the contribution of Nα-terminal acetylation to the locked-in active site structure, which prevents the formation of CP-SO3H, has not been clearly revealed.
Our far-UV CD spectroscopy results support the stabilization of the hPrx II structure by Nα-terminal acetylation. This structural stabilization is not because of the formation of oligomers because we could not detect any obvious changes in molecular size between hPrx II and rPrx II using size-exclusion chromatography and multiangle light scattering techniques (DAWN-EOS multiangle laser light scattering detector, Wyatt Technology; data not shown). Our data support the notion that Nα-terminal acetylation induces a change in secondary structure and that the resulting structural change increases the thermal stability of hPrx II. However, the precise mechanics of this structural change in facilitating the reversible hyperoxidation or protecting against irreversible hyperoxidation remain to be elucidated.
Distinct physiological roles for sulfinic (CP-SO2H) and sulfonic (CP-SO3H) derivatives of 2-Cys Prxs are not well understood. However, through irreversible sulfonic hyperoxidation, cells can remove two additional molecules of hydrogen peroxide without consuming cellular reducing equivalents. In addition, cells gain a molecular chaperone function by trading peroxidase catalytic activity (13, 42, 59, 60). Switching of the 2-Cys Prxs to a molecular chaperone by isotype-specific hyperoxidation has not been studied in detail; however, redox status-dependent oligomeric structure changes and corresponding chaperone activity changes have been reported (13, 42, 59). The redox status of CP is controlled neatly by the cellular environment as well as the oligomeric structure of Prx. Consequently, the chaperone activity of Prxs is precisely regulated by changes in the cellular environment. To study the redox-controlled cellular chaperone function of 2-Cys Prxs, evaluation of a chaperone activity with a defined population of redox states (CP-SH, CP-S-S-CR, CP-SOH, CP-SO2H, and CP-SO3H) is needed instead of evaluation with a mixture of various redox forms. Fine-tuning of isotype-specific oxidation conditions and isolation methods for specific redox forms remains to be established.
In summary, the results presented here provide evidence that hPrx II in HeLa cells is more sensitive to hyperoxidative inactivation than hPrx I. In contrast to hPrx I, the hyperoxidative inactivation of hPrx II is mostly reversible. Nα-terminal acetylation exclusively exists in hPrx II and is responsible for protection against irreversible hyperoxidation. Evidence of N-acetylation in a fraction of the hPrx I population is provided as well. In addition, studies with routine N-terminal tagging of hPrx II or C-terminal tagging of hPrx I should be cautiously interpreted because this modification may prevent Nα-terminal acetylation of Prx II and affect the N-acetylation of Prx I. As a consequence, these results could be physiologically irrelevant.
Acknowledgments
We thank Drs. Zheng You and Rod Levine for the help of LC/MS analysis and Merry Peters for carefully reading the manuscript.
This work was authored, in whole or in part, by National Institutes of Health staff. This work was supported by Korea Research Foundation Grant KRF-2005-070-C00081, Chonnam National University Grant CNU-2008-1005, Korea Science and Engineering Foundation Grant R01-2008-000-21072-02008 (to H. Z. C.), and Second Stage BK21 Project from the Ministry of Education, Science and Technology of Korea.
Footnotes
The abbreviations used are: Prx, peroxiredoxin; hPrx, human peroxiredoxin; rPrx, recombinant peroxiredoxin; CP, peroxidatic cysteine; CR, resolving cysteine; HPLC, high performance liquid chromatography; CHX, cycloheximide; ABC, ammonium bicarbonate; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; DTT, dithiothreitol; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; MS, mass spectrometry; RPLC/ESI-MS/MS, reverse-phase liquid chromatography/electrospray ionization tandem mass spectrometry; AspN, peptidyl-Asp metalloendopeptidase; ACN, acetonitrile; FBS, fetal bovine serum; Ac, acetylation; rTrx, rat thioredoxin; ACTH, adrenocorticotropic hormone.
