Abstract
Focal adhesions are specialized sites of cell attachment to the extracellular matrix where integrin receptors link extracellular matrix to the actin cytoskeleton, and they are constantly remodeled during cell migration. Focal adhesion kinase (FAK) is an important regulator of focal adhesion remodeling. AGAP2 is an Arf GTPase-activating protein that regulates endosomal trafficking and is overexpressed in different human cancers. Here we examined the regulation of the FAK activity and the focal adhesion remodeling by AGAP2. Our results show that FAK binds the pleckstrin homology domain of AGAP2, and the binding is independent of FAK activation following epidermal growth factor receptor stimulation. Overexpression of AGAP2 augments the activity of FAK, and concordantly, the knockdown of AGAP2 expression with RNA interference attenuates the FAK activity stimulated by epidermal growth factor or platelet-derived growth factor receptors. AGAP2 is localized to the focal adhesions, and its overexpression results in dissolution of the focal adhesions, whereas knockdown of its expression stabilizes them. The AGAP2-induced dissolution of the focal adhesions is independent of its GTPase-activating protein activity but may involve its N-terminal G protein-like domain. Our results indicate that AGAP2 regulates the FAK activity and the focal adhesion disassembly during cell migration.
Focal adhesions are macromolecular structures that connect actin cytoskeleton to the extracellular matrix and play an important role in cell migration (1). Components of focal adhesions include signaling proteins such as focal adhesion kinase (FAK),3 c-Src, and paxillin, as well as structural proteins such as talin and vinculin (2, 3). Focal adhesions are constantly formed and disassembled (i.e. remodeled) at the leading edge of migrating cells, and they are disassembled at the trailing edge during the cell migration (4, 5). Available evidence demonstrates that the remodeling of focal adhesions is regulated by FAK (6) and Arf-directed GTPase-activating proteins (Arf GAPs) (7).
FAK is a member of the Src family nonreceptor tyrosine kinases whose activities are regulated by intra-molecular phosphorylation (8). Autophosphorylation of FAK on tyrosine residue 397 provides docking sites for Src homology 2 domain-containing proteins, including c-Src. After binding to FAK, c-Src phosphorylates FAK on Tyr-576 and Tyr-577 to activate fully the intrinsic kinase activity of FAK (9). Cellular functions of FAK are many and include cell migration, survival, and proliferation; and activation of FAK occurs upon integrin clustering or stimulation of cell surface receptors such as the epidermal growth factor (EGF) or platelet-derived growth factor (PDGF) receptors. FAK activation following integrin clustering results in recruitment of structural and signaling proteins that collectively contribute to the formation of the focal adhesions (10). In FAK null cells, focal adhesions are formed but cannot disassemble (11), suggesting that FAK is required for the focal adhesion disassembly.
ADP-ribosylation factors (Arfs) are GTP-binding proteins that lack detectable intrinsic GTPase activities. Therefore, hydrolysis of GTP bound to Arf is mediated by Arf GAPs (12, 13). The AZAP family of Arf GAPs are multidomain proteins that contain a catalytic core of pleckstrin homology (PH), Arf GAP, and ankyrin repeat domains (14), and each subgroup possesses characteristic domain(s). For example, ASAPs have a BAR (Bin, Amphiphysin, Rvs) domain at their N termini and a Src homology 3 domain at their C termini; ARAPs have a Rho GAP domain and five PH domains; and AGAPs have a G protein-like domain (GLD) at their N termini and their PH domains are split, i.e. there is an insert of 80–100 amino acids between the β5 strand and β6 strand. The Arf GAPs regulate membrane trafficking and remodeling of the actin cytoskeleton (7, 15), but the molecular mechanisms underlying the contribution of individual Arf GAPs to membrane trafficking and actin remodeling are being defined. We have reported that AGAP2 binds the clathrin adaptor protein AP-1 and regulates the AP-1/Rab4-dependent endosomal trafficking (16). Studies from other groups have indicated that AGAP2 is overexpressed in different human cancers, including glioblastoma, and that AGAP2 enhances the invasion of glioblastoma cells (17, 18).
In this study, we tested the hypothesis that AGAP2 regulates focal adhesion remodeling and cell migration. We find that AGAP2 forms a complex with FAK, increases the FAK activity, and provokes the focal adhesion disassembly that may lead to increased cell migration. Some Arf GAPs have been shown to regulate focal adhesions, and each Arf GAP seems to regulate the focal adhesions by a distinct mechanism. Our results introduce a new way to regulate the focal adhesions by the Arf GAP AGAP2, i.e. through the regulation of FAK activity. These observations support the idea that various Arf GAPs function coordinately to provide temporal and spatial regulation of the focal adhesions during cell migration.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies—Cloning of AGAP1 and AGAP2 (wild-type and mutant forms) with a FLAG tag (DYKDDDDK) into pCI and pSI vectors for mammalian expression or into pGEX4T-1 vector for bacterial expression was described before (16). For stable expression of AGAP2 and PZA2 (see Fig. 1B for AGAP2 structure), the cDNA fragments were subcloned into XhoI and BamHI sites of pcDNA3.1(–) (Invitrogen). Antibodies were obtained as follows: polyclonal and monoclonal anti-FLAG and monoclonal anti-vinculin antibodies from Sigma; anti-FAK, anti-paxillin, and anti-EGF receptor antibodies from BD Biosciences; anti-Erk from Santa Cruz Biotechnology; anti-Akt, anti-pAkt, and anti-pErk antibodies from Cell Signaling; anti-pFAK from Invitrogen; anti-phosphotyrosine from Millipore; and fluorescein isothiocyanate- and rhodamine-conjugated secondary antibodies from Jackson ImmunoResearch. Preparation of the polyclonal anti-AGAP2 was described before (16). EGF and PDGF were from Invitrogen.
FIGURE 1.
