Abstract
During transcription elongation the nascent RNA remains base-paired to the template strand of the DNA before it is displaced and the two strands of the DNA reanneal, resulting in the formation of a transcription “bubble” of ∼10 bp. To examine how the length of the RNA-DNA hybrid is maintained, we assembled transcription elongation complexes on synthetic nucleic acid scaffolds that mimic the situation in which transcript displacement is compromised and the polymerase synthesizes an extended hybrid. We found that in such complexes bacterial RNA polymerase exhibit an intrinsic endonucleolytic cleavage activity that restores the hybrid to its normal length. Mutations in the region of the RNA polymerase near the site of RNA-DNA separation result in altered RNA displacement and translocation functions and as a consequence in different patterns of proofreading activities. Our data corroborate structural findings concerning the elements involved in the maintenance of the length of the RNA-DNA hybrid and suggest interplay between polymerase translocation, DNA strand separation, and intrinsic endonucleolytic activity.
One of the key features of DNA-dependent RNA polymerases (RNAPs)2 that distinguish them from other members of polynucleotide synthetases is the ability to displace the nascent product (RNA) from the DNA template and to maintain the size of the transcription bubble during elongation. These are the crucial tasks in transcription elongation as the newly synthesized RNA must be immediately available for further processing. Moreover overextended (>9-bp) or abnormally short (<7-bp) RNA-DNA hybrids result in complexes with a compromised stability and termination of transcription (1–5). Although RNA displacement is controlled by reannealing of the DNA template and non-template strands at the trailing edge of the transcription bubble, a number of reports suggest an active role of prokaryotic and eukaryotic RNAP in strand separation as well as in maintenance of the length of RNA-DNA hybrid (6–8).
Structural and biochemical studies of multisubunit RNAPs have implicated structurally conserved motifs located at the upstream end of the RNA-DNA duplex that serve as a steric barrier for the growing heteroduplex. In both yeast and bacterial RNAPs a “lid” element that contains conserved aromatic residues (yeast RNAP) or aliphatic residues (bacterial RNAP) stacks upon the last base pair of the RNA-DNA hybrid and is thought to serve as a wedge in displacing the RNA from the heteroduplex (6, 9). The role of this structural element was investigated before high resolution structures of the bacterial elongation complex (EC) became available (7, 8). It was found that lid deletion mutants did not exhibit defects in most transcription assays (including termination) and showed a clear phenotype only when transcribing single-stranded DNA templates. In this case, lid deletion mutants, in contrast to the wild type (WT) RNAP that was able to extend a transcript only by 5–7 nt, could complete runoff transcription by forming an extended RNA-DNA hybrid. In addition to the lid, several other elements, such as “rudder,” “fork loop” (yeast RNAP), and “switch 3” loop (bacterial RNAP), in multisubunit RNAPs have been implicated in RNA displacement. These other structural elements are not directly involved in RNA displacement but rather are thought to prevent reassociation of RNA after it has been displaced from the template DNA and to form an extensive network of interactions resulting in stabilization of the nucleic acid components of the EC (6, 9, 10).
Elongation complexes assembled on nucleic acid scaffolds mimic promoter-originated ECs and have proven useful in numerous studies of transcription elongation (1, 4, 11–14). The scaffold design in most ECs includes two conjugated nucleic acid duplexes: the 8–9-bp RNA-DNA hybrid and a 15–20-bp downstream DNA duplex. The lack of a complementary non-template (NT) DNA strand in the RNA-DNA region results in a compromised displacement of the nascent RNA from the RNA-DNA hybrid in both prokaryotic and eukaryotic RNAPs as well as in a single subunit T7 RNAP (1, 7, 8). This phenomenon is a universal feature in single subunit and multisubunit RNAPs as these enzymes form an overextended RNA-DNA hybrid and stall after incorporation of 5–7 nt into the RNA during primer extension. Although addition of a fully complementary NT DNA strand to the scaffold EC restores its ability to displace RNA, the efficiency of formation of such assemblies is low (4). In this work, using ECs assembled on single-stranded DNA scaffolds with WT and mutant RNAPs, we demonstrate that bacterial RNAP can maintain the length of the RNA-DNA hybrid as a result of an intrinsic endonuclease activity. Our data corroborate structural findings concerning the elements involved in the maintenance of RNA-DNA hybrid and suggest interplay between RNAP translocation, DNA strand separation, and endonucleolytic activity.
EXPERIMENTAL PROCEDURES
Purification of RNAP and Polymerase Activity Assay—WT Thermus thermophilus core RNAP was purified from cell biomass obtained from the HB8 strain as described previously (15). His-tagged WT Escherichia coli core polymerase (pIA299) was purified from cell biomass obtained as described previously (16). An additional purification step using a Superdex 200 size exclusion column (600 mm) was used. The purity of these RNAPs was greater then 99.8% as judged by SDS-PAGE. Polymerase activity was measured by the ability to extend a 32P-labeled RNA primer by one nucleotide in a reaction in which the concentrations of nucleic acid scaffold (R9/TS35/NT35) and RNAP were equimolar (see transcription conditions below). Only RNAP preparations that extended the RNA primer with close to 100% efficiency within 5 min of incubation at 37 °C (E. coli core) or at 60 °C (T. thermophilus core) were used.
