Abstract
The light-driven pump Halorhodopsin (hR) uses the energy stored in an initial meta-stable state (K), in which the bound retinal chromophore has been photoisomerized from all-trans to 13-cis, to drive the translocation of one chloride anion across the membrane. Thus far it was unclear whether retinal twisting or charge separation between the positive Schiff base of the retinal and the chloride anion is the primary mechanism of energy storage. Here, combined quantum mechanical/molecular mechanical (QM/MM) simulations show that the energy is predominantly stored by charge separation. However, a large variability in retinal twisting is observed, thus reconciling the contradictory hypotheses for storage. Surprisingly, the energy stored in the K-state amounts to only one-fifth of the photon energy. We explain why the protein cannot store more: even though this would accelerate chloride pumping, raising the K-state also reduces the relative energy barriers against unproductive decay, in particular via the premature cis to trans back-isomerization. Indeed, the protein has maximized its storage so that the back-isomerization barriers are just high enough (≥18 kcal/mol) to keep the decay rate (1/100 ms) slower than the remaining photocycle (1/20 ms). This need to stabilize the captured photon-energy until it can be used in subsequent steps is inherent to light-driven proteins.
Light driven pumps such as halorhodopsin (hR)2 stand apart from other energy-driven transporters. To them, the primary source of energy is not permanently available, unlike energy stored in the form of chemical bonds (for example, ATP in the case of the Na+/K+-ATPase (1)) or in electrochemical transmembrane gradients (like the Na+/glucose symport transporter (2)). Thus, they must be able to first harvest the energy of the photon, before converting that energy into work. The harvesting of the fleeting photon necessitates converting the photon energy into a meta-stable form of molecular energy, and to prevent the dissipation of this molecular energy until it can be subsequently used to power the pumping mechanism. In other words, the protein must be able to simultaneously store and stabilize the energy. For the absorption of light, proteins employ a chromophoric ligand or group, which changes from one state to another upon photon absorption. This transition is unfavorable and slow in the electronic ground-state, but is favored when the chromophore has been excited by the photon. After the return of the system to the electronic ground-state (within picoseconds in the case of hR (3)), the chromophore relaxes either to the starting ground-state conformation (thus reducing the quantum yield) or to a meta-stable high-energy state (with a quantum yield of 34% in the case of hR (3)). The energy stored in this meta-stable form relative to the ground-state form is what powers the subsequent pumping steps (Fig. 1A). Understanding how this energy is stored thus constitutes an essential aspect of the mechanism and has been the focus of studies on visual rhodopsins (4) and bacteriorhodopsin (5). In the case of hR, the free energy available in the initial metastable state (called the K-state) corresponds to about 10 kcal/mol (6), essentially all from the enthalpic component (7). Interestingly, this is only ⅕ of the photon energy, raising the question of why the protein does not store more of the photon's energy (∼50 kcal/photon at 580 nm). Indeed, the rate of pumping depends (among other things) on the amount of energy initially stored, so it would be advantageous for the protein to raise the energy level of the initial metastable state as much as possible. However, this state must be stabilized well enough to prevent unproductive thermal decay back to the ground-state form. To achieve this, the protein sets up energy barriers against the unproductive decay routes. Raising the energy level of the initial metastable state eventually lowers the relative height of these confinement barriers. Thus there exists a trade-off between maximizing the energy level of the metastable state and keeping this state sufficiently stable. We have examined whether this trade-off is optimally achieved in halorhodopsin by getting a quantitative understanding of the manner in which the photon energy is both stored and stabilized in the initial K-state.
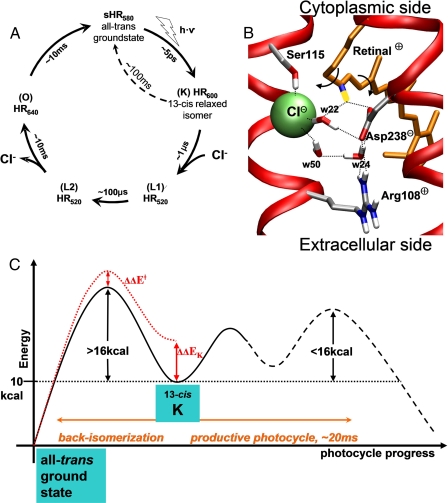
FIGURE 1.
A, photocycle of hR from H. salinarum. Subscript numbers indicate the absorption wavelength of the accumulating intermediates (their common denomination in parentheses). Unproductive decay from K by premature cis → trans back-isomerization is shown as a dotted arrow. B, environment of the PSB in the all-trans ground-state crystal. The two arrows indicate the bond torsions that allow the 13-cis ↔ all-trans motion. The proton of the PSB is in yellow, carbons in gray, chloride in green, nitrogens in blue, and oxygens in red. Water molecules are numbered as in the crystal. C, schematic energy profile of the electronic ground-state around the initial metastable K-state. Dotted lines are used for hypothetical energy levels.