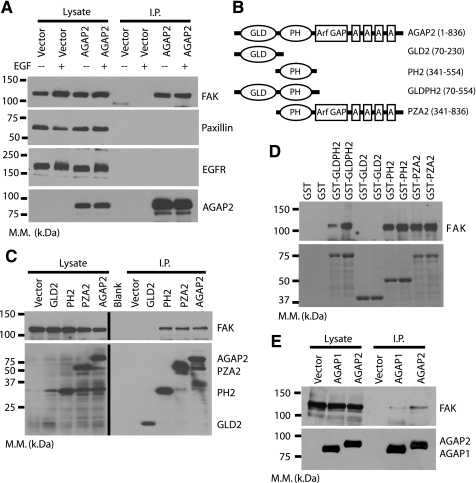
AGAP2 forms a complex with focal adhesion kinase. A, co-immunoprecipitation of FAK with AGAP2. HEK293 cells were transfected with cDNAs encoding empty vector or FLAG-AGAP2. The AGAP2 was immunoprecipitated (I.P.) with anti-FLAG antibody and eluted with FLAG peptide. The eluted proteins were resolved by SDS-PAGE, and AGAP2 was detected by Western blotting using polyclonal anti-FLAG antibody. The endogenous FAK, EGFR, and paxillin proteins that co-precipitated with AGAP2 were detected using appropriate antibodies. B, schematic of AGAP2 and its deletion mutants. GLD, G protein-like domain; PH, pleckstrin homology domain; GAP, GTPase-activating protein (containing a zinc finger); A, ankyrin repeats. C, FAK co-immunoprecipitates with PH2 domain of AGAP2. The cDNAs encoding FLAG-tagged wild-type and deletion mutants of AGAP2 were transfected into HEK293 cells and immunoprecipitated. Co-immunoprecipitation of FAK was examined by Western blotting. D, pulldown of FAK with GST-AGAP2 fusion proteins. Purified GST fusion proteins of AGAP2 deletion mutants were incubated with U87 cell lysates, and pulldown of FAK was examined by Western blotting (top panel). Bottom panel shows Coomassie blue staining of the GST fusion proteins. E, selective binding of AGAP2 with FAK. HEK293 cells were transfected with cDNAs encoding empty vector, FLAG-AGAP1, or FLAG-AGAP2, and the proteins were immunoprecipitated as described above. Co-immunoprecipitation of endogenous FAK was examined by Western blotting. M.M., molecular mass.
Cell Culture, Transfection, and Immunofluorescence—HEK293 and U87 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C with 5% CO2. Cells were transfected with Lipofectamine 2000 (Invitrogen) for 24–48 h and harvested for immunoprecipitation and immunoblotting. For stable overexpression of AGAP2, cells were transfected with cDNAs encoding empty vector or FLAG-AGAP2 in pcDNA3.1(–) vector, selected with G418 at 500–800 μg/ml for 2 weeks, and maintained with G418 at 200 μg/ml. Monoclonal cells were pooled, and the expression of AGAP2 was confirmed by Western blotting. For immunofluorescence staining, U87 cells were seeded on glass coverslips coated with fibronectin (25 μg/ml) for 5 h for the staining of focal adhesions or for the indicated times for cell spreading. Cells were fixed with 4% formaldehyde for 10 min at room temperature followed by staining with the indicated antibodies. Focal adhesions were visualized by staining with anti-paxillin, anti-vinculin, or anti-phosphotyrosine antibodies. Actin was visualized by staining with rhodamine-conjugated phalloidin. Confocal microscopy was performed at the MCG Imaging Core Facility with a Zeiss LSM 510 Meta system mounted on an Axiovert 200 M microscope (Carl Zeiss) using a 63 × 1.4 NA Zeiss Plan-Apochromat oil immersion objective.
Immunoprecipitation and GST Pulldown Assays—Cells were washed with PBS and harvested in lysis buffer containing 25 mm Tris, pH 8.0, 100 mm NaCl, 1% (v/v) Triton X-100, 10% (v/v) glycerol, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 2 μg/ml pepstatin A. For detection of phosphorylated proteins, phosphatase inhibitor mixture (P1 and P2, Sigma, at 1:100 dilution) and 1 mm sodium orthovanadate were included in the lysis buffer. The cell lysates were cleared by centrifugation at 10,000 × g for 15 min at 4 °C and incubated with anti-FLAG antibody, and proteins were precipitated and eluted as we described previously (16). For pulldown assays, the GST fusion proteins were expressed in BL21 cells and purified using glutathione-Sepharose 4B beads. The protein-bound beads were washed with PBS and incubated with cell lysates overnight at 4 °C, washed with PBS, and boiled in sample buffer. The precipitated proteins were resolved by SDS-PAGE and detected by immunoblotting.
Knockdown of AGAP2 by siRNA and shRNA—Two siRNA duplexes with symmetric 3′-UU overhangs were synthesized at Dharmacon. The siRNA sequences targeting AGAP2 were 5′-AGA CAC AUC UGG UGC UAA U-3′ and 5′-GUA AUG GCU UUC UAC UCU A-3′. Transient transfection of U87 and HEK293 cells with the mixture of both siRNAs (50 nm each) was performed with Lipofectamine RNAiMax (Invitrogen), using the nontargeting siRNA (Ambion) as a control. The efficiency of AGAP2 knockdown was examined by immunoblotting 72 h after transfection. For stable knockdown of AGAP2 by shRNA, we used five lentiviral DNA expression vectors that contain 21 nucleotide shRNA duplexes that target AGAP2 (OpenBiosystems). The vectors were co-transfected with equal concentrations of VSVG and Δ8.9 vector into HEK293T cells using FuGENE 6 (Roche Applied Science). As a negative control, HEK293T cells were transfected with the same amount of expression vector containing the green fluorescent protein targeting sequence 5′-GCA AGC TGA CCC TGA AGT TCA T-3′. Virus-containing medium from transfected HEK293T cells was collected 24 h after transfection and mixed with 5 μl of Polybrene solution for infection of U87 cells. HEK293T cells were replenished with fresh medium for another 24 h, and the medium was collected for second infection. Cells were infected for 2 h, and the medium was changed to Dulbecco's modified Eagle's medium containing 10% fetal bovine serum for 48 h before selection with puromycin (2 μg/ml), using uninfected U87 cells as a selection control. Cells were maintained with puromycin (1 μg/ml), and the efficiency of AGAP2 knockdown was determined by Western blotting.