Construction and Purification of Mutant RNAPs—Lid loop deletion β′Δ252–263 and switch 3 loop deletion βΔ1250–1259 His-tagged mutant E. coli RNAPs were obtained by site-directed mutagenesis (QuikChange, Stratagene) starting with plasmid pIA299 (16). DNA primers (Integrated DNA Technologies, Inc.) used for mutagenesis were (5′ to 3′) CCAGATCTGCGTCCGGGTGACCTGAACGATCTG (β′Δ252–263) and CGCGCGTTCCACCGGTGGTGGTAAGGCACAG (βΔ1250–1259). After mutagenesis, genes encoding all RNAP subunits in pIA299 were sequenced to confirm absence of any unintended substitutions. Mutant RNAPs were purified by a combination of His-Trap nickel-agarose, heparin-Sepharose, Q FF (Q-Sepharose Fast Flow), and Superdex 200 chromatography.
RNA and DNA Oligonucleotides—The following synthetic oligonucleotides were used (all sequences are 5′ to 3′): RNA oligomers (Dharmacon), GCGGCGAU (R8), GCGGCGAUA (R9), GCGGCGAUAUC (R11), GAGUCUGCGGCGAUA (R15), and GAGUCUGCGGCGAU (R14); DNA oligomers (Integrated DNA Technologies, Inc.), CCTGTCTGAATCGATATCGCCGC (TS35), CCTGTCTGAATCGATATCGCC (TS70), CCTGTCTGAATCTATATCGCCGC (TS35A), CCTGTCTGAATCTATATCGCC (TS71), CGCCGCTATTGCTAAGTCTGTCCC (TS39), TCGATTCAGACAGG (NT35), GATTCAGACAGG (NT358), and ACGATTCAGACAGGG (NT39).
Assembly of ECs and Transcription Conditions—Nucleic acid scaffolds were assembled by annealing equimolar concentrations of complementary RNA and DNA oligomers as described previously (17). RNA oligomers were labeled at their 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) prior to assembly. To assemble elongation complexes, core RNAP (0.2–1 μm) was incubated with an equimolar concentration of scaffold for 5 min at room temperature in 10 μl of transcription buffer (40 mm Tris, pH 7.9 at 25 °C, 100 mm NaCl, 5 mm MgCl2, and 5 mm 2-mercaptoethanol). Primer extension was achieved by incubation of these complexes with substrate NTPs (100 μm) for 1–60 min at 60 °C (T. thermophilus RNAP) or 37 °C (E. coli RNAP). Exo/endonucleolytic activity was probed by incubation of complexes for 0–60 min at 60 °C (T. thermophilus RNAP) or 37 °C (E. coli RNAP). Reactions were stopped by the addition of an equal volume of 95% formamide, 0.05 m EDTA, and the products were resolved by 20% PAGE in the presence of 6 m urea and visualized by a PhosphorImager (GE Healthcare).
To probe the nature of RNA hydrolysis in complexes having overextended RNA-DNA hybrids, we first incorporated [32P]AMP at the 3′ end of the RNA primer in EC assembled using R14/TS35/NT35 scaffold. The 3′-labeled RNA (15 nt, or R15) was then purified by extraction from 20% denaturing PAGE, annealed with the DNA template (TS53) to produce 11-bp RNA-DNA duplex, and incubated with RNAP in exo/endonucleolytic assays as described above.
RESULTS AND DISCUSSION
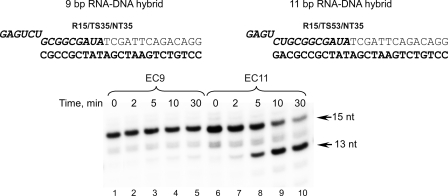
In our earlier experiments with T. thermophilus RNAP we noticed that elongation complexes assembled on scaffolds having a long (>10-bp) RNA-DNA hybrid exhibited a substantial intrinsic cleavage activity (12). Most notably, extension of the RNA primer in an EC containing a 9-bp RNA-DNA (EC9) by 2 nt resulted in a complex that was hypersensitive to intrinsic endonuclease activity presumably due to the formation of an 11-bp hybrid. The extended RNA primer in this complex was immediately hydrolyzed to yield a 9-bp EC (12). ECs having 11-bp hybrids can be assembled on an RNA-DNA hybrid directly (Fig. 1). As in similar experiments described previously (12), we observed hydrolysis of the RNA primer resulting in formation of an endonuclease-resistant EC9. Although the rate of the RNA hydrolysis in the EC11 assembled in this manner was lower compared with complexes obtained by primer extension (12) (likely because of the extra time required for the binding of nucleic acids to the polymerase and formation of an EC), we observed almost complete primer conversion within 30 min of incubation time. In view of this, we speculated that RNAP endonucleolytic activity in ECs assembled on scaffolds having extended RNA-DNA hybrids may, in part, be responsible for the transcript-assisted proofreading activity recently described in Thermus aquaticus ECs having a mismatched 3′ end (18). There the scaffold ECs contained 11-bp RNA-DNA hybrids that may have stimulated RNAP backtracking and as a consequence endonuclease cleavage in complexes having both matched or mismatched 3′ RNA bases (18). To investigate the relation of two physiologically important RNAP functions in transcription elongation (maintenance of the length of the RNA-DNA duplex and proofreading activity) we used scaffolds with different hybrid lengths to assemble ECs with WT E. coli RNAP and with mutant RNAPs in which deletions of elements implicated in RNA displacement had been made.
