Abstract
A delicate balance between cell death and survival pathways maintains normal physiology, which is altered in many cancers, shifting the balance toward increased survival. Several studies have established a close connection between the Wnt/β-catenin pathway and tumorigenesis, aberrant activation of which might contribute toward increased cancer cell growth and survival. Extensive research is underway to identify therapeutic agents that can induce apoptosis specifically in cancer cells with minimal collateral damage to normal cells. Although tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) can induce apoptosis specifically in tumor cells, many cancer cells develop resistance, which can be overcome by combinatorial treatment with other agents: for example, peroxisome proliferator-activated receptorγ (PPARγ) ligands. To identify the molecular target mediating combinatorial drug-induced apoptosis, we focused on β-catenin, a protein implicated in oncogenesis. Our results show that co-treatment of TRAIL-resistant cancer cells with TRAIL and the PPARγ ligand troglitazone leads to a reduction ofβ-catenin expression, coinciding with maximal apoptosis. Modulation of β-catenin levels via ectopic overexpression or small interference RNA-mediated gene silencing modulates drug-induced apoptosis, indicating involvement of β-catenin in regulating this pathway. More in-depth studies indicated a post-translational mechanism, independent of glycogen synthase kinase-3β activity regulating β-catenin expression following combinatorial drug treatment. Furthermore, TRAIL- and troglitazone-induced apoptosis was preceded by a cleavage of β-catenin, which was complete in a fully apoptotic population, and was mediated by caspases-3 and -8. These results demonstrate β-catenin as a promising new target of drug-induced apoptosis, which can be targeted to sensitize apoptosis-resistant cancer cells.
Apoptosis is a form of cell death that permits the removal of damaged, senescent, or unwanted cells in multicellular organisms, without damage to the cellular microenvironment. Alterations of cellular machinery that lead to inactivation or evasion of apoptosis represents a major causative factor in the development and progression of cancer. Therapeutic approaches that can restore cancer cell apoptosis are expected to provide an effective means of treating various forms of cancer. Induction of cancer cell apoptosis via TNF2-related apoptosis-inducing ligand (TRAIL/apo2L) is an attractive novel form of cancer therapy, because TRAIL targets transformed cells with minimal damage to the normal cells (1, 2). TRAIL belongs to the tumor necrosis factor α (TNF-α) superfamily of cytokines, which include TNFα and Fas ligand (FasL)/CD95L all of which function via binding to their corresponding death receptors (3) leading to activation of caspases (4). Stimulation by TRAIL leads to its binding to death receptors DR4 and DR5, resulting in activation of initiator caspases (caspases-8 and -10), executioner caspase (caspase-3) (5, 6), and finally apoptosis via extrinsic pathway. Activated caspase-8 can also amplify death signal via an intrinsic pathway, through cleavage of proapoptotic BID resulting in its translocation to the mitochondria and release of cytochrome c in the cytoplasm. Cytochrome c in the presence of dATP and apoptotic protease-activating factor 1 activates caspase-9, which in turn activates caspase-3 further leading to more apoptosis (7).
Results from preclinical studies with recombinant TRAIL have demonstrated its significant anti-tumor activities, indicating the potential of utilizing TRAIL as an anticancer agent (8). Despite this proapoptotic role of TRAIL in the transformed cells, many cancer cells develop resistance toward TRAIL-induced apoptosis (9, 10). One potential reason for this resistance could be due to the presence of non-signaling decoy receptors for TRAIL, DcR1, DcR2 (11), and osteoprotegerin (12). Sensitivity of the tumor cells toward TRAIL-mediated apoptosis have also been linked with the activity of various pro- and anti-apoptotic proteins (10, 13) as well as loss of caspase-8 activity (14). Identification of drugs or agents that can overcome TRAIL resistance and sensitize cancer cells toward TRAIL-induced apoptosis is thus critically important for targeting TRAIL-resistant cancer cells. In earlier studies, a combination of TRAIL with radiation and some chemotherapeutic agents (9, 15) have provided limited success, because this combination also resulted in an increase in systemic toxicity.