Cell Migration Assay—Cells were starved overnight in Opti-MEM, washed with PBS, detached using Versene (Invitrogen), centrifuged, and resuspended in Opti-MEM and counted. A total of 1.0 × 105 cells in 100 μl of Opti-MEM was added to the migration chamber and placed on the feeder tray that contains 150 μl of Opti-MEM supplemented, or not, with EGF (50 nm). Cells migrating to the underside were detached, lysed, and stained with the fluorescent dye provided in the cell migration kit (Chemicon). Fluorescence intensities were measured with a plate reader using 485/520 filter set (POLARstar OPTIMA, BMG Labtech) that reflected the concentration of nucleic acids of the migrated cells.
Statistics—Data are presented as the mean ± S.E. from at least three independent experiments. The data were analyzed by Student's t test or analysis of variance and followed by the Tukey post-test to determine the statistical significance of differences. All statistical analyses were performed, and all graphs were generated using GraphPad Prism 3.0 (GraphPad).
RESULTS
AGAP2 Forms a Complex with FAK—AGAP2 was shown to express at elevated levels in different human cancers, including malignant glioma, and the overexpressed AGAP2 increased the invasion of malignant glioma cells. Cell migration plays a critical role in the invasion and metastasis of cancer cells, and we hypothesized that AGAP2 regulates focal adhesions and cell migration. To test this idea, we examined the possible interaction of AGAP2 with the known regulators of the focal adhesions FAK and paxillin. FLAG epitope-tagged AGAP2 was expressed in HEK293 cells and immunoprecipitated using the M2 anti-FLAG antibody and visualized by immunoblotting using polyclonal anti-FLAG antibody (Fig. 1A). The presence of FAK in the lysates and immunoprecipitates was examined using anti-FAK antibody (Fig. 1A). The AGAP2 co-precipitated FAK, and the binding of AGAP2 to FAK was independent of the FAK activation by EGF receptors; the stimulation with EGF did not impact AGAP2-FAK complex formation (Fig. 1A). To test if AGAP2 also bound other components of the focal adhesions, we examined the possible co-immunoprecipitation of paxillin with AGAP2. Under the same conditions that evidenced the AGAP2-FAK complex, paxillin was not detected (Fig. 1A). Because FAK is activated following EGF stimulation, we next examined whether AGAP2 forms a complex with the EGF receptors. Under conditions used to co-immunoprecipitate FAK with AGAP2, we did not observe the co-immunoprecipitation of the EGF receptors (Fig. 1A).
Next, we mapped the domain of AGAP2 that is responsible for the binding to FAK. Different deletion mutants of AGAP2, including GLD2, PH2, PZA2, as well as the full-length AGAP2 (Fig. 1B), were expressed as FLAG epitope-tagged proteins in HEK293 cells. Cleared cell lysates were subjected to immunoprecipitation using the M2 anti-FLAG antibody, and co-immunoprecipitation of FAK was examined as described above. The results show that FAK co-immunoprecipitated with the PH2, PZA2, and full-length AGAP2 but not the isolated GLD2 domain (Fig. 1C), suggesting that the PH2 domain of AGAP2 is the binding site for FAK. To confirm the PH2 domain mediated binding of AGAP2 to FAK, we performed GST pulldown assays using GST fusion proteins of the deletion mutants of AGAP2. Similar amounts of purified GST, GST-GLD2, GST-GLDPH2, GST-PH2, and GST-PZA2 were incubated with lysates from the glioblastoma U87 cells, and the presence of endogenous FAK in the precipitates was examined by Western blotting. As shown in Fig. 1D, GST fusion proteins containing the PH2 domain of AGAP2 successfully precipitated FAK. The binding of FAK to AGAP2 was independent of cell type as similar results were obtained using HEK293 and the breast carcinoma MCF-7 cells (not shown). The immunoprecipitation and GST pulldown results indicate that the AGAP2 PH2 domain is the binding site for FAK. To further examine the specificity of the interaction between AGAP2 and FAK, we compared the binding of AGAP1 and AGAP2 to FAK by co-immunoprecipitation. FLAG-tagged AGAP1 and AGAP2 were expressed in HEK293 cells to a similar level and immunoprecipitated using the M2 anti-FLAG antibody, and co-immunoprecipitation of FAK was examined as described above. As shown in Fig. 1E, both AGAP1 and AGAP2 formed a complex with FAK, but AGAP2 was more efficient in precipitating the endogenous FAK.
AGAP2 Regulates Activity of FAK—FAK activity is controlled, in part, by its interaction with partner proteins, and we established the direct interaction between AGAP2 and FAK (Fig. 1), prompting us to examine whether AGAP2 regulates the FAK activity. To this end, we overexpressed FLAG-AGAP2 in HEK293 cells (Fig. 2A, bottom panel), and we compared the phosphorylation signal on Tyr-397 residue of FAK, an indicator for its activation. In cells with and without AGAP2 overexpression, the stimulation with EGF increased the phosphorylation of Tyr-397 of FAK (Fig. 2A, upper panel). However, overexpression of AGAP2 augmented the FAK activation, whereas total cellular levels of FAK were not affected by the AGAP2 overexpression or stimulation with EGF (Fig. 2A, middle panel). This result suggests that AGAP2 regulates the function of FAK.
FIGURE 2.