FIGURE 1.
Intrinsic cleavage activity in T. thermophilus ECs having different lengths of the RNA-DNA hybrid. The complexes containing 5′-32P-labeled RNA primers were assembled as described under “Experimental Procedures” and incubated for 2–30 min at 60 °C. The sequence of the TS DNA strand is shown in bold; RNA is in bold italics.
In our transcript cleavage assays, we used E. coli RNAP preparations that were additionally purified by means of gel filtration on Superdex 200 to remove traces of exogenous exo/endonuclease activity not associated with RNAP or GreA/GreB factors (see “Experimental Procedures”). Incubation of a purified RNAP with 5′-32P-labeled RNA or RNA-DNA scaffold (R9/TS35/NT35) for an extended period of time (20 h) revealed no RNA degradation suggesting that the final RNAP preparations were essentially free of exogenous nuclease activity (data not shown). As in the experiments with T. thermophilus RNAP, we observed the same trend in the efficiency of intrinsic cleavage activity with E. coli RNAP ECs (Fig. 2A). Thus, ECs having 11-bp RNA-DNA hybrids (EC11) were readily converted to stable (resistant to further degradation) ECs with 9-bp hybrids within 40–60 min (lanes 1–7). At the same time, EC9 that was determined previously to be mostly in the post-translocated state (12) remained intact as no RNA hydrolysis was detected (lanes 15–21). Similar results were obtained when EC11 was produced by extension of the RNA in EC9 by 2 nt (data not shown). These data are consistent with our prior observations obtained with T. thermophilus RNAP ECs and confirm that bacterial RNAP can “sense” an overextended hybrid and correct it by backtracking and trimming the 3′ end of the RNA.
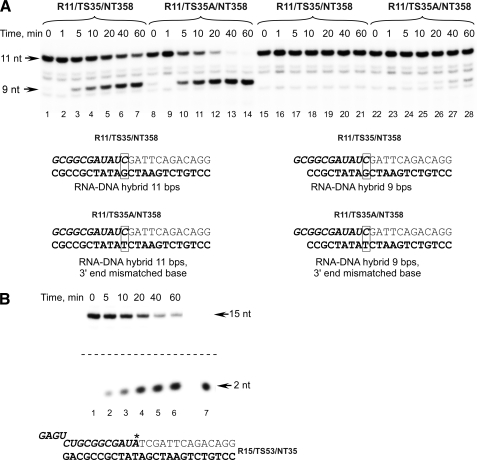
FIGURE 2.
Intrinsic cleavage activity in E. coli ECs having different lengths of the RNA-DNA hybrid. A, intrinsic cleavage activity of WT E. coli RNAP in ECs assembled on 11-bp (lanes 1–7), 3′ end-mismatched 11-bp (lanes 8–14), 9-bp (lanes 15–21), and 3′ mismatched 9-bp (lanes 22–28) RNA-DNA hybrid scaffolds. E. coli core RNAP was mixed with the scaffold template, and the mixture was incubated at 37 °C for the time indicated. Note that in the scaffolds used the RNA sequence is preserved to exclude its effect on rates of RNA hydrolysis. B, restoration of a normal RNA-DNA hybrid length is due to intrinsic endonucleolytic activity of RNAP. E. coli ECs containing 3′ end-32P-labeled RNA primer (indicated by an asterisk) were obtained as described under “Experimental Procedures” and incubated at 37 °C for the time indicated, and the products of the reaction were analyzed by 25% PAGE containing 6 m urea (lanes 1–6). RNA dinucleotide primer (ApU) that was 5′ end-labeled with T4 polynucleotide kinase was used as a size marker in this experiment (lane 7).