Peroxisome proliferator-activated receptor γ (PPARγ) belongs to the nuclear receptor superfamily, which is involved in regulating various cellular processes including proliferation and apoptosis (16). Multiple artificial ligands of PPARγ have been reported so far, which belong to the thiazolidinedione family of insulin sensitizers and include troglitazone (TZD), ciglitazone, pioglitazone, and rosiglitazone. Our earlier studies have demonstrated that TZD can induce cell cycle arrest in proliferating liver cells, via targeting cell cycle regulators (17). At high doses these ligands can inhibit tumor growth in vivo via regulating cellular proliferation and apoptosis (18, 19). More recent studies have also demonstrated that PPARγ ligands can sensitize various cancer cells toward TRAIL-induced apoptosis (13, 20–24) thus raising the possibility of utilizing the TRAIL-PPARγ ligand combination for treating cancer cells. However, the detailed mechanism how this drug combination increases TRAIL sensitivity in TRAIL-resistant cancer cells is still unclear. Although one study demonstrates the cell cycle regulator cyclin D3 as a target of TRAIL-PPARγ agonist-mediated apoptosis (21), others suggest the anti-apoptotic protein cellular FLICE inhibitory protein as the potential target (22–24). Identification of molecular effectors, which can be targeted to increase TRAIL sensitivity in resistant cancer cells, is an important first step toward the development of effective therapeutic approaches.
In an attempt to identify additional targets contributing toward TRAIL resistance in the cancer cells, we focused on β-catenin due to a close link of β-catenin activation with cancer pathways (25, 26). Recent studies have also established a connection of β-catenin in mediating cell survival (27, 28). β-Catenin is a multifunctional protein, the expression of which is tightly regulated in normal cells and aberrant activation of which can lead to tumorigenesis (25). In the conventional pathway of β-catenin degradation, the serine threonine kinase glycogen synthase kinase 3β (GSK3β) in the presence of axin and functionally active adenomatous polyposis coli (APC) phosphorylates specific N-terminal residues of β-catenin and targets it toward degradation (29). Mutations of either APC or β-catenin itself (30, 31) or activation of the Wnt signaling pathway (25) can lead to inhibition of β-catenin degradation, resulting in increased cytoplasmic pools of β-catenin. Other pathways of β-catenin degradation have also been reported recently (32–34). Stabilization of β-catenin results in its translocation into the nucleus, interaction with transcription factors of the T cell factor/lymphoid enhancer factor (TCF/LEF) family to activate target gene transcription (25). A significant percentage of the human and mouse liver tumors harbor oncogenic mutations of the β-catenin gene (30), (35), whereas inactivating mutations of APC are responsible for β-catenin stabilization in the colorectal adenocarcinomas (29, 37).
In the present study we determined whether combinatorial treatment with TRAIL and TZD induces apoptosis in TRAIL-resistant cancer cells via targeting β-catenin. Our studies indicate that TRAIL- and TZD-mediated apoptosis is associated with a dramatic reduction of full-length β-catenin expression, coinciding with the degree of apoptosis. Ectopic overexpression of β-catenin antagonizes this drug-induced apoptosis, whereas small interference RNA (siRNA)-mediated knock-down of endogenous β-catenin initiates spontaneous apoptosis in cancer cells. TRAIL-TZD-mediated attenuation of β-catenin expression involved a GSK3β independent post-translational mechanism. Reduction of full-length β-catenin expression was also associated with a cleavage of C-terminal β-catenin mediated by caspases-3 and -8. These results indicate the promising possibility of targeting β-catenin as an effective means of overcoming TRAIL resistance.