References
- 1.Rhee, S. G., Chae, H. Z., and Kim, K. (2005) Free Radic. Biol. Med. 38 1543–1552 [DOI] [PubMed] [Google Scholar]
- 2.Wood, Z. A., Schröder, E., Harris, R. J., and Poole, L. B. (2003) Trends Biochem. Sci. 28 32–40 [DOI] [PubMed] [Google Scholar]
- 3.Hofmann, B., Hecht, H.-J., and Flohé, L. (2002) Biol. Chem. 383 347–364 [DOI] [PubMed] [Google Scholar]
- 4.Chae, H. Z., Robison, K., Poole, L. B., Church, G., Storz, G., and Rhee, S. G. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 7017–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee, S. G., Kang, S. W., Chang, T.-S., Jeong, W., and Kim, K. (2001) IUBMB Life 52 35–41 [DOI] [PubMed] [Google Scholar]
- 6.Bryk, R., Griffin, P., and Nathan, C. (2000) Nature 407 211–215 [DOI] [PubMed] [Google Scholar]
- 7.Trujillo, M., Ferrer-Sueta, G., and Radi, R. (2008) Methods Enzymol. 441 173–196 [DOI] [PubMed] [Google Scholar]
- 8.Trujillo, M., Ferrer-Sueta, G., Thomson, L., Flohé, L., and Radi, R. (2007) Subcell. Biochem. 44 83–113 [DOI] [PubMed] [Google Scholar]
- 9.Poole, L. B. (2007) Subcell. Biochem. 44 61–81 [DOI] [PubMed] [Google Scholar]
- 10.Wood, Z. A., Poole, L. B., and Karplus, P. A. (2003) Science 300 650–653 [DOI] [PubMed] [Google Scholar]
- 11.Karplus, P. A., and Hall, A. (2007) Subcell. Biochem. 44 41–60 [DOI] [PubMed] [Google Scholar]
- 12.Yang, K. S., Kang, S. W., Woo, H. A., Hwang, S. C., Chae, H. Z., Kim, K., and Rhee, S. G. (2002) J. Biol. Chem. 277 38029–38036 [DOI] [PubMed] [Google Scholar]
- 13.Lim, J. C., Choi, H.-I., Park, Y. S., Nam, H. W., Woo, H. A., Kwon, K.-S., Kim, Y. S., Rhee, S. G., Kim, K., and Chae, H. Z. (2008) J. Biol. Chem. 283 28873–28880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo, H. A., Kang, S. W., Kim, H. K., Yang, K. S., Chae, H. Z., and Rhee, S. G. (2003) J. Biol. Chem. 278 47361–47364 [DOI] [PubMed] [Google Scholar]
- 15.Mitsumoto, A., Takanezawa, Y., Okawa, K., Iwamatsu, A., and Nakagawa, Y. (2001) Free Radic. Biol. Med. 30 625–635 [DOI] [PubMed] [Google Scholar]
- 16.Rabilloud, T., Heller, M., Gasnier, F., Luche, S., Rey, C., Aebersold, R., Benahmed, M., Louisot, P., and Lunardi, J. (2002) J. Biol. Chem. 277 19396–19401 [DOI] [PubMed] [Google Scholar]
- 17.Wagner, E., Luche, S., Penna, L., Chevallet, M., Dorsselaer, A. V., Leize-Wagner, E., and Rabilloud, T. (2002) Biochem. J. 366 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo, H. A., Chae, H. Z., Hwang, S. C., Yang, K. S., Kang, S. W., Kim, K., and Rhee, S. G. (2003) Science 300 653–656 [DOI] [PubMed] [Google Scholar]
- 19.Biteau, B., Labarre, J., and Toledano, M. B. (2003) Nature 425 980–984 [DOI] [PubMed] [Google Scholar]
- 20.Budanov, A. V., Sablina, A. A., Feinstein, E., Koonin, E. V., and Chumakov, P. M. (2004) Science 304 596–600 [DOI] [PubMed] [Google Scholar]
- 21.Chang, T. S., Jeong, W., Woo, H. A., Lee, S. M., Park, S., and Rhee, S. G. (2004) J. Biol. Chem. 279 50994–51001 [DOI] [PubMed] [Google Scholar]