Halorhodopsin uses one photon to drive the transfer of one chloride anion from the extracellular to the intracellular side of the membrane against an electrochemical potential to maintain osmotic pressure during cell growth in its natural high-salt environment (8, 9). It is a member of the family of retinal proteins, which have a similar membrane domain (10): seven-transmembrane helices, in the middle of which the retinal chromophore is covalently bound via a Schiff base (SB) link to a lysine (Fig. 2) (11). In hR, the SB is protonated (i.e. it carries a net charge of +1) throughout the normal photocycle. In the ground-state of light-adapted hR, the retinal has all the bonds of its polyene chain in trans. The crystal structure of the ground-state shows a chloride anion bound in close proximity to the proton of the protonated SB (PSB) (Fig. 1B) (12), allowing for very favorable charge-charge interactions. These are modulated by the proximity of Asp238- and Arg108+. Photoexcitation of the conjugated π-system of retinal to the first electronic excited state promotes the fast isomerization of retinal around the C13 = C14 double bond. The return to the electronic ground-state and vibrational relaxation of retinal dissipate about ⅘ of the absorbed photon energy. The remaining energy is stored in the K-state, in which the retinal is in a 13-cis form (11, 12). Using time-resolved laser-induced optoacoustic spectroscopy the enthalpy of the K-state has been measured at 11 ± 3 kcal/mol above the ground-state (hR of Natronobacterium pharaonis) (7). The high-resolution crystal structures of the ground- and K-states are very similar. This is not surprising, given the very short time (5 ps) for conversion between them, too fast to allow significant rearrangements in the protein main chain. The resolution is not sufficient to unambiguously decide on the amount of twisting of the 13-cis retinal, which has been the subject of much debate also in the closely related proton-pump bacteriorhodopsin (13, 14). It is not clear in hR whether the energy in this state is predominantly stored in the twisting of the retinal chain and/or in the electrostatic interactions between the cationic PSB and the surrounding charges (5, 15). Because the essential characteristic of the K-state is the cis conformation of the C13 = C14 double bond, this configuration must be stabilized against a premature back-isomerization to all-trans. A lower bound for the back-isomerization barrier can be deduced from the time scale of the remaining photocycle. Indeed, for the relaxation of the K-state to occur predominantly via the productive photocycle, any other non-productive relaxation pathway must be slower than this time scale (see schematic energy diagram in Fig. 1C). Because the remaining photocycle takes on the order of 10 ms (8), the barrier against premature back-isomerization must be at least 16 kcal/mol high (using Eyring's transition state law, rate = kBT·exp(-ΔG‡/RT)/h). The barrier of back-isomerization around the C13=C14 bond is in the range of 25 kcal/mol for retinal in vacuum (16). In the protein, the barrier is unknown and may depend on whether the rotation is clockwise or counterclockwise.
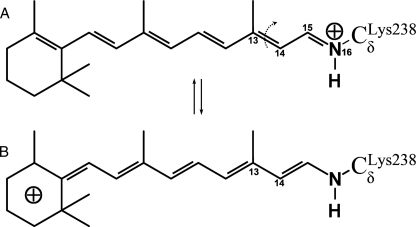
FIGURE 2.
Resonance states of protonated retinal. A, state most populated in planar retinal (all-trans or 13-cis), with the positive charge on the Schiff base. B, state most populated when the C13–C14 bond is fully twisted (90°), with the positive charge de-localized toward the ionone ring.
Thus the relevant questions are: 1) how is the energy stored in the meta-stable K-state (electrostatically or via retinal strain) and how twisted is the retinal? 2) What is the height of the barriers that stabilize the K-state against thermal cis → trans back-isomerization and what is the origin of these barriers? 3) Could more energy be stored in the K-state without risking unproductive back-isomerization? To answer these, we use combined quantum mechanical/molecular mechanical (QM/MM) simulations in which the behavior of retinal is accurately described using quantum methods and the flexibility of the surrounding protein is fully accounted for by a classical force-field. The dynamics of the ground- and K-states are characterized with molecular dynamics, and the stability of the K-state is determined by minimum energy path (MEP) calculations of retinal isomerization. This reveals that: 1) in the K-state retinal can adopt planar as well as partially twisted conformations, but the dominant energy storage comes from the chloride/PSB charge separation. 2) The 13-cis configuration is stabilized against thermal decay to all-trans by an 18 kcal/mol energy barrier, coming mostly from the torsion energy of the C13=C14 double bond. 3) This barrier is close to the C13=C14 barrier in vacuum, showing that the protein already fully exploits this intrinsic torsion barrier and therefore could not stabilize the K-state enough if the relative barrier to back-isomerization were lowered by an elevation of the K-state. It suggests that halorhodopsin stores the maximal fraction of the photon energy that is allowed by the retinal.
EXPERIMENTAL PROCEDURES
Protein coordinates for the all-trans ground-state were taken from the HR578 crystal structure of hR from Halobacterium salinarum (PDB entry 1E12) (12), retaining its trimeric arrangement. The K-state was modeled by placing a retinal in the 13-cis/15-anti form into the hR ground-state crystal structure and performing geometry optimization. This results in protein coordinates nearly identical to those of the K-state crystal structure.3 The HBUILD facility of the program CHARMM (22) was used to place the hydrogens. Based on pK1/2 calculations (23), Asp238 was taken as deprotonated. Only the carboxylic acid groups of Asp128 and the co-crystallized palmitate lipids were protonated. Fourier transform infrared data shows that the protonation state of aspartic residues in halorhodopsin does not change upon chloride binding (24). Atoms far away from the retinal were kept fixed, as described previously (25).