Several studies indicate that bacterial RNAPs in ECs having a mismatched base pair at the growing end of the RNA-DNA hybrid exhibit proofreading activity that results in the removal of the incorrect nucleotide (18–20). Because both the mismatch and RNA-DNA hybrid length correction reactions described above appeared to have the same nature (intrinsic cleavage activity), we decided to investigate how RNA-DNA hybrid length contributes to the RNAP proofreading activity. We first looked at how mismatched bases affect intrinsic cleavage activity in ECs having different RNA-DNA hybrid lengths (Fig. 2A, lanes 8–14 and 22–28). We found that whereas a mismatched bp placed at the 3′ end of a 9-bp EC stimulated cleavage activity only slightly, a 3′ mismatched EC11 was rapidly converted to EC9. Importantly as evident from Fig. 2A, an 11-bp RNA-DNA hybrid by itself contributes to the RNA hydrolysis. Although we found that sequence at the 3′ end of the RNA affects the efficiency of its hydrolysis in EC11 (data not shown), it is possible that the hybrid length correction reaction might have contributed to RNA hydrolysis in experiments involving mismatched ECs (18).
To verify that both reactions are due to the same type of intrinsic RNAP activity, we labeled the RNA by incorporation of [32P]AMP at its 3′ end (Fig. 2B). Incubation of 3′-labeled EC in transcription buffer resulted in accumulation of a labeled dinucleotide cleavage product (pUpA), indicating that the cleavage activity observed in this complex was due to the E. coli RNAP endonuclease activity and thus has the same mechanism (intrinsic endonucleolytic cleavage) as the hydrolysis observed in T. aquaticus ECs having a mismatched base at the 3′ end of the transcript (18).
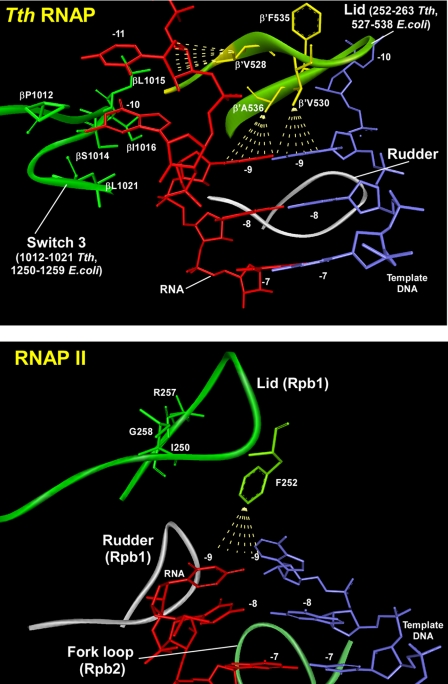
Analysis of the crystal structure of T. thermophilus RNAP EC suggests that several regions may be involved in maintenance of the length of the RNA-DNA hybrid (9, 12). The lid element (β′ residues 250–270) provides a physical barrier to the growing end of the RNA-DNA hybrid and may play a role similar to that of the lid loop in yeast RNAP II (6) (Fig. 3). Another loop in the RNAP β subunit (switch 3, residues 1240–1260) forms a hydrophobic cleft that accommodates the unpaired RNA base (–10) just after its separation from the RNA-DNA hybrid (9). To address the role of these structural elements in RNA-DNA strand separation and particularly in maintaining the length of the RNA-DNA hybrid during elongation, we made deletion mutants in E. coli core RNAP that involve these regions. The lid deletion mutation (β′Δ252–263; corresponds to the interval 527–538 in T. thermophilus RNAP) was similar to one described in earlier publications (7, 8). In another construct, the hydrophobic region (βΔ1250–1259; corresponds to the interval 1012–1021 in T. thermophilus RNAP) in the switch 3 element of E. coli RNAP β subunit was deleted (Fig. 3A).
FIGURE 3.
Close-up of the RNA displacement region in T. thermophilus RNAP EC, PDB code 2O5I (top), and in yeast RNAP II EC, PDB code 1R9R (bottom). Residues implicated in RNA displacement in the switch 3 element of T. thermophilus (Tth) RNAP and equivalent residues of the lid (separation loop) of T. thermophilus and yeast RNAP II are highlighted. Regions in the lid and switch 3 elements deleted in mutant E. coli RNAPs generated in this work are indicated.