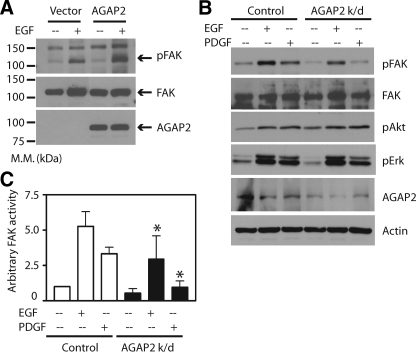
AGAP2 regulates FAK phosphorylation. A, overexpression of AGAP2 increases FAK phosphorylation. The cDNAs encoding empty vector or FLAG-AGAP2 were transfected into HEK293 cells for 48 h. Cleared cell lysates were resolved by SDS-PAGE and followed by Western blotting to detect phosphorylated FAK (pFAK), total FAK, and AGAP2. M.M., molecular mass. B, knockdown of AGAP2 attenuates FAK phosphorylation. Expression of AGAP2 in U87 cells was suppressed with siRNA, and nontargeting siRNA was used as a control. Knockdown of AGAP2 was confirmed by Western blotting using anti-AGAP2 antibody. Cells were treated with or without EGF (50 nm) or PDGF (50 ng/ml) for 5 min. Cleared cell lysates were resolved on SDS-PAGE and analyzed by immunoblotting. C, quantitation of FAK phosphorylation. Relative level of pFAK signals was measured by densitometry using Scion Image software. FAK phosphorylation in control cells with no treatment was defined as 1 arbitrary unit. *, p < 0.05 compared with control.
To gain confidence in the role of AGAP2 in FAK signaling, we tested if the AGAP2 expression was required for FAK activation. We used the glioblastoma cell line U87 that was previously reported to express a relatively high level of endogenous AGAP2 (16). Expression of endogenous AGAP2 was suppressed by siRNA, and nontargeting siRNA was transfected and used as a control. Knockdown of AGAP2 was examined by Western blotting using the anti-AGAP2 antibody that we described before (16), and the level of actin was measured as a protein loading control. As shown in Fig. 2B, siRNA successfully reduced expression of AGAP2 in the U87 cells on average by 75%, compared with control siRNA-expressing cells.
Cells with and without AGAP2 knockdown were starved overnight and treated with EGF or PDGF. In control cells, EGF and PDGF induced activation of FAK as detected by phosphorylation of Tyr-397 (Fig. 2B). Reduction of endogenous AGAP2 levels resulted in a substantial decrease in FAK activation by EGF or PDGF stimulation (Fig. 2B). The relative level of FAK activity under different conditions was measured by densitometry, and the basal level of control cells was taken as 1 arbitrary unit (Fig. 2C). Knockdown of AGAP2 decreased the agonist-induced FAK activity by 50% (Fig. 2C). The filters used for Tyr-397 phosphorylation were stripped and re-probed with an anti-FAK antibody. As shown in Fig. 2B, cellular levels of FAK remained unchanged with or without AGAP2 knockdown, suggesting that AGAP2 affects the FAK activation. The results from both overexpression and knockdown of AGAP2 indicate that AGAP2 regulates the FAK activity and not expression levels.
AGAP2 was reported to bind Akt and to increase the activity of Akt (18), and we examined if AGAP2 expression affects the activation of Akt. In U87 cells, phosphorylation of Akt was induced by treatment with EGF and PDGF (Fig. 2B). However, unlike the FAK activation, we did not observe measurable differences in the Akt activation status in cells with and without AGAP2 knockdown (Fig. 2B). To further test selectivity of AGAP2 effect on FAK, we examined the activation status of Erk, another protein kinase that is activated by both EGF and PDGF. Again, the phosphorylation of Erk induced by EGF or PDGF did not show consistent difference in cells with and without AGAP2 knockdown (Fig. 2B). These results suggest the selective effect of AGAP2 on FAK activation.
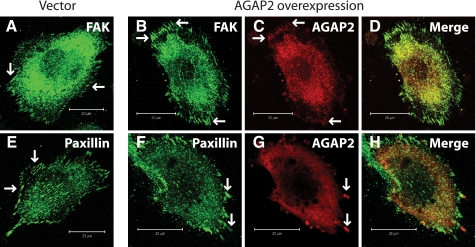
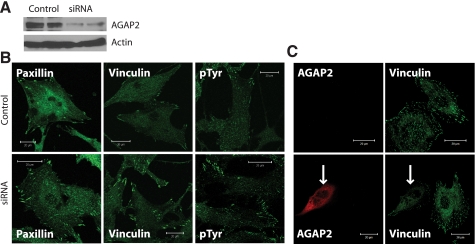
AGAP2 Is Localized in Focal Adhesions—A principal function of FAK is to regulate the formation and disassembly of focal adhesions. We examined the possible effect of AGAP2 on focal adhesions in U87 cells. First, we tested if AGAP2 and FAK co-localized in the focal adhesions. We used the U87 cells that stably express FLAG-AGAP2 at a moderate level because transient overexpression of AGAP2 at high levels disrupts the focal adhesions (see below). In control cells transfected with cDNAs encoding the empty vectors, endogenous FAK was present in linear structures at the periphery of the cells, presumably focal adhesions (Fig. 3A). Overexpressed AGAP2 was also observed in the focal adhesions and co-localized with endogenous FAK (Fig. 3, B–D). To provide further support that AGAP2 is localized in focal adhesions, we examined its co-localization with a second marker for focal adhesions, the paxillin. The staining of paxillin appeared as linear structures (Fig. 3E), similar to those stained with anti-FAK antibodies (Fig. 3A); and overexpressed AGAP2 co-localized with paxillin in the focal adhesions (Fig. 3, F–H). These results evidence localization of AGAP2 in focal adhesions.
FIGURE 3.
AGAP2 co-localizes with FAK and paxillin. U87 cells stably expressing empty vector or FLAG-AGAP2 were plated on fibronectin-coated glass coverslips for 5 h, fixed in 4% formaldehyde, processed for immunofluorescence staining, and examined by confocal microscopy. AGAP2 was stained using anti-FLAG antibody. Endogenous FAK and paxillin proteins were stained using anti-FAK and anti-paxillin antibodies. Staining of endogenous FAK in control cells (A) and in AGAP2-overexpressing cells (B–D) is shown in the top panels. Staining of endogenous paxillin in control cells (E) and AGAP2-overexpressing cells (F–H) is shown in the bottom panels. Arrows indicate the presence of FAK, paxillin, and FLAG-AGAP2 in focal adhesions. Scale bar, 20 μm.