The protein was partitioned into QM and MM parts. Retinal and the side chain of Lys242 were treated quantum mechanically (79 atoms) in a minimal QM region (QM = retinal). Alternatively, the side chains of Asp238, Ser115, and the water molecules 22, 24, 50 (near the Schiff base) were also included in the quantum region (QM = extended). The link atom approach was used to satisfy the valence requirements of the quantum part when the frontier between the QM and MM parts crosses a covalent bond (26). This occurs at the Cβ-Cγ covalent bond in Asp238 and Ser115 and at the bond Cγ-Cδ for Lys242. The energy for the MM region used the CHARMM force-field with parameter set 22 for aromatic side chains (27) and parameter set 19 (28) for the rest of the opsin. Nonbonded interactions were switched off between 7 and 14 Å. Bulk solvent screening effects were accounted for by applying non-uniform charge scaling (29). The QM region was treated with the self-consistent charge density functional tight binding (DFTB) method, which has been shown to accurately describe retinal behavior (30). The geometries of stationary points were optimized using DFTB. To evaluate the dependence on the choice of the quantum method and the influence of chloride polarization, the energy along the pathways was calculated without geometry re-optimization using density functional theory at the B3LYP/6–31G* level, including the chloride anion in the QM region. This showed (Table 1 and Fig. 5A) that DFTB with only retinal in the QM region gives reliable results for the purpose of the present study.
TABLE 1.
Comparison of quantum regions and methods
| DFTB (QM = retinal)a | DFTB (QM = extended)b | DFTB(ext)//DFT(ext)c | DFTB(ext)//DFT(ext+Cl–)d | |
|---|---|---|---|---|
| Planar Ke | 10.7 | 10.5 | 12.2 | 12.1 |
| ‡(Asp238) barrierf | 17.9 | 18.7 | 21.2 | 21.3 |
| ‡(Ser115) barrierf | 22.8 | 20.9 | 24.8 | 25.1 |
QM region = retinal, optimized with DFTB
Opsin = protein including chloride anion and water, but excluding retinal
Retinal only
Interaction energy between retinal and opsin
Energy relative to the all-trans ground state
Rate-limiting saddle-point along the 13-cis → all-trans pathways for clockwise (‡Asp238) or counter-clockwise (‡Ser115) rotation. The barrier is relative to Kp
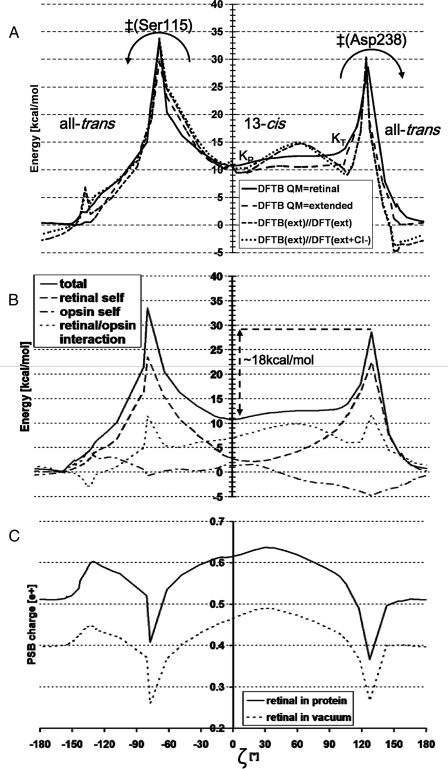
FIGURE 5.
MEP along a 360° rotation of the PSB, trans → 13-cis (via Ser115) → trans (via Asp238). The arrows show the two possible back-isomerization routes. See Fig. 3 for structures along this path and a definition of ζ. The angle ζ is not the driving coordinate, but has been measured in structures taken along the MEP. A, total energy. Optimized energy with DFTB: QM region is retinal only (—) or extended (–––) (see Table 1 for definition). Geometries optimized with DFTB (extended QM region): energy from B3LYP/6–31G* with (···) or without (- - -) the chloride in the QM region. B, energy decomposition (DFTB, QM region is retinal). The total energy (continuous line) is the sum of: the retinal self-energy (dashes), the opsin self-energy (everything excluding retinal, in dash dots), and the interaction between retinal and the opsin (in dots). C, charge (in fraction of one proton charge) on the protonated Schiff base group (see “Experimental Procedures”). Mulliken charges calculated in the presence (continuous line) or absence (dotted line) of the electrostatic field from the protein.
Minimum energy paths were computed using the conjugate peak refinement algorithm (31) as implemented in the TReK module of CHARMM. This method has been used already to determine the mechanisms of several complex reactions in hR (25) and bacteriorhodopsin (32). Conjugate peak refinement starts from an initial guess of the path that consists of a series of structures connecting the reactant to the product. Here, the energy minimized all-trans crystal structure was taken as the reactant state, the K-state as the product. The clockwise (on the Asp238 side) and counterclockwise (on the Ser115 side) initial pathways were seeded by adding one intermediate structure in which the C13=C14 bond was rotated rigidly. Conjugate peak refinement refines the initial path by automatically adding, removing, and optimizing path intermediates until all energy maxima along the connecting path segments are first-order saddle points of the surface defined by the energy function. Low energy regions of the MEP were further optimized with the synchronous chain minimization algorithm implemented in TReK.