EXPERIMENTAL PROCEDURES
Reagents—Dulbecco's modified Eagle's medium/F-12, Dulbecco's modified Eagle's medium, RPMI, McCoy's 5A tissue culture media, geneticin sulfate, and Lipofectamine 2000 were purchased from Invitrogen; troglitazone, TRAIL, cycloheximide, Z-VAD-fmk, Z-DEVD-fmk, IEDT-fmk, MG-132, lactacystin, and GSK3β inhibitor VIII (AR-A014418) were purchased from Calbiochem; the ELISAPLUS kit and in situ Cell Death Detection Kit (fluorescein) were purchased from Roche Applied Sciences; and 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) was from Vector Laboratories (Burlingame, CA). The antibodies were obtained from the following sources: poly(ADP-ribose) polymerase (PARP), caspase-3, cleaved caspase-3, caspase-8, GSK3β, phospho-GSK3βSer9, Akt, and phospho-glycogen synthaseSer641 from Cell Signaling Technology (Danvers, MA); β-catenin from Zymed Laboratories Inc., Invitrogen, and BD Biosciences (San Jose, CA); glycogen synthase from Invitrogen; cyclin-D1 from Neomarkers (Fremont, CA); Bcl-xL from Exalpha Biologicals Inc. (Watertown, MA); GAPDH from Ambion Inc. (Austin, TX). The Huh-7 cells were obtained from Dr. Robert E. Lanford (University of Texas Health Science Center, San Antonio, TX) (38) and the RKO and RKO-β-catenin cells were obtained from Dr. Wallace K. MacNaughton and Hongying Wang (University of Calgary, Calgary, Canada) (39). The LNCaP and SKOV3 cells were obtained commercially from ATCC. The wild type (WT) and GSK3β knock-out (-/-) mouse embryonic fibroblasts (MEFs) were obtained from Dr. James R. Woodgett (University of Toronto, Toronto, Canada) (40).
Cell Culture—Huh-7 and RKO cells were grown in Dulbecco's modified Eagle's/F-12 medium supplemented with 10% fetal bovine serum and RKO-β-catenin cells were maintained in a similar medium containing 500 μg/ml Geneticin. LNCaP cells were maintained in RPMI containing 10 mm HEPES, 4.5 g/liter glucose, and 10% fetal bovine serum and SKOV3 cells were grown in modified McCoy's 5A medium supplemented with 10% fetal bovine serum. WT-GSK3β (+/+) and GSK3β knock-out (-/-) MEFs were maintained in Dulbecco's modified Eagle's medium containing 4.5 g/liter glucose, 4 mm glutamine, and 10% fetal bovine serum. In TRAIL and TZD experiments, cells were treated with 100 ng/ml TRAIL or 50 μm TZD (unless indicated otherwise) alone or in combination for various lengths of time followed by either apoptosis assays or Western blot analysis. In the studies with GSK3β inhibitor (AR-A014418) or caspase inhibitors, cells were pretreated for 1 h with 10 μm AR-A014418 or 50 μm Z-VAD-fmk or 25 μm DEVD-fmk or 25 μm IETD-fmk, respectively, followed by TRAIL-TZD treatment.
Apoptosis Detection by Fluorescence Microscopy—This was performed utilizing protocols described earlier (41). Briefly, subconfluent cells were plated in 2-well chamber slides and were treated with 50 μm TZD and 100 ng/ml TRAIL either alone or in combination. To determine the optimal drug concentrations required to induce maximal apoptosis in cancer cells, increasing concentrations of TZD or TRAIL were utilized in pilot experiments and apoptosis assays were performed. Following treatment with TZD and TRAIL, cells were fixed with 4% paraformaldehyde, permeabilized with phosphate-buffered saline containing 0.1% Triton X-100, and stained with DAPI. DAPI-stained nuclei were analyzed for apoptotic morphology by fluorescence microscopy (Axiovert 200 inverted microscope, Zeiss). A minimum of 200 nuclei were analyzed per treatment. The percentage of apoptotic cells was calculated as the number of cells with condensed/fragmented nuclear morphology divided by the total number of cells analyzed. To determine the effect of various agents on apoptotic morphology, cells were also photographed by inverted phase-contrast microscopy.
Apoptosis Detection by Cell Death Detection ELISA—This assay was performed utilizing the cell death detection ELISAPLUS kit (Roche Applied Sciences) following the manufacturer's specification as described previously (42), with slight modifications. This assay utilizes a quantitative sandwich immunoassay principle to detect and quantitate the mono- and oligonucleosomes specifically released in the cytoplasm during apoptotic cell death. Cells plated on 6-well plates were treated with TZD or TRAIL, following which both adherent and floating (apoptotic) populations were harvested. Cells were lysed in Nonidet P-40 lysis buffer and the supernatant was collected following centrifugation at 3000 × g for 10 min. Nucleosomes were detected photometrically at 405 nm in an ELISA plate reader (SpectraMax 190, Molecular Devices) by measuring the peroxidase activity of 2,2′-azino-di(3-ethylbenzthiazolinsulfonate) as substrate. The readings were expressed as degree of apoptosis considering the corresponding untreated control as 1.