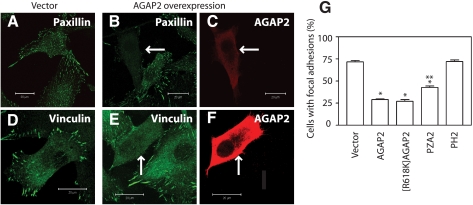
Overexpressed AGAP2 Disrupts Focal Adhesions—Binding with FAK and localization in focal adhesions raised the possibility that AGAP2 may affect the dynamics of focal adhesions, and we tested this idea by transiently overexpressing AGAP2. In control U87 cells transfected with cDNAs encoding empty vector, focal adhesions were present in 75% of the cells (Fig. 4G) as examined by staining with anti-paxillin antibody (Fig. 4A) or anti-vinculin antibody (Fig. 4D). Overexpression of AGAP2 disrupted the focal adhesions that were evident either by staining with anti-paxillin (Fig. 4, B and C) or anti-vinculin antibodies (Fig. 4, E and F). Following AGAP2 overexpression, focal adhesions were observed in only 25% of the cells (Fig. 4G).
FIGURE 4.
AGAP2 disrupts focal adhesions. U87 cells were transfected with cDNAs encoding empty vector or FLAG-AGAP2 for 48 h and replated on fibronectin-coated glass coverslips for 5 h. Cells were stained with anti-paxillin (A and B) or anti-vinculin (D and E) antibodies to visualize focal adhesions. Cells overexpressing AGAP2 were identified by staining with anti-FLAG antibody (C and F). Focal adhesion staining in control cells is shown in A (for paxillin) and D (for vinculin). Focal adhesion staining in AGAP2-expressing cells is shown in B (for paxillin) and E (for vinculin). Arrows indicate cells overexpressing FLAG-AGAP2. Scale bar, 20 μm. G, quantitation of focal adhesion (as visualized with anti-vinculin antibody) bearing cells following transfection with cDNAs encoding empty vector, FLAG-AGAP2, FLAG-[R618K]AGAP2, FLAG-PZA2, or FLAG-PH2. Fifty to 100 cells were counted for each sample for the presence of focal adhesions. *, p < 0.05 compared with vector control; **, p < 0.05 compared with wild-type AGAP2-expressing cells.
We next examined if the GAP activity of AGAP2 was required for the dissolution of focal adhesions. The GAP-deficient [R618K]AGAP2 was expressed in U87 cells, and its effect on focal adhesions was examined. Unexpectedly, [R618K]AGAP2 was as effective as the wild-type AGAP2 in inducing the focal adhesion dissolution (Fig. 4G), suggesting the effect on focal adhesions is independent of the AGAP2 GTPase activity. To further investigate the structural requirement of AGAP2 for this effect, we examined the possible role of the GLD2 domain. To this end, we examined the effect of PZA2 that lacked the GLD2 domain (Fig. 1B). Overexpressed PZA2 promoted dissolution of the focal adhesions in a manner similar to the wild-type AGAP2 (Fig. 4G). However, PZA2 was not as effective as wild-type AGAP2 (Fig. 4G), suggesting the GLD2 domain contributes to this effect of AGAP2. Because the PH2 domain is the major binding site for FAK, we next examined if the isolated PH2 domain affected focal adhesions. Expression of FLAG-PH2 in U87 cells did not affect the formation of focal adhesions (Fig. 4G), suggesting that other domains of AGAP2 are also required to regulate focal adhesions.
Knockdown of AGAP2 Stabilizes Focal Adhesions—The decrease in focal adhesions in U87 cells following AGAP2 overexpression could result from a reduction in the formation or an increase in the disassembly of focal adhesions. To differentiate these two possibilities, we suppressed the expression of endogenous AGAP2 in U87 cells by siRNA, and we used the nontargeting siRNA as a control. The knockdown of AGAP2 was confirmed by Western blot analysis (Fig. 5A). Following AGAP2 knockdown, U87 cells exhibited an increase in the number of focal adhesions that appear longer in size than those in the control cells (Fig. 5B). This effect of AGAP2 was observed when focal adhesions were visualized by staining with anti-paxillin, anti-vinculin, or anti-phosphotyrosine antibodies (Fig. 5B). Thus, the effect of AGAP2 is through the regulation of the structure of focal adhesions rather than through regulation of a particular component of the focal adhesions.
FIGURE 5.
Knockdown of AGAP2 stabilizes focal adhesions. A, expression of endogenous AGAP2 in U87 cells was suppressed using siRNA, and the nontargeting siRNA was used as a control. Knockdown of AGAP2 was confirmed by Western blotting, and actin was examined as the protein loading control. B, visualization of focal adhesions in U87 cells with and without AGAP2 knockdown. Focal adhesions were stained using anti-paxillin, anti-vinculin, or anti-phosphotyrosine (pTyr) antibodies in control (top panels) and AGAP2 knockdown (bottom panel) cells. Scale bar, 20 μm. C, rescue of AGAP2 knockdown. Expression of endogenous AGAP2 in U87 cells was suppressed by shRNA targeting the 3′-untranslated region. Plasmids encoding empty vector (top panels) or FLAG-AGAP2 (bottom panels) were transfected into U87 cells with stable knockdown of AGAP2. Focal adhesions were examined by staining with anti-vinculin antibody. Arrow indicates the cell expressing FLAG-AGAP2.
To gain confidence in the observed effect of AGAP2 knockdown on the focal adhesions, U87 cells with stable AGAP2 knockdown (by the shRNA targeting the 3′-untranslated region of AGAP2) were generated and transfected with a cDNA encoding AGAP2 (i.e. insensitive to the shRNA effect). Cells with stable AGAP2 knockdown showed an increase in the number and average size of focal adhesions (Fig. 5C, top right panel), similar to those observed in transient knockdown of AGAP2 by siRNA (Fig. 5B). Rescued expression of AGAP2 significantly reduced the number and average size of focal adhesions (Fig. 5C, bottom panels), suggesting that the remodeling of focal adhesions is directly linked to the expression level of AGAP2. These results, together with the effect of overexpressed AGAP2 to reduce focal adhesions, suggest that AGAP2 increases the focal adhesion disassembly.