Decomposition of energies was achieved by calculating point energies in a reduced system while maintaining the geometries obtained for the full system: the retinal self-energy is defined here as the quantum energy of the retinal in the absence of the rest of the system (=opsin). The opsin self-energy is obtained in absence of retinal. The retinal/opsin interaction energy is the sum of the classical van der Waals interaction and the quantum electrostatic interaction between opsin and retinal (the latter was computed by subtracting the retinal self-energy from the total quantum energy). Decomposition by individual residues of the retinal/opsin interaction was done by subtracting from the full system the QM/MM interaction energies of the system lacking a given residue. The changes of charge localization on the retinal were computed with B3LYP/6–31G*, by adding the Mulliken charges of the atoms of the protonated Schiff base group (atoms C15, H15, N16, H16, CγLys-242, H1(CγLys-242), H2(CγLys-242)).
RESULTS
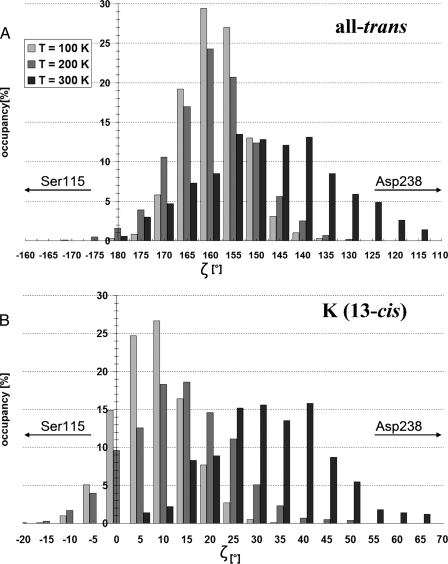
Energy Storage in the K-state—Geometry optimization of the all-trans ground-state (Fig. 3A) or the 13-cis K-state both result in protein coordinates around the retinal very similar to those of the crystal structures. The K-state conformation of lowest energy has a nearly planar retinal (Fig. 3D), with the PSB pointing toward the cytoplasmic side (ζ =+5°, see caption of Fig. 3 for a description of ζ), designated here as KP. A second, weakly stable conformation of 13-cis has its PSB strongly twisted toward Asp238 (ζ =+100°, not shown), designated here as KT. KT lies in a shallow energy basin that is only 1 kcal/mol higher than KP, separated from KP by a small 1 kcal/mol energy barrier (based on density functional theory optimized geometries of KT,KP, and the transition state between them, see “Experimental Procedures”). This is consistent with experiments that have trapped an early twisted K-state at low (T = 75 K) and a late planar K-state at high (T = 150 K) temperature (17). Molecular dynamics simulations were performed to examine the twisting behavior of the retinal at different temperatures. In all-trans, the orientation of the PSB (as measured by the angle ζ) oscillates with an amplitude of 30° at 100 K (Fig. 4A). At 300 K this amplitude increases to 65°, ranging from an orientation toward the extracellular side (ζ = 180°) to an orientation toward the Asp238 (ζ = 115°). In 13-cis, the average PSB orientation shifts from pointing almost straight toward the cytoplasmic side (ζ = 8°) at 100 K to pointing nearly in the direction of Asp238 (ζ = 35°) at 300 K (Fig. 4B). The large amplitude of accessible orientations (ranging from ζ = 0° to ζ = 70°) at 300 K reflects the fact that the energy profile in that range is quite flat in the K-state, as can be seen in Fig. 5A.
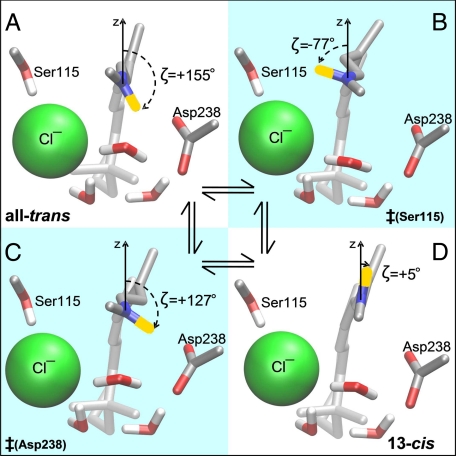
FIGURE 3.
Stationary states of the C13=C14 torsion pathway. A, all-trans ground-state. B, counterclockwise transition state ‡(Ser115). C, clockwise transition state ‡(Asp238). D, 13-cis planar K-state. The angle ζ is defined as the angle between the N–H bond of the PSB and the z axis (orthogonal to the membrane plane). ζ = 0° has the hydrogen of the PSB pointing toward the cytoplasmic side. The sign of ζ is defined as positive when the N-H vector is pointing to the Asp238 side of the plane formed by the retinal chain, as negative when pointing to the Ser115 side. The same coloring as described in the legend to Fig. 1B was used. Molecular movies showing the transitions between these four states are available in the supplemental material.
FIGURE 4.
Schiff base orientations during molecular dynamics. A, in the all-trans ground-state and B, in the 13-cis K-state, at different temperatures (QM region: retinal with DFTB). See Fig. 3 for description of ζ.