Apoptosis Detection by TUNEL Assay—Terminal deoxynucleotidyl transferase (TdT)-mediated deoxyuridine triphosphate (dUTP) nick end-labeling (TUNEL) assay was performed utilizing the in situ Cell Death Detection Kit (fluorescein), from Roche Applied Sciences according to the manufacturer's instructions. Briefly, Huh-7 cells plated in 2-well chamber slides were treated with various agents, fixed in freshly prepared 4% paraformaldehyde, and then permeabilized in 0.1% Triton X-100. Cells were then washed twice with phosphate-buffered saline and subjected to the TUNEL reaction at 37 °C in a humidified atmosphere in the dark for 60 min. At the end of the incubation cells were counterstained with DAPI. The fluorescent signal, emitted by fluorescein-labeled dUTP incorporated into fragmented DNA, was visualized by fluorescence microscopy (Axiovert 200 inverted microscope, Zeiss), interfaced with a camera (Axiocam), and image analyzer software (Axiovision, Zeiss).
Small Interference RNA (siRNA)—The β-catenin siRNA oligonucleotides with the sequence (sense 5′-AGCUGAUAUUGAUGGACAG-3′) along with the corresponding antisense oligonucleotide were synthesized from Dharmacon (Lafayette, CO) as described (43). A negative control siRNA (Santa Cruz Biotechnology) was used as control siRNA. Caspase-8 siRNA was obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and caspase-3 siRNA was obtained from Cell Signaling Technology (Danvers, MA). siRNA transfection was performed using Lipofectamine 2000 as per the manufacturer's instructions. Briefly, subconfluent Huh-7 cells were plated in 6-well plates in regular growth medium. The next day they were transfected with either 100 nm control siRNA or the target protein siRNA for 24 h followed by recovery in serum containing medium. After 72 h of siRNA transfection, the cells were treated with either DMSO or a combination of TZD and TRAIL for an additional 24 h followed by apoptosis assays.
Western Blot Analysis—Western blot analysis was performed following treatment of the cells with various agents and at different time intervals following the procedures described previously (34). Equal amounts of total protein were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, followed by Western blotting with the antibodies indicated. In the studies with cycloheximide, cells were treated with (20 μg/ml) cycloheximide in the presence or absence of TRAIL-TZD for various lengths of time up to a maximum of 8 h.
RESULTS
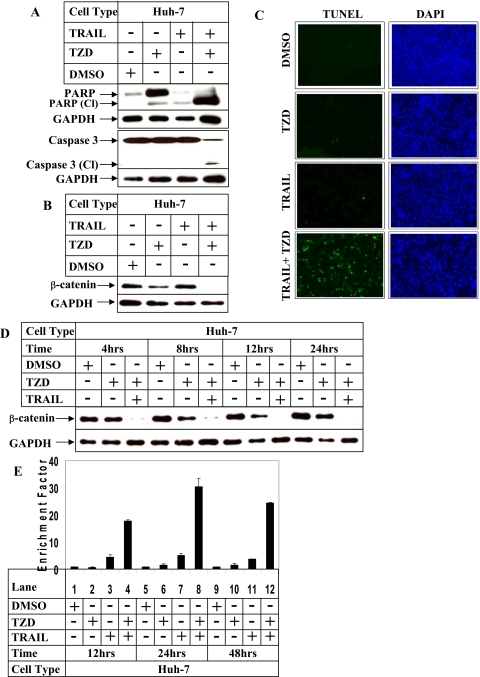
TRAIL and TZD-mediated Apoptosis in Hepatocellular Carcinoma Cells Is Associated with a Reduction of β-Catenin Expression—Earlier studies have reported that combinatorial treatment of TRAIL-resistant cells with TRAIL and PPARγ ligand (21) sensitizes them toward apoptosis. To identify a specific target mediating this apoptosis, we used the hepatocellular carcinoma cell line Huh-7, because they show resistance to apoptosis when treated with TRAIL alone (44). Western blot analysis indicated an activation of PARP and caspase-3 (indicated by the presence of the corresponding cleaved fragments) following incubation with the TRAIL and TZD combination (Fig. 1A, fourth lane). Apoptosis assays designed under similar conditions also showed an increase in Huh-7 cell apoptosis with the TRAIL and TZD combination, compared with either agent alone, as shown in Fig. 1C and supplemental Fig. S1A. To gain further insight into the effectors involved in mediating the pathway of apoptosis following combinatorial drug treatment, we estimated changes of β-catenin protein expression following treatment with TRAIL and TZD. Treatment of Huh-7 cells with the TRAIL and TZD combination resulted in a complete loss of β-catenin expression (Fig. 1B, fourth lane), compared with treatment with either drug alone, indicating a possible involvement of β-catenin in this apoptotic pathway.