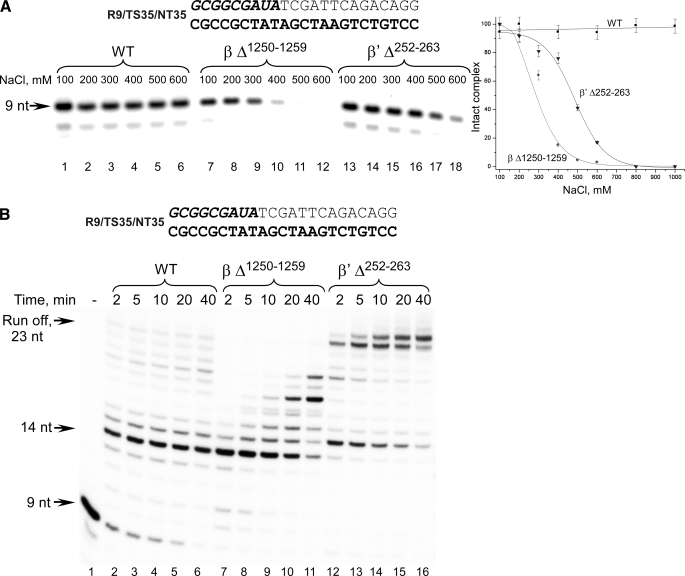
To characterize the mutants we first examined the stability of ECs in salt challenge experiments (Fig. 4A). After incubation of RNAPs with the scaffolds, the ECs were immobilized on nickel-agarose beads and incubated with different concentrations of salt. The complexes were washed to remove the dissociated components and analyzed by PAGE. As expected based on the results of the previous experiments, ECs formed with WT E. coli RNAP were stable in the presence of 1 m NaCl (4, 21). At the same time, both deletion mutants formed ECs that were sensitive to high salt concentrations. Thus, consistent with earlier data (8), 50% of ECs assembled using the lid deletion mutant dissociated at 500 mm NaCl, whereas ECs formed with βΔ1250–1259 RNAP withstood challenge only up to 300 mm NaCl. These findings are consistent with the T. thermophilus EC crystal structure that indicates that a number of ionic interactions are formed between the upstream part of the RNA-DNA hybrid and RNAP that may be disrupted by salt (9). Nevertheless ECs formed with all mutant RNAPs were stable in transcription buffer (i.e. in the presence of 100 mm NaCl) for over 80 min (supplemental Fig. S1), and therefore these conditions were used in all further assays involving these mutant RNAPs.
FIGURE 4.
Characterization of E. coli RNAPs having mutations in the RNA displacement region. A, mutant E. coli RNAPs form salt-sensitive ECs. ECs formed with the WT (lanes 1–6), βΔ1250–1259 (lanes 7–12), and β′Δ252–263 (lanes 13–18) mutant E. coli RNAPs were immobilized on nickel-agarose beads, incubated with different NaCl concentrations, washed with the transcription buffer, and analyzed by 20% PAGE containing 6 m urea. Dissociation of ECs is plotted as a function of salt concentration. Error bars represent S.D. calculated in three independent experiments. B, mutant E. coli RNAPs demonstrate different primer extension properties. ECs were formed using R9/TS35/NT35 scaffold. Runoff transcription assays were performed in the presence of 100 μm NTPs at 37 °C for the times indicated and analyzed as described above.
As mentioned above, WT E. coli RNAP can extend the RNA-DNA hybrid by incorporation of only 5–7 substrate NTPs when scaffolds lacking a fully complementary NT strand are used to assemble an EC (7, 8, 18, 22). We examined whether the mutant RNAPs exhibited a different ability to extend the RNA primer. As expected, WT RNAP stalled after incorporation of 5 nt into the transcript, producing very little runoff product (23 nt; Fig. 4B). In contrast, the β′Δ252–263 mutant, although pausing upon incorporation of 5 nt, was able to complete transcription and produced a full-length product that extended to the end of the template with high efficiency. This suggests that removal of the lid eliminates the physical barrier for the growing RNA-DNA hybrid that causes instability in EC in full agreement with the previous studies (7, 8). Transcription by the βΔ1250–1259 mutant paused after incorporation of 6 nt (Fig. 4). Although this mutant was able to recover from the apparent pause, runoff products were not detected even after 40 min of incubation, suggesting that this RNAP region is also involved in maintenance of the RNA-DNA hybrid length in the absence of the NT DNA strand.
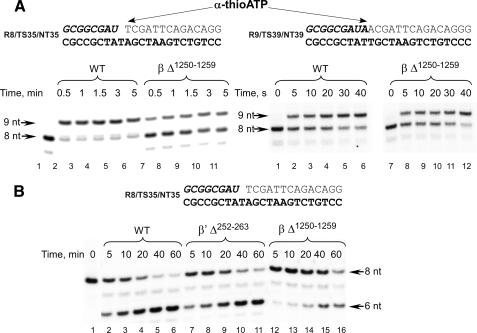
We next examined the ability of the mutant RNAPs to extend the RNA primer by incorporation of a single substrate NTP. Although WT and mutant RNAPs were catalytically competent and fully extended the RNA primer in a standard activity assay (see “Experimental Procedures”), we noticed that βΔ1250–1259 RNAP could complete the reaction only within 2–5 min, whereas less than 15 s were required for WT and lid deletion mutant RNAPs (data not shown). Although fast flow kinetic measurements would be needed to determine the precise rates of substrate incorporation, we took advantage of a slower rate of incorporation of α-thio-NTP derivatives to examine this phenomenon (Fig. 5A). Two scaffold complexes were used: EC8 and EC9, which as we previously reported exist in mostly the pretranslocated or the post-translocated conformation, respectively (9, 12). Here the difference in substrate incorporation rates was much more apparent: in the pretranslocated EC8, only 30% of initial RNA primer was extended within 5 min by βΔ1250–1259 RNAP, whereas WT RNAP completed extension within 0.5–1 min (Fig. 5A, left panel). Interestingly when primer extension was analyzed using post-translocated EC9, the difference in the rate of substrate incorporation between WT and βΔ1250–1259 RNAP was negligible (Fig. 5A, right panel). The observed difference likely reflects defects during the translocation stage of the nucleotide addition cycle because translocation is required for substrate incorporation in the pretranslocated EC8 but not post-translocated EC9. Although the allosteric effect of the switch 3 deletion on RNAP catalytic activity cannot be completely excluded, the chemistry step of NTP incorporation in βΔ1250–1259 RNAP seems to be unaffected (at least in the time scale of these experiments) because this mutant incorporated both ATP and α-thio-ATP as fast as the WT RNAP in the post-translocated EC9 (Fig. 5A).