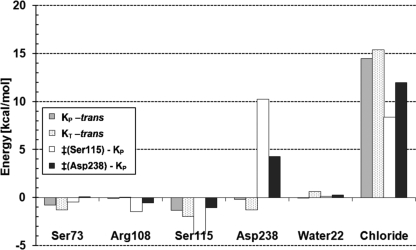
The energy of K is 10.7–12.7 kcal/mol above the all-trans ground-state (Table 1). To understand the mechanism of this energy storage, the energy difference between the optimized ground-state and the K-state conformations was decomposed into contributions from (i) changes in the retinal self-energy (i.e. the retinal strain), (ii) the opsin self-energy (opsin refers here to the entire system minus the retinal), and (iii) the retinal/opsin interaction energy (Table 2 summarizes this data). There is 7.2 kcal/mol of retinal strain in the KT form, as expected from its strong twisting, but only 2.9 kcal/mol in the quasi-planar KP form, close to the 2 kcal/mol energy difference between the fully planar 13-cis and all-trans forms adopted by unconstrained retinal in vacuum (with DFTB (16)). The opsin self-energy decreases by 2.3 kcal/mol in KT and increases by 1.3 kcal/mol in KP, indicating that opsin by itself plays little role in the energy storage. The dominant contribution in both KT and KP is the significant loss of favorable interaction between retinal and the opsin (6.5–7.8 kcal/mol). Further decomposition into contributions from individual opsin groups reveals that this stems almost entirely from the charge separation between the Schiff base NH+ group and the chloride anion: Fig. 6 shows that both in KP and KT the retinal/chloride interactions are ∼15 kcal/mol less favorable than in the ground-state. Other groups contribute little to energy storage from the retinal/opsin interaction (Fig. 6). In summary, chloride/PSB charge separation is the main mechanism of storing energy in the most populated conformers of the K-state.
TABLE 2.
K-state storage energy and decay barriers
| Totala | Opsin selfb | Retinal selfc | Retinal/opsind | |
|---|---|---|---|---|
| Planar Ke | 10.7 | 1.3 | 2.9 | 6.5 |
| Twisted Ke | 12.7 | –2.3 | 7.2 | 7.8 |
| ‡(Asp238) barrierf | 17.9 | –5.9 | 19.4 | 4.4 |
| ‡(Ser115) barrierf | 22.8 | –3.9 | 20.4 | 6.3 |
QM region = retinal, optimized with DFTB
Extended QM region (=retinal, Asp238, Ser115, water 22, 24, and 50), optimized with DFTB
Geometries optimized as in b, energy calculated with B3LYP-6–31G*
Geometries optimized as in b, energy calculated with B3LYP-6–31G* for the extended QM region and B3LYP-6–31G** for the chloride anion
Energy relative to the all-trans ground state
Rate-limiting saddle-point along the 13-cis → all-trans pathways for clockwise (‡Asp238) or counter-clockwise (‡Ser115) rotation. The barrier is relative to Kp
FIGURE 6.
Interactions between retinal and opsin residues. The largest contributions to energy storage are shown for the KP (gray) and the KT (hatched) forms (relative to the all-trans ground-state). Contributions to the 13-cis → all-trans barrier (relative to KP) are shown for the route via Ser115 (white) and via Asp238 (black). See Fig. 3, B and C, for structures of the corresponding transition states. Energies calculated with DFTB, the QM region is retinal.
Barriers against Unproductive Decay of K—To determine the energy barriers against thermal 13-cis→all-trans back-isomerization, MEP calculations were performed, taking into consideration the two possible directions of rotation around the C13=C14 bond, namely clockwise when viewed as in Fig. 3 (with the PSB proton pointing toward Asp238), or counterclockwise (with the PSB pointing toward Ser115). This enabled us to identify the rate-limiting transition states of these two routes, designated here as ‡(Asp238) and ‡(Ser115), and to build the energy profile along a 360° rotation of the PSB: all-trans → ‡(Ser115) → KP → KT → ‡(Asp238) → all-trans, shown in Fig. 5. Note that because KT can rapidly (compared with the time scale of back-isomerization) convert into KP (see above), all back-isomerization barriers are taken here relative to KP. Fig. 5A and supplemental Fig. S1 show that extending the QM treatment to the chloride and groups near retinal (see “Experimental Procedures”) yields results very similar to the ones obtained with only retinal in the QM region. Thus, the following discussion refers to the latter (supplemental Movie M2) values.
Supplemental molecular movies of the MEP show that the isomerization motion is a simple rigid body rotation of the planar -C14-C15H15=N16H16-moiety (see Fig. 2A for nomenclature), enabled by the simultaneous torsion of the C13=C14 and the N16-C Lys-242γ bonds (arrows in Fig. 1B). The counter-clockwise route (supplemental Movie M1) has its transition state ‡(Ser115) at ζ =-77° (Fig. 3B), where the PSB forms a hydrogen bond with the Ser115 oxygen. The energy barrier for back-isomerization along this pathway is 22.8 kcal/mol. The clockwise route (supplemental Movie M2) has its transition state ‡(Asp238) at ζ =+127° (Fig. 3C), where the PSB can make a salt bridge with the Asp238 carboxyl group. This pathway has a lower barrier of 17.9 kcal/mol, and thus is the preferred route for 13-cis → trans back-isomerization.