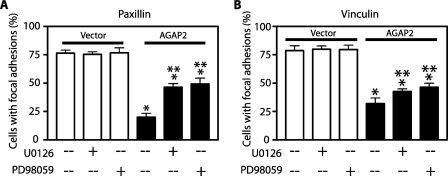
Activation of FAK is known to induce focal adhesion disassembly (4), and our results with overexpression and knockdown of AGAP2 on focal adhesions highly suggest that FAK mediates the AGAP2 effect on the focal adhesions. To test this possibility, we interfered with selective downstream effectors of FAK and tested whether inhibition of Erk activity, for example, could affect the AGAP2-induced dissolution of focal adhesions. Erk activation was blocked by the treatment with the MEK inhibitors U0126 or PD98059. Treatment of control cells with either inhibitor had no effect on the focal adhesion formation as visualized by staining with anti-paxillin (Fig. 6A) or anti-vinculin (Fig. 6B) antibodies. However, in the presence of either inhibitor, overexpressed AGAP2 was less effective in promoting the focal adhesion disassembly as examined by staining with anti-paxillin (Fig. 6A) or anti-vinculin (Fig. 6B) antibodies. These results affirm that AGAP2 affects focal adhesion dynamics through activation of FAK.
FIGURE 6.
Effect of MEK inhibitors on AGAP2-induced reduction of focal adhesions. U87 cells were transfected with cDNAs encoding empty vector or FLAG-AGAP2, treated with U0126 (30 μm) or PD98059 (50 μm) for 1 h, trypsinized, and replated on fibronectin-coated glass coverslips in the presence or absence of U0126 or PD98059 for 5 h. Focal adhesions were visualized by staining with anti-paxillin (A) or anti-vinculin (B) antibodies, and cells overexpressing AGAP2 were identified by staining with anti-FLAG antibodies. Fifty to 100 cells were counted for each sample for the presence of focal adhesions. *, p < 0.05 compared with vector control; **, p < 0.05 compared with no MEK inhibitor.
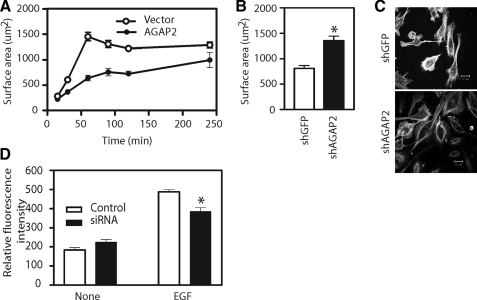
AGAP2 Regulates Cell Spreading and Migration—Focal adhesions are dynamic cellular structures involved in signal transduction that controls the cell migration. Regulation of focal adhesion disassembly by AGAP2 implied a role for AGAP2 in the directional cell migration. To provide support to this idea, we first examined a possible effect of AGAP2 on cell spreading. Overexpression of AGAP2 slowed U87 cell spreading up to 2 h following replating, and the surface areas of the cells overexpressing AGAP2 were smaller than those of the control cells expressing empty vectors (Fig. 7A). Distinctly, the knockdown of endogenous AGAP2 by shRNA accelerated spreading of the U87 cells. The average sizes of the U87 cells with AGAP2 knockdown were larger than those of the control cells (Fig. 7, B and C). Next, we compared the migration of U87 cells with and without AGAP2 knockdown. The knockdown of AGAP2 expression did not affect the basal level of U87 cell migration but slowed the migration toward EGF (Fig. 7D), suggesting that AGAP2 is involved in the regulation of directed migration of the U87 cells.
FIGURE 7.
Effect of AGAP2 on cell spreading and migration. A, forced overexpression of AGAP2 slows the cell spreading. U87 cells were transfected with cDNAs encoding empty vector or FLAG-AGAP2 for 48 h and replated on fibronectin-coated glass coverslips in Opti-MEM for the indicated times. Cells were fixed and processed for immunofluorescence analysis. Rhodamine-conjugated phalloidin was used to stain actin, and anti-FLAG antibody was used to identify AGAP2-overexpressing cells. Cells were examined using confocal microscopy, and the surface areas were measured using the ImagePro software. B and C, AGAP2 knockdown increases cell spreading. Endogenous AGAP2 was knocked down by shRNA, and the shRNA targeting green fluorescent protein was used as a control. Knockdown of AGAP2 was confirmed by Western blotting. Cells with and without AGAP2 knockdown were replated on fibronectin-coated glass coverslips for 5 h and fixed, and cell surface areas were measured as described above. Representative images in C show cells with and without AGAP2 knockdown. Scale bar, 20 μm. D, AGAP2 knockdown retards directed cell migration. U87 cells were transfected with AGAP2 siRNA and nontargeting siRNA was used as a control. Knockdown of AGAP2 was confirmed by Western blotting. Cell migration toward Opti-MEM supplemented, or not, with EGF (50 nm) was measured using the QCM chemotaxis cell migration system. Fluorescence intensities were measured with a plate reader using 485/520 filter set. *, p < 0.05 compared with control.
DISCUSSION
Focal adhesions physically anchor the cells to the extracellular matrix and transduce binary signals between the cells and their extracellular milieu. Focal adhesion remodeling is critical for cell migration that controls both physiologic and pathologic processes, including development, wound healing, and cancer cell invasion and metastasis (19, 20). Remodeling of the focal adhesions is highly controlled, and known regulators include Rho GTPase and Arf GAPs. Here we show the regulation of focal adhesion remodeling and cell migration by the Arf GAP AGAP2. We find that AGAP2 binds FAK and regulates its activity following activation of EGF or PDGF receptors. Overexpression of AGAP2 induced dissolution of the focal adhesions, and reduction of AGAP2 expression with RNAi stabilized the focal adhesions, similar to the effect of FAK knock-out on focal adhesions (11). Collectively, our results suggest that AGAP2 regulates FAK activity to increase focal adhesion remodeling and cell migration.