FIGURE 5.
βΔ1250–1259 mutant RNAP is defective in forward and backward translocation. A, RNA primer extension was carried out by incubation of the pretranslocated EC8 (left panel, lanes 1–11) or the post-translocated EC9 (right panel, lanes 1–12) with α-thio-ATP (100 μm) at room temperature for the times indicated. B, GreB-induced hydrolysis of RNA in the pretranslocated ECs. E. coli GreB (1 μm) was added to the complexes assembled with the WT (lanes 1–6), β′Δ252–263 (lanes 7–11), and βΔ1250–1259 (lanes 12–16) RNAPs and incubated at 37 °C for the times indicated.
We further explored the translocation defects by monitoring activity that requires backward translocation of RNAP, i.e. GreB-stimulated endonuclease hydrolysis (Fig. 5B). This activity is more efficient in the pretranslocated EC8 than in the post-translocated E. coli EC9 (12). As in the primer extension experiments, hydrolysis of the RNA in the GreB-stimulated reaction by βΔ1250–1259 RNAP was notably slower when compared with WT RNAP or the lid deletion mutant (Fig. 5B). These data together with the primer extension experiment described above (Fig. 5A) suggest that the switch 3 element may play an important role in the translocation event because EC8 is in a pretranslocated state and translocation is required for both substrate incorporation (forward translocation) and endonucleolytic activity (backward translocation). This conclusion also suggests an interesting analogy with yeast RNAP EC where it has been proposed that the formation of a strand/loop network at the upstream end of the RNA-DNA hybrid affects the position of the 3′ end of the transcript and thus the conformation of the EC (6).
To monitor an intrinsic endonucleolytic activity of the mutant RNAPs, we used ECs assembled on an 11-bp RNA-DNA hybrid scaffold (Fig. 6). As in the previous experiments (Fig. 2A), we observed RNA hydrolysis by WT RNAP. At the same time, RNA hydrolysis in ECs assembled using mutant βΔ1250–1259 and β′Δ252–263 RNAPs was not observed (Fig. 6A). We speculate that the basis of such behavior is different for each of these mutants. As shown above (see Fig. 4B) and demonstrated in previously published experiments (7, 8), the lid deletion mutant that exhibits no translocation or catalytic defects is not destabilized by the presence of the overextended RNA-DNA hybrid and can produce runoff products when all substrates are present in the transcription reaction. Thus, this mutant RNAP is unable to sense the overextended RNA-DNA hybrid and therefore does not need to backtrack and correct its length by the endonucleolytic activity. In contrast, both forward and backward translocation in βΔ1250–1259 mutant RNAP is compromised, impeding its ability to hydrolyze the RNA primer in EC11.
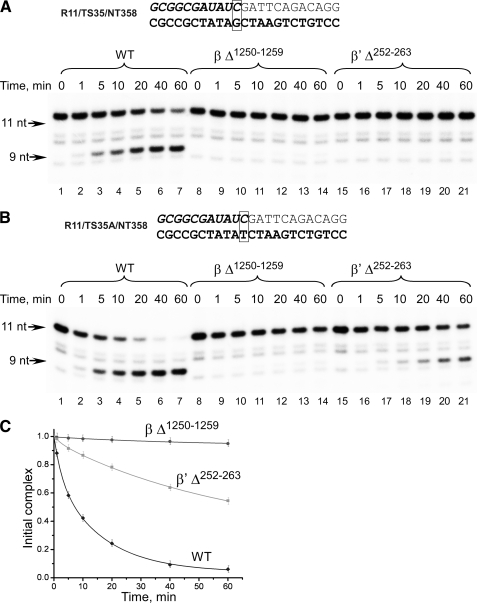
FIGURE 6.
Endonucleolytic activity of mutant RNAPs on scaffolds having matched or mismatched bases at the 3′ end of the RNA. A, mutant RNAPs are defective in RNA-DNA hybrid length restoration. The ECs were assembled using 11-bp RNA-DNA hybrid scaffolds using WT (lanes 1–7), βΔ1250–1259 (lanes 8–14), and β′Δ252–263 (lanes 14–21) E. coli RNAPs and incubated at 37 °C for the times indicated. B and C, endonuclease activity of mutant RNAPs on 3′ end-mismatched scaffold. The ECs, having mismatched bases at the 3′ end of the RNA, were assembled using 11-bp RNA-DNA hybrid scaffolds using WT (lanes 1–7), βΔ1250–1259 (lanes 8–14), and β′Δ252–263 (lanes 15–21) RNAPs and incubated at 37 °C for the times indicated. The fraction of RNA primer hydrolyzed is plotted as a function of time. Error bars represent S.D. calculated in three independent experiments.