- 22.Rhee, S. G., Jeong, W., Chang, T. S., and Woo, H. A. (2007) Kidney Int. 106 S3–S8 [DOI] [PubMed] [Google Scholar]
- 23.Jönsson, T. J., and Lowther, W. T. (2007) Subcell. Biochem. 44 115–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jönsson, T. J., Johnson, L. C., and Lowther, W. T. (2008) Nature 451 98–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae, H. Z., Kim, H. J., Kang, S. W., and Rhee, S. G. (1999) Diabetes Res. Clin. Pract. 45 101–112 [DOI] [PubMed] [Google Scholar]
- 26.Kang, S. W., Chae, H. Z., Seo, M. S., Kim, K., Baines, I. C., and Rhee, S. G. (1998) J. Biol. Chem. 273 6297–6302 [DOI] [PubMed] [Google Scholar]
- 27.Peskin, A. V., Low, F. M., Paton, L. N., Maghzal, G. J., Hampton, M. B., and Winterbourn, C. C. (2007) J. Biol. Chem. 282 11885–11892 [DOI] [PubMed] [Google Scholar]
- 28.Ogusucu, R., Rettori, D., Munhoz, D. C., Netto, L. E. S., and Augusto, O. (2007) Free Radic. Biol. Med. 42 326–334 [DOI] [PubMed] [Google Scholar]
- 29.Wen, S. T., and Van Etten, R. A. (1997) Genes Dev. 11 2456–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu, Z. M., Yin, X. Y., and Prochownik, E. V. (2002) J. Biol. Chem. 277 43175–43184 [DOI] [PubMed] [Google Scholar]
- 31.Jung, H., Kim, T., Chae, H. Z., Kim, K. T., and Ha, H. (2001) J. Biol. Chem. 276 15504–15510 [DOI] [PubMed] [Google Scholar]
- 32.Park, S.-Y., Yu, X., Ip, C., Mohler, J. L., Bogner, P. N., and Park, Y.-M. (2007) Cancer Res. 67 9294–9303 [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. Y., Kim, T. J., and Lee, K.-Y. (2008) FEBS Lett. 582 1913–1918 [DOI] [PubMed] [Google Scholar]
- 34.Neumann, C. A., and Fang, Q. (2007) Curr. Opin. Pharmacol. 7 375–380 [DOI] [PubMed] [Google Scholar]
- 35.Choi, M. H., Lee, I. K., Kim, G. W., Kim, B. U., Han, Y. H., Yu, D. Y., Park, H. S., Kim, K. Y., Lee, J. S., Choi, C., Bae, Y. S., Lee, B. I., Rhee, S. G., and Kang, S. W. (2005) Nature 435 347–353 [DOI] [PubMed] [Google Scholar]
- 36.Xiao, N., Du, G., and Frohman, M. A. (2005) FEBS J. 272 3929–3937 [DOI] [PubMed] [Google Scholar]
- 37.Neumann, C. A., Krause, D. S., Carman, C. V., Das, S., Dubey, D. P., Abraham, J. L., Bronson, R. T., Fujiwara, Y., Orkin, S. H., and Van Etten, R. A. (2003) Nature 424 561–565 [DOI] [PubMed] [Google Scholar]
- 38.Egler, R. A., Fernandes, E., Rothermund, K., Sereika, S., de Souza-Pinto, N., Jaruga, P., Dizdaroglu, M., and Prochownik, E. V. (2005) Oncogene 24 8038–8050 [DOI] [PubMed] [Google Scholar]
- 39.Lee, T. H., Kim, S. U., Yu, S. L., Kim, S. H., Park, D. S., Moon, H. B., Dho, S. H., Kwon, K. S., Kwon, H. J., Han, Y. H., Jeong, S., Kang, S. W., Shin, H. S., Lee, K. K., Rhee, S. G., and Yu, D. Y. (2003) Blood 101 5033–5038 [DOI] [PubMed] [Google Scholar]
- 40.Sarafian, T. A., Verity, M. A., Vinters, H. V., Shih, C. C., Shi, L., Ji, X. D., Dong, L., and Shau, H. (1999) J. Neurosci. Res. 56 206–212 [PubMed] [Google Scholar]
- 41.Lee, K., Park, J.-S., Kim, Y.-J., Lee, Y. S., Hwang, T. S., Kim, D.-J., Park, E.-M., and Park, Y.-M. (2002) Biochem. Biophys. Res. Commun. 296 337–342 [DOI] [PubMed] [Google Scholar]