In addition to AGAP2, several members of the AZAP subfamily, including ASAP1 (21), ASAP3 (22), and ARAP2 (23), have been implicated in the regulation of focal adhesions, suggesting a redundant role for Arf GAPs in the regulation of focal adhesion dynamics. Alternatively, the different Arf GAPs may provide distinct regulatory effect on focal adhesion remodeling. Our data, together with published results (21–25), support the idea that Arf GAPs are specific regulators of the focal adhesions. First, the knockdown of AGAP2 with RNAi in U87 cells prevents the focal adhesion disassembly, which is not compensated by the other Arf GAPs that are expressed in these cells (7). Second, the effect of AGAP2 knockdown on focal adhesions in glioblastoma cell lines is polar opposite to the effect of ARAP2 knockdown (23), and although the knockdown of AGAP2 expression stabilizes focal adhesions, the knockdown of ARAP2 expression reduces the formation of the focal adhesions (23). Third, different Arf GAPs use distinct mechanisms to regulate the focal adhesions. For instance, we showed that AGAP2 affects focal adhesion remodeling through interaction with FAK, whereas the ARAP2 and ASAP1 seem to regulate the trafficking of focal adhesion components (23–25). Hence, these observations reinforce the idea that different Arf GAPs function coordinately to regulate specific facets of the focal adhesion dynamics during the complex process of cell migration.
We showed in this study that AGAP2 regulates the FAK activity and the remodeling of focal adhesions. The mechanism(s) underlying this effect of AGAP2 await further investigation. One possibility is that AGAP2 functions as a scaffold to bring FAK and its activators into close proximity. The second possibility is that AGAP2 traffics FAK and/or its activators to their site of action. The third possibility is that the binding of AGAP2 may induce a conformational change in FAK resulting in the FAK activation. To our knowledge, this is the first report showing that an Arf GAP directly regulates the function of a Src family protein kinase.
The PH domains of AZAP family Arf GAPs bind phosphoinositides that result in their partition to the membrane and activation (26). The PH2 domain of AGAP2 is distinct from those of other AZAP family Arf GAPs. First, AGAP2 harbors an insert of about 100 amino acids in its PH2 domain (16). Second, AGAP2 shows low phosphoinositide specificity in terms of its GAP activity; phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate activate AGAP2 equally well (16), which is in contrast, for example, to the specific activation of ASAP1 (27) and ARAP1 (28) by phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, respectively. Third, the PH domains of the AGAP subfamily bind clathrin adaptor proteins AP-1 and AP-3, whereas a function in protein binding has not been observed for the PH domains of the other AZAP family Arf GAPs. In this study, we report the binding of FAK to the AGAP2 PH domain, further supporting the notion that the PH2 domain is a protein-binding module.
We have proposed that AZAP family Arf GAPs function as coat proteins, and their binding partners serve as cargoes for the transport intermediates (15). Binding of FAK to AGAP2 raises the possibility that AGAP2 regulates the intracellular trafficking of FAK. Consistent with this idea, we observed co-localization of AGAP2 with the focal adhesion markers paxillin and vinculin in punctate structures in cells with dissolution of focal adhesions (data not shown). We speculate that AGAP2 regulates focal adhesion dynamics by at least two different mechanisms as follows: the activation of FAK to disassemble focal adhesions, and the trafficking of focal adhesion components or regulators to (or from) the focal adhesions while they are being remodeled.
Regulation of focal adhesions and cell migration by AGAP2 suggests that AGAP2 may play a role in cancer cell invasion and metastasis. Several Arf GAPs, including AGAP2, have been implicated in human cancers (29–31). AGAP2 is encoded by a gene located at chromosome band 12q13.3, close to the amplicon containing cyclin-dependent kinase 4 (32). This region is frequently amplified, and elevated expression of AGAP2 protein was observed in human cancers, including malignant glioma (32, 33). Expression of AGAP2 also promotes the invasion of glioblastoma cells (18). We observed that AGAP2 interacts with FAK, whose expression and activity are also elevated in malignant human glioma. Therefore, interaction of AGAP2 and FAK may contribute to the increased migratory and invasive behavior that is a prominent feature of glioma cells.
In summary, we provide evidence that the Arf GAP AGAP2 forms a complex with FAK and regulates its activity and the focal adhesion disassembly. Expression level of AGAP2 correlated with the rate of spreading and directional migration of U87 glioma cells. We propose that disrupting the interaction between AGAP2 and FAK may help control the increased migration and invasion of glioblastoma cells that overexpress both proteins.
Acknowledgments
We thank Dr. P. Randazzo for insightful discussions and continued support.
This work was supported, in whole or in part, by National Institutes of Health Grants CA129155 and CA131988 (to Y. D.). This work was also supported by start-up funds from the Department of Pathology at the Medical College of Georgia (to Z. N.).
Footnotes
The abbreviations used are: FAK, focal adhesion kinase; EGF, epidermal growth factor; Erk, extracellular signal-regulated kinase; GAP, GTPase-activating protein; GLD, G protein-like domain; ASAP, Arf GAP with Src homology 3, ankyrin repeat, and PH domain; ARAP, Arf GAP with Rho GAP, ankyrin repeat, and PH domain; AGAP, Arf GAP with G protein-like domain (GLD), ankyrin repeat, and PH domain; GST, glutathione S-transferase; PDGF, platelet-derived growth factor; PH, pleckstrin homology; PZA, pleckstrin homology, Arf GAP (containing a zinc finger motif) and ankyrin repeats; PBS, phosphate-buffered saline; shRNA, short hairpin RNA; siRNA, small interfering RNA; RNAi, RNA interference.