What happens when a mismatched EC11 is assembled using mutant RNAPs? Considering that both RNA-DNA hybrid length and mismatch correction reactions contribute to the endonucleolytic cleavage, one would expect a decrease in overall rates of RNA hydrolysis in ECs formed by mutant RNAPs as it can now only be a result of a transcript-assisted proofreading activity (18). Indeed the endonuclease activity was observed in the mismatched EC11 assembled with β′Δ252–263 mutant although at a reduced rate (Fig. 6, B and C). Because the lid deletion prevents functioning of the hybrid length correction mechanism, the observed endonucleolytic activity likely comes from a transcript-assisted proofreading activity as described previously (18). On the other hand, no endonuclease activity was detected in the mismatched EC11 assembled with βΔ1250–1259 mutant RNAP. This result is in good agreement with our hypothesis that βΔ1250–1259 RNAP is defective in translocation and therefore cannot efficiently perform RNA hydrolysis whether it is caused by an extended RNA-DNA hybrid or by the presence of the mismatched base at the 3′ end of the transcript.
As demonstrated in recent studies, the presence of the NT DNA strand is sufficient to displace RNA from the RNA-DNA hybrid (7, 8, 18, 22). A mechanistic model analogous to a strung bow might best describe the situation when RNAP transcribes templates where the NT strand is lacking or damaged. In this scenario, RNAP can incorporate several nucleotides, producing a long RNA-DNA hybrid up to 15–17 bp in length. This will likely change the pathway of the 5′ end of the RNA toward the exit pore, forcing it to follow the DNA template strand and “string” the bow by displacing the elements (such as lid and switch 3) at the upstream end of the RNA-DNA hybrid. When the flexibility of RNAP to accommodate this situation is exceeded, the overextended RNA-DNA hybrid (“arrow”) is pushed forward due to the tendency of lid and switch 3 elements to resume their normal positions, causing RNAP backtracking and extrusion of the 3′ end of the RNA, thus relaxing the bow. Subsequently endonucleolytic activity of RNAP restores the length of the RNA-DNA hybrid. When the lid is removed, charging of the bow does not occur because there would be no barrier for the growing RNA-DNA hybrid, or as suggested earlier a new opening may now be formed (8). As a consequence, no backtracking and therefore no endonucleolytic activity is observed in complexes with the extended RNA-DNA hybrid. On the other hand, in the absence of switch 3, which provides a binding pocket for the 5′ end of the RNA transcript and thus holds the arrow (RNA-DNA hybrid), the string is loose, and the bow cannot be strung (at least all the way), affecting both forward and backward translocation of RNAP and as a consequence diminishing endonucleolytic activity.
Our experiments model the situation when the RNA-DNA hybrid exceeds its normal length by 2–3 bp and thus induces stress in a ternary complex, resulting in backtracking of RNAP along the DNA template. However, two alternative scenarios are possible when an overextended RNA-DNA hybrid escapes correction during the initial stages of its formation and reaches the length of 16–20 bp. In the first scenario, an overextended hybrid destabilizes the ternary complex causing RNAP dissociation from the DNA template, preventing its further transcription (1, 8). In the second scenario, RNAP “backslides” along a single-stranded DNA template without extrusion of the 3′ end of a single-stranded transcript and forms a transcription-incompetent priming complex resistant to GreB-stimulated cleavage (22). This mechanism allows M13 phage to prime its replication by using host RNAP and, as argued by authors, can also be used during RNAP-catalyzed priming of replication on double-stranded replicons such as ColE1 (22). Additional experiments will be required to better understand instances in which overextended RNA-DNA hybrids are formed and what role they play in transcription regulation.
Transcription of DNA templates that lack an NT strand or have nicks or gaps in the NT strand may result in the formation of an extended RNA-DNA hybrid (23). At the same time, NT DNA damage increases the chance of misincorporation because of template strand misalignment (24, 25). It is therefore tempting to speculate that, by restoring the RNA-DNA hybrid to its normal 8–9-bp length, cellular RNAPs backtrack and excise the 3′ end region of transcript containing misincorporated bases. Thus, the mechanism we observe in in vitro experiments may be used by RNAP during in vivo transcription where it carries out proofreading either alone or in combination with transcript cleavage factors such as GreA/GreB in prokaryotes or TFIIS in eukaryotes.