To understand how the protein stabilizes the K-state against back-isomerization, the two barriers were decomposed into energy terms (see Table 2), in the same way as described above for the energy storage. This was performed also on every structure along the MEP, as shown in Fig. 5B. It reveals that the energy profile is dominated by the shape of the retinal self-energy term (Fig. 5B, dashed curve), whose behavior reflects the degree of twisting of the C13=C14 double bond. This term contributes 20.4 kcal/mol and 19.4 kcal to the ‡(Ser115) and ‡(Asp238) barriers, respectively. In contrast, the opsin self-energy somewhat lowers the barriers, by 3.9 and 5.9 kcal/mol relative to KP, respectively, for the ‡(Ser115) and ‡(Asp238) barriers. The latter is due to an electrostatically favorable change in distances to the chloride anion, which moves slightly away from Asp238- and closer to Arg108+. The retinal/opsin interaction energy contributes 6.3 and 4.4 kcal/mol to the ‡(Ser115) and ‡(Asp238) barriers, respectively. Further decomposition of the retinal interaction energy (Fig. 6) shows that this is dominated by unfavorable contributions from the charged groups in contact with the PSB, namely the chloride and Asp238-. In particular at ‡(Asp238), the retinal/Asp238 interaction can be seen to raise the barrier by 4.3 kcal/mol. This is surprising, because the PSB and Asp238 form a salt bridge in the ‡(Asp238) structure (Fig. 3C), which would be expected to lower the barrier. Equally surprising is the 8.4 kcal/mol contribution from the retinal/chloride interaction to the ‡(Ser115) barrier, even though the PSB proton has moved toward the chloride anion in this transition state (Fig. 3B). These two unexpected effects arise from charge shifts out of the PSB, as explained below.
Charge Shifts along the Polyene Chain of Retinal—Interestingly, the otherwise smooth profile of the retinal/opsin interaction (Fig. 5B, over dotted curve) makes a sharp peak at each transition state, locally peaking (at ζ =-77° and ζ =+127°) by about 5 kcal/mol above the rest of the curve. Energy decomposition (data not shown) reveals that these peaks are not due to van der Waals clashes, but are attributable to an unfavorable change in the electrostatic interactions with the PSB, even though the distances between the PSB and nearby charged opsin groups become more favorable as the pathways cross the transitions states. The reason for this unusual behavior is that the distribution of the +1 charge along the retinal polyene chain changes sharply when the C13=C14 bond becomes highly twisted. Indeed, the conjugated double bond system of retinal allows many possible resonance states (two of which are shown in Fig. 2), so that the positive charge can delocalize along the polyene chain (18). A large fraction of the charge is located at the Schiff base when the C13=C14 bond is planar and has double bond characteristics (Fig. 2A). Twisting this bond gives it a single bond character, so that the other resonance state (Fig. 2B) becomes more populated and the charge moves out of the PSB. As a result, the net charge of the Schiff base group drops by more than 0.2e+ during torsion of the C13=C14 bond, see Fig. 5C (for example, from 0.62 e+ at ζ = 0° to 0.37e+ at ζ =+127°). This effect, which had been observed also in bacteriorhodopsin (19), is not due to the protein environment, but is inherent to retinal (i.e. when the electrostatic field of the protein is switched off, see dotted curve in Fig. 5C). The sharp local decrease (∼0.15e+) seen around transition states, which amounts to a ∼25% reduction relative to the PSB charge in the ground-state, significantly weakens the favorable interaction of the PSB with the nearby anionic groups (Asp238-, chloride). This explains the observed peaks in the retinal/opsin interaction at the two transition states. It also explains the unexpected raising (rather than decreasing) of the barriers from the retinal interactions with Cl- and Asp238- observed in Fig. 6 (mentioned above). Charge shifting also affects the energy storage in the K-state: Fig. 5C shows that the PSB charge is 0.1e+ higher in K (0.62–0.64e+) compared with the all-trans ground-state (0.52e+). Thus, charge shifting somewhat lowers (by ∼15%) the energy storage that can be achieved by the charge separation between the PSB and the chloride.
DISCUSSION
The values found here for the energy storage in the K-state and the height of the barriers against its unproductive decay are consistent with experiments: 1) the 11 ± 1 kcal/mol potential energy level computed for the K-state agrees perfectly with the 11 ± 3 kcal/mol enthalpy measured by laser-induced optoacoustic spectroscopy (7). 2) When hR is made to pump against an electrostatic membrane potential, the chloride pumping rate slows down. Pumping stops when the potential reaches the so-called “reverse potential” VR, which has been extrapolated at -400 mV (for N. pharaonis hR expressed in Xenopus oocytes (6)). At this value, the transfer of one chloride across the membrane corresponds to a work of 9.2 kcal/mol (ΔG = zCl·FVR, where zCl is the anion valence and F, Faraday's constant). This represents a lower bound on the value of the energy that must be stored in the protein before chloride motion starts (i.e. the K-state), consistent with the 11 kcal/mol computed here. It follows that the remaining part of the pumping cycle (K → L1→ etc., Fig. 1A) is quite efficient: ∼84% (=9.2/11) of the energy stored in K is converted into productive work and only 16% dissipated in heat. 3) The lifetime of the 13-cis conformation has been determined while blocking anion transport by high chloride concentrations (20). In 5 m Cl- solvent conditions, hR no longer transfers anions after photoexcitation, but relaxes from the 13-cis back to the all-trans form in about 100 ms (dotted arrow in Fig. 1A). This time scale corresponds to an energy barrier of 18 kcal/mol, in perfect agreement with the barrier calculated here for the cis → trans back-isomerization pathway via Asp238.