References
- 1.Sastry, S. K., and Burridge, K. (2000) Exp. Cell Res. 261 25–36 [DOI] [PubMed] [Google Scholar]
- 2.Carragher, N. O., and Frame, M. C. (2004) Trends Cell Biol. 14 241–249 [DOI] [PubMed] [Google Scholar]
- 3.Lo, S. H. (2006) Dev. Biol. 294 280–291 [DOI] [PubMed] [Google Scholar]
- 4.Broussard, J. A., Webb, D. J., and Kaverina, I. (2008) Curr. Opin. Cell Biol. 20 85–90 [DOI] [PubMed] [Google Scholar]
- 5.Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T., and Horwitz, A. R. (2003) Science 302 1704–1709 [DOI] [PubMed] [Google Scholar]
- 6.Cohen, L. A., and Guan, J. L. (2005) Curr. Cancer Drug Targets 5 629–643 [DOI] [PubMed] [Google Scholar]
- 7.Randazzo, P. A., Inoue, H., and Bharti, S. (2007) Biol. Cell 99 583–600 [DOI] [PubMed] [Google Scholar]
- 8.McLean, G. W., Carragher, N. O., Avizienyte, E., Evans, J., Brunton, V. G., and Frame, M. C. (2005) Nat. Rev. Cancer 5 505–515 [DOI] [PubMed] [Google Scholar]
- 9.Mitra, S. K., Hanson, D. A., and Schlaepfer, D. D. (2005) Nat. Rev. Mol. Cell Biol. 6 56–68 [DOI] [PubMed] [Google Scholar]
- 10.Mitra, S. K., and Schlaepfer, D. D. (2006) Curr. Opin. Cell Biol. 18 516–523 [DOI] [PubMed] [Google Scholar]
- 11.Ilic, D., Furuta, T., Kanazawa, S., Takeda, N., Sobue, K., Nakatsuji, N., Nomura, S., Fujimoto, J., Okada, M., Yamamoto, T., and Aizawa, S. (1995) Nature 377 539–544 [DOI] [PubMed] [Google Scholar]
- 12.Donaldson, J. G., and Honda, A. (2005) Biochem. Soc. Trans. 33 639–642 [DOI] [PubMed] [Google Scholar]
- 13.D'Souza-Schorey, C., and Chavrier, P. (2006) Nat. Rev. Mol. Cell Biol. 7 347–358 [DOI] [PubMed] [Google Scholar]
- 14.Randazzo, P. A., and Hirsch, D. S. (2004) Cell. Signal. 16 401–413 [DOI] [PubMed] [Google Scholar]
- 15.Nie, Z. Z., and Randazzo, P. A. (2006) J. Cell Sci. 119 1203–1211 [DOI] [PubMed] [Google Scholar]
- 16.Nie, Z. Z., Fei, J. J., Premont, R. T., and Randazzo, P. A. (2005) J. Cell Sci. 118 3555–3566 [DOI] [PubMed] [Google Scholar]
- 17.Liu, X., Hu, Y., Hao, C., Rempel, S. A., and Ye, K. (2007) Oncogene 26 4918–4927 [DOI] [PubMed] [Google Scholar]
- 18.Ahn, J. Y., Rong, R., Kroll, T. G., Van Meir, E. G., Snyder, S. H., and Ye, K. Q. (2004) J. Biol. Chem. 279 16441–16451 [DOI] [PubMed] [Google Scholar]
- 19.Wozniak, M. A., Modzelewska, K., Kwong, L., and Keely, P. J. (2004) Biochim. Biophys. Acta 1692 103–119 [DOI] [PubMed] [Google Scholar]
- 20.Schlaepfer, D. D., Mitra, S. K., and Ilic, D. (2004) Biochim. Biophys. Acta 1692 77–102 [DOI] [PubMed] [Google Scholar]
- 21.Randazzo, P. A., Andrade, J., Miura, K., Brown, M. T., Long, Y. Q., Stauffer, S., Roller, P., and Cooper, J. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha, V. L., Bharti, S., Inoue, H., Vass, W. C., Campa, F., Nie, Z. Z., de Gramont, A., Ward, Y., and Randazzo, P. A. (2008) J. Biol. Chem. 283 14915–14926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon, H. Y., Miura, K., Cuthbert, E. J., Davis, K. K., Ahvazi, B., Casanova, J. E., and Randazzo, P. A. (2006) J. Cell Sci. 119 4650–4666 [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y. H., Loijens, J. C., Martin, K. H., Karginov, A. V., and Parsons, J. T. (2002) Mol. Biol. Cell 13 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Y. H., Yerushalmi, G. M., Grigera, P. R., and Parsons, J. T. (2005) J. Biol. Chem. 280 8884–8892 [DOI] [PubMed] [Google Scholar]
- 26.Che, M. M., Boja, E. S., Yoon, H. Y., Gruschus, J., Jaffe, H., Stauffer, S., Schuck, P., Fales, H. M., and Randazzo, P. A. (2005) Cell. Signal. 17 1276–1288 [DOI] [PubMed] [Google Scholar]
- 27.Kam, J. L., Miura, K., Jackson, T. R., Gruschus, J., Roller, P., Stauffer, S., Clark, J., Aneja, R., and Randazzo, P. A. (2000) J. Biol. Chem. 275 9653–9663 [DOI] [PubMed] [Google Scholar]
- 28.Miura, K., Jacques, K. M., Stauffer, S., Kubosaki, A., Zhu, K. J., Hirsch, D. S., Resau, J., Zheng, Y., and Randazzo, P. A. (2002) Mol. Cell 9 109–119 [DOI] [PubMed] [Google Scholar]
- 29.Ehlers, J. P., Worley, L., Onken, M. D., and Harbour, J. W. (2005) Clin. Cancer Res. 11 3609–3613 [DOI] [PubMed] [Google Scholar]
- 30.Lin, D., Watahiki, A., Bayani, J., Zhang, F., Liu, L., Ling, V., Sadar, M. D., English, J., Fazli, L., So, A., Gout, P. W., Gleave, M., Squire, J. A., and Wang, Y. Z. (2008) Cancer Res. 68 4352–4359 [DOI] [PubMed] [Google Scholar]
- 31.Okabe, H., Furukawa, Y., Kato, T., Hasegawa, S., Yamaoka, Y., and Nakamura, Y. (2004) Int. J. Oncol. 24 43–48 [PubMed] [Google Scholar]
- 32.Elkahloun, A. G., Krizman, D. B., Wang, Z. L., Hofmann, T. A., Roe, B., and Meltzer, P. S. (1997) Genomics 42 295–301 [DOI] [PubMed] [Google Scholar]
- 33.Ahn, J. Y., Hu, Y. X., Kroll, T. G., Allard, P., and Ye, K. Q. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6993–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]