Supplementary Material
Acknowledgments
Dr. I. Artsimovitch is acknowledged for the generous gift of E. coli GreB. Dr. D. Vassylyev is acknowledged for sharing coordinates of bacterial EC prior to publication and helpful suggestions. We are thankful to Brian Lebude for technical assistance and to Dr. William McAllister and Dr. Nikolay Zenkin for fruitful discussion and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM38147 (to W. T. McAllister) and GM74252 (to D. G. Vassylyev). This work was also supported by the University of Medicine and Dentistry of New Jersey Foundation (to D. T.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: RNAP, RNA polymerase; EC, elongation complex; WT, wild type; nt, nucleotides; NT, non-template.
References
- 1.Kireeva, M. L., Komissarova, N., and Kashlev, M. (2000) J. Mol. Biol. 299 325–335 [DOI] [PubMed] [Google Scholar]
- 2.Kashlev, M., and Komissarova, N. (2002) J. Biol. Chem. 277 14501–14508 [DOI] [PubMed] [Google Scholar]
- 3.Korzheva, N., and Mustaev, A. (2001) Curr. Opin. Microbiol. 4 119–125 [DOI] [PubMed] [Google Scholar]
- 4.Sidorenkov, I., Komissarova, N., and Kashlev, M. (1998) Mol. Cell 2 55–64 [DOI] [PubMed] [Google Scholar]
- 5.Gusarov, I., and Nudler, E. (1999) Mol. Cell 3 495–504 [DOI] [PubMed] [Google Scholar]
- 6.Westover, K. D., Bushnell, D. A., and Kornberg, R. D. (2004) Science 303 1014–1016 [DOI] [PubMed] [Google Scholar]
- 7.Toulokhonov, I., and Landick, R. (2006) J. Mol. Biol. 361 644–658 [DOI] [PubMed] [Google Scholar]
- 8.Naryshkina, T., Kuznedelov, K., and Severinov, K. (2006) J. Mol. Biol. 361 634–643 [DOI] [PubMed] [Google Scholar]
- 9.Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H., and Artsimovitch, I. (2007) Nature 448 157–162 [DOI] [PubMed] [Google Scholar]
- 10.Kuznedelov, K., Korzheva, N., Mustaev, A., and Severinov, K. (2002) EMBO J. 21 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Nahum, G., Epshtein, V., Ruckenstein, A. E., Rafikov, R., Mustaev, A., and Nudler, E. (2005) Cell 120 183–193 [DOI] [PubMed] [Google Scholar]
- 12.Kashkina, E., Anikin, M., Tahirov, T. H., Kochetkov, S. N., Vassylyev, D. G., and Temiakov, D. (2006) Nucleic Acids Res. 34 4036–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korzheva, N., Mustaev, A., Kozlov, M., Malhotra, A., Nikiforov, V., Goldfarb, A., and Darst, S. A. (2000) Science 289 619–625 [DOI] [PubMed] [Google Scholar]
- 14.Kyzer, S., Ha, K. S., Landick, R., and Palangat, M. (2007) J. Biol. Chem. 282 19020–19028 [DOI] [PubMed] [Google Scholar]
- 15.Vassylyeva, M. N., Lee, J., Sekine, S. I., Laptenko, O., Kuramitsu, S., Shibata, T., Inoue, Y., Borukhov, S., Vassylyev, D. G., and Yokoyama, S. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1497–1500 [DOI] [PubMed] [Google Scholar]
- 16.Artsimovitch, I., Svetlov, V., Murakami, K. S., and Landick, R. (2003) J. Biol. Chem. 278 12344–12355 [DOI] [PubMed] [Google Scholar]
- 17.Temiakov, D., Anikin, M., and McAllister, W. T. (2002) J. Biol. Chem. 277 47035–47043 [DOI] [PubMed] [Google Scholar]
- 18.Zenkin, N., Yuzenkova, Y., and Severinov, K. (2006) Science 313 518–520 [DOI] [PubMed] [Google Scholar]
- 19.Laptenko, O., and Borukhov, S. (2003) Methods Enzymol. 371 219–232 [DOI] [PubMed] [Google Scholar]
- 20.Erie, D. A., Hajiseyedjavadi, O., Young, M. C., and von Hippel, P. H. (1993) Science 262 867–873 [DOI] [PubMed] [Google Scholar]
- 21.Korzheva, N., Mustaev, A., Nudler, E., Nikiforov, V., and Goldfarb, A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63 337–345 [DOI] [PubMed] [Google Scholar]
- 22.Zenkin, N., Naryshkina, T., Kuznedelov, K., and Severinov, K. (2006) Nature 439 617–620 [DOI] [PubMed] [Google Scholar]
- 23.Jiang, M., Ma, N., Vassylyev, D. G., and McAllister, W. T. (2004) Mol. Cell 15 777–788 [DOI] [PubMed] [Google Scholar]
- 24.Pomerantz, R. T., Temiakov, D., Anikin, M., Vassylyev, D. G., and McAllister, W. T. (2006) Mol. Cell 24 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashkina, E., Anikin, M., Brueckner, F., Pomerantz, R. T., McAllister, W. T., Cramer, P., and Temiakov, D. (2006) Mol. Cell 24 257–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.