Charge separation between the PSB and the chloride anion is the dominant mechanism of the energy storage in the K-state. However, the torsional energy profile of retinal in the K-state is very flat (Fig. 5A), i.e. a wide range of Schiff base orientation angles result in a similar energy. It allows for large fluctuations at 300 K, ranging from a planar retinal conformation to one with the PSB highly twisted toward Asp238 (Fig. 4B). This flatness of the energy profile results from compensation between the unfavorable retinal C13=C14 torsion and the electrostatically favorable interaction between the PSB and Asp238-, as reflected by the anti-correlated behavior between the retinal-self and the retinal/opsin interaction terms in the ζ =-20° to 110° range (dashed and dotted curves in Fig. 5B), allowing an exchange of up to 3 kcal/mol between the two terms. This shows that an interplay between retinal twisting and charge separation is involved in the mechanism of energy storage. Given the high similarity between the structures of hR and bacteriorhodopsin near the PSB, an analogous behavior may occur in bacteriorhodopsin, thus explaining and reconciling the contradictory experimental data pleading in favor of a planar versus twisted retinal in the early metastable states of the bacteriorhodopsin photocycle (13, 14).
Charge separation and energy storage in the K-state depend on the 13-cis conformation of retinal. Thus it is important for the protein to stabilize the C13=C14 bond against premature back-isomerization into 13-trans. The protein achieves this by keeping the lowest barrier to unproductive decay (18 kcal/mol for the clockwise rotation via Asp238, Fig. 5B) high enough to keep this process slower (1/100 ms) than the rate of the remaining pumping cycle (1/20 ms). This insures that the relaxation of the K-state to the ground-state predominantly occurs via the pumping pathway. Decomposition of the back-isomerization barrier shows that the barrier is mainly due to the inherent penalty for twisting the C13=C14 double bond (see Fig. 5B, dashed curve).
We have shown how the back-isomerization barrier is significantly increased by charge de-localization along the retinal chain, which causes about 25% of the positive charge to move out of the Schiff base group (Fig. 5C), thus weakening its favorable interaction with the nearby chloride and Asp238 anions. It is possible to speculate how this charge shift might play a role also later in the photocycle (L2 →→, Fig. 1A), after the chloride has moved away from the PSB and into the cytoplasm. At this stage, the retinal needs to be restored to its all-trans configuration, preparing for the next photocycle. If this late 13-cis → all-trans back-isomerization were to have a rate as slow as in the K-state (1/100 ms), then this process would become rate-limiting for the pumping cycle and significantly deteriorate the pumping rate. Based on the present results, the late back-isomerization barrier is expected to be significantly lower than in the K-state, because the effect of the PSB charge shift (i.e. weakening the favorable PSB+/CL- interactions) is not contributing to the barrier in the absence of chloride near the PSB. This lowers the late 13-cis → all-trans barrier, facilitating a fast return to the all-trans configuration. Indeed, Fourier transform infrared experiments indicate that the 13-cis → all-trans isomerization late in the productive photocycle is fast and non-rate-limiting (21). Thus, hR uses the inherent dependence between polyene charge delocalization and twisting to its advantages in two ways: (i) to raise the confinement barrier of the K-state early in the photocycle and (ii) to lower the barrier of all-trans restoration after the chloride has been transferred.
Interestingly, the 11 kcal/mol stored in the meta-stable K-state are much larger than the work that would be necessary to power chloride transfer against the largest membrane potential encountered physiologically (5 kcal/mol for Vmb ≤ -200 mV, a higher potential leading to membrane rupture (6)). Why does hR store more energy than necessary, incurring the risk of unproductive decay? The answer is that successive energy barriers encountered during the photocycle can be overcome faster if the starting energy level is higher (because it allows to raise the level of intermediates, thus lowering the relative barriers of escape from these intermediates). Indeed, experiments show that the photocycle turnover rate decreases linearly with an increase in opposing electrostatic membrane potential (6). In other words, hR can pump faster than it would if it had stored only as much as strictly necessary for working against the physiological membrane potential. This increased flux per protein is beneficial for the halobacterium, because fewer hR proteins need to be synthesized to maintain the osmotic pressure.
Insofar as a higher initial energy level allows for faster pumping, why did hR not evolve to store even more than 11 kcal/mol in the K-state (i.e. storing a larger fraction of the 50 kcal/mol photon)? To do so (ΔΔEK in Fig. 1C) would require raising the energy level of the transition states of the premature 13-cis → all-trans back-isomerization (ΔΔE‡ in Fig. 1C), so that the corresponding barrier remains high enough relative to the time scale of the photocycle (i.e. >16 kcal/mol, Fig. 1C). As shown here, the energy level of these transition states (at ζ =-77° and ζ =+127° in Fig. 5) is determined by properties inherent to the retinal, namely its intrinsic torsion barrier (Fig. 5B) and charge delocalization behavior (Fig. 5C). Thus, it appears that hR has little scope to further raise the energy level of the K-state. The present results suggest that this light-driven protein has been optimized to maximize energy storage under the constraint of maintaining high enough barriers against unproductive decay pathways. The same sort of finely tuned balance between maximizing energy storage versus stabilizing the first photointer-mediate has been shown to be achieved in the light-driven proton pump bacteriorhodopsin (5).
Supplementary Material
Acknowledgments
We are grateful to Professor Dieter Oesterhelt for access to the K-state crystal structure of halorhodopsin.
This work was supported by Deutsche Forschungsgemeinschaft Grant SF63/7.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Movies M1 and M2.
Footnotes
The abbreviations used are: hR, halorhodopsin; DFTB, density functional tight binding; SB, Schiff base; PSB, protonated Schiff base; QM, quantum mechanical; MM, molecular mechanical; MEP, minimum energy path.