- 42.Lee, W., Choi, K.-S., Tiddell, J., Ip, C., Ghosh, D., Park, J.-H., and Park, Y.-M. (2007) J. Biol. Chem. 282 22011–22022 [DOI] [PubMed] [Google Scholar]
- 43.Matsumura, T., Okamoto, K., Iwahara, S., Hori, H., Takahashi, Y., Nishino, T., and Abe, Y. (2008) J. Biol. Chem. 283 284–293 [DOI] [PubMed] [Google Scholar]
- 44.Chae, H. Z., Kang, S. W., and Rhee, S. G. (1999) Methods Enzymol. 300 219–226 [DOI] [PubMed] [Google Scholar]
- 45.Luthman, M., and Holmgren, A. (1982) Biochemistry 21 6628–6633 [DOI] [PubMed] [Google Scholar]
- 46.Taggart, C., Cervantes-Laurean, D., Kim, G., McElvaney, N. G., Wehr, N., Moss, J., and Levine, R. L. (2000) J. Biol. Chem. 275 27258–27265 [DOI] [PubMed] [Google Scholar]
- 47.Chevallet, M., Wagner, E., Luche, S., van Dorsselaer, A., Leize-Wagner, E., and Rabilloud, T. (2003) J. Biol. Chem. 278 37146–37153 [DOI] [PubMed] [Google Scholar]
- 48.Parmigiani, R. B., Xu, W. S., Venta-Perez, G., Erdjument-Bromage, H., Yaneva, M., Tempst, P., and Marks, P. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9633–9638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monnot, M., Gilles, A.-M., Girons, I. S., Michelson, S., Bârzu, O., and Fermandjian, S. (1987) J. Biol. Chem. 262 2502–2506 [PubMed] [Google Scholar]
- 50.Ogawa, K., Nishimura, S., Doi, M., Kyogoku, Y., Hayashi, M., and Kobayashi, Y. (1994) Eur. J. Biochem. 222 659–666 [DOI] [PubMed] [Google Scholar]
- 51.Polevoda, B., and Sherman, F. (2000) J. Biol. Chem. 275 36479–36482 [DOI] [PubMed] [Google Scholar]
- 52.Bradshaw, R. A., Brickey, W. W., and Walker, K. W. (1998) Trends Biochem. Sci. 23 263–267 [DOI] [PubMed] [Google Scholar]
- 53.Schröder, E., Littlechild, J. A., Lebedev, A. A., Errington, N., Vagin, A. A., and Isupov, M. N. (2000) Structure (Lond.) 8 605–615 [DOI] [PubMed] [Google Scholar]
- 54.Stark, G. R. (1972) Methods Enzymol. 25 579–584 [DOI] [PubMed] [Google Scholar]
- 55.Ok, E., Basnakian, A. G., Apostolov, E. O., Barri, Y. M., and Shah, S. V. (2005) Kidney Int. 68 173–178 [DOI] [PubMed] [Google Scholar]
- 56.Kraus, L. M., and Kraus, A. P. Jr. (2001) Kidney Int. 78. S102–S107 [DOI] [PubMed] [Google Scholar]
- 57.Lapko, V. N., Smith, D. L., and Smith, J. B. (2001) Protein Sci. 10 1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy, J., Hopwood, F., Oxley, D., Laver, M., Castagna, A., Righetti, P. G., Williams, K., and Herbert, B. (2003) J. Proteome Res. 2 239–242 [DOI] [PubMed] [Google Scholar]
- 59.Jang, H. H., Lee, K. O., Chi, Y. H., Jung, B. G., Park, S. K., Park, J. H., Lee, J. R., Lee, S. S., Moon, J. C., Yun, J. W., Choi, Y. O., Kim, W. Y., Kang, J. S., Cheong, G. W., Yun, D. J., Rhee, S. G., Cho, M. J., and Lee, S. Y. (2004) Cell 117 625–635 [DOI] [PubMed] [Google Scholar]
- 60.Moon, J. C., Hah, Y. S., Kim, W. Y., Jung, B. G., Jang, H. H., Lee, J. R., Kim, S. Y., Lee, Y. M., Jeon, M. G., Kim, C. W., Cho, M. J., and Lee, S. Y. (2005) J. Biol. Chem. 280 28775–28784 [DOI] [PubMed] [Google Scholar]