D. Oesterheldt, personal communication.
References
- 1.Skou, J. C. (1957) Biochim. Biophys. Acta 23 394-401 [DOI] [PubMed] [Google Scholar]
- 2.Wright, E. M. (2001) Am. J. Physiol. 280 F10-F18 [DOI] [PubMed] [Google Scholar]
- 3.Alshuth, T., Stockburger, M., Hegemann, P., and Oesterhelt, D. (1985) FEBS Lett. 179 55-59 [Google Scholar]
- 4.Gascon, J. A., and Batista, V. S. (2004) Biophys. J. 87 2931-2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondar, N., Fischer, S., Suhai, S., and Smith, J. (2005) J. Phys. Chem. 109 14786-14788 [DOI] [PubMed] [Google Scholar]
- 6.Seki, A., Miyauchi, S., Hayashi, S., Kikukawa, T., Kubo, M., Demura, M., Ganapathy, V., and Kamo, N. (2007) Biophys J. 92 2559-2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Losi, A., Wegener, A. A., Engelhard, M., and Braslavsky, S. E. (2001) Photochem. Photobiol. 74 495-503 [DOI] [PubMed] [Google Scholar]
- 8.Váró, G., Needleman, R., and Lanyi, J. K. (1995) Biochemistry 34 14500-14507 [DOI] [PubMed] [Google Scholar]
- 9.Oesterhelt, D. (1995) Isr. J. Chem. 35 475-494 [Google Scholar]
- 10.Mukohata, Y., Ihara, K., Tamura, T., and Sugyiama, Y. (1999) J. Biochem. (Tokyo) 125 649-657 [DOI] [PubMed] [Google Scholar]
- 11.Spudich, J. L. (2000) Science 288 1358-1359 [DOI] [PubMed] [Google Scholar]
- 12.Kolbe, M., Besir, H., Essen, L. O., and Oesterhelt, D. (2000) Science 288 1390-1396 [DOI] [PubMed] [Google Scholar]
- 13.Schobert, B., Cupp-Vickery, J., Hornak, V., Smith, S. O., and Lanyi, J. K. (2002) J. Mol. Biol. 321 715-726 [DOI] [PubMed] [Google Scholar]
- 14.Neutze, R., Pebay-Peyroula, E., Edman, K., Royant, A., Navarro, J., and Landau, E. M. (2002) Biochim. Biophys. Acta 1565 144-167 [DOI] [PubMed] [Google Scholar]
- 15.Dér, A., Száraz, S., and Keszthelyi, L. (1992) J. Photochem. Photobiol. B Biol. 15 299-306 [Google Scholar]
- 16.Zhou, H., Tajkhorshid, E., Frauenheim, T., Shuai, S., and Elstner, M. (2002) Chem. Phys. 277 91-103 [Google Scholar]
- 17.Dioumaev, A. K., and Braiman, M. S. (1997) Photochem. Photobiol. 66 755-763 [DOI] [PubMed] [Google Scholar]
- 18.Herzfeld, J., and Tounge, B. (2000) Biochim. Biophys. Acta 1460 95-105 [DOI] [PubMed] [Google Scholar]
- 19.Paizs, B., Tajkhorshid, E., and Suhai, S. (1999) J. Phys. Chem. B 103 5388-5395 [Google Scholar]
- 20.Muneyuki, E., Shibazaki, C., Wada, Y., and Yakushizin, M. (2002) Biophys. J. 83 1749-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guijarro, J., Engelhard, M., and Siebert, F. (2006) Biochemistry 45 11578-11588 [DOI] [PubMed] [Google Scholar]
- 22.Brooks, B. R. (1983) J. Comp. Chem. 4 187-217 [Google Scholar]
- 23.Kloppmann, E., Becker, T., and Ullmann, G. M. (2005) Structure 61 953-965 [DOI] [PubMed] [Google Scholar]
- 24.Rothschild, K. J., Bousché, O., Braiman, M. S., Hasselbacher, C. A., and Spudich, J. L. (1988) Biochemistry 27 2420-2424 [DOI] [PubMed] [Google Scholar]
- 25.Gruia, A. D., Bondar, A. N., Smith, J. C., and Fischer, S. (2005) Structure 13 617-627 [DOI] [PubMed] [Google Scholar]
- 26.Field, M. J., Bash, P. A., and Karplus, M. (1990) J. Comput. Chem. 11 700-733 [Google Scholar]
- 27.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, S., Gao, J., Guo, H., Ha, S., Joseph-McCarthy, D., Kuchnir, L., Kuczera, K., Lau, F. T. K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D. T., Prodhom, B., Reiher, W. E., Roux, B., Schlenkrich, M., Smith, J. C., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 28.Neria, E., Fischer, S., and Karplus, M. (1996) J. Chem. Phys. 105 1902-1921 [Google Scholar]
- 29.Schwarzl, S. M., Huang, D., Smith, J. C., and Fischer, S. (2005) J. Comput. Chem. 26 1359-1371 [DOI] [PubMed] [Google Scholar]
- 30.Elstner, M., Porezag, D., Jungnickel, G., Elsner, J., Haugk, M., Frauenheim, T., Suhai, S., and Seifert, G. (1998) Phys. Rev. B 58 7260-7726 [Google Scholar]
- 31.Fischer, S., and Karplus, M. (1992) Chem. Phys. Lett. 194 252-261 [Google Scholar]
- 32.Bondar, N., Elstner, M., Suhai, S., Smith, J., and Fischer, S. (2004) Structure 12 1281-1288 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.