FIGURE 1.
Effect of combinatorial treatment with TRAIL and TZD on apoptosis and β-catenin expression in Huh-7 hepatocellular carcinoma cells. A and B, Huh-7 cells were treated with 50 μm TZD and 100 ng/ml TRAIL either alone or in combination for 24 h, following which cells were harvested and total protein was extracted. Western blot analysis of the cell extracts was then performed with the indicated antibodies specific for apoptosis (A) or with antibodies against β-catenin and GAPDH (as control) (B). C, Huh-7 cells were treated as in A, and analyzed by TUNEL assay. The fluorescent signal, emitted by fluorescein-labeled dUTP incorporated into fragmented DNA (green), and from DAPI (blue) were visualized and photographed utilizing fluorescence microscopy (Axiovert 200 inverted microscope, Zeiss), interfaced with the camera (Axiocam). D, Huh-7 cells were treated with 50 μm TZD or 100 ng/ml TRAIL either alone or in combination for different periods of time. At the end of the incubation total protein was extracted and Western blot analysis was performed with antibodies against β-catenin and GAPDH (as control). E, Huh-7 cells were treated with the two agents for different periods of time. At the indicated times cells were harvested and apoptosis assays were performed using the cell death detection ELISAPLUS kit. The data in each set represent the mean ± S.D. of two independent experiments.
TRAIL and TZD Combination Decrease β-Catenin Expression and Induce Apoptosis in a Time- and Dose-dependent Manner—To establish a correlation between the decrease of β-catenin expression and increase in apoptosis, time course studies with the drug combination were designed next. These indicated that a decrease in β-catenin expression is maximal at 24 h (Fig. 1D), which coincided with maximal apoptosis (Fig. 1E and supplemental Fig. S1B). To determine the optimal concentration of TRAIL and TZD required for maximal β-catenin reduction, Western blot analyses were performed with Huh-7 cell extracts following treatment with increasing concentrations of either TZD or TRAIL. The results indicated that 50–100 μm TZD (Fig. 2A) and 50–100 ng/ml TRAIL (Fig. 2C) were optimal for reducing β-catenin expression maximally. This drug concentration also correlated with apoptosis, which was maximum with 50–100 μm TZD (Fig. 2B and supplemental Fig. S2A), and 100 ng/ml TRAIL (Fig. 2D and supplemental Fig. S2B). Based on these results, the 50 μm TZD and 100 ng/ml TRAIL combination was utilized in all future studies. These results indicated a close correlation between down-regulation of β-catenin expression and induction of apoptosis.
FIGURE 2.
Effect of increasing concentrations of TRAIL or TZD on apoptosis and β-catenin expression in Huh-7 hepatocellular carcinoma cells. A and B, Huh-7 cells were treated with either DMSO or a combination of 100 ng/ml TRAIL and increasing concentrations of TZD for 24 h. At the end of incubation, apoptotic cells were analyzed either via Western blot analysis with the indicated antibodies (A) or counted via DAPI staining (B). The data in each set represent the mean ± S.D. of two independent experiments. C and D, Huh-7 cells were treated with either DMSO or a combination of 50 μm TZD and increasing concentrations of TRAIL. At the end of the incubation, apoptotic cells were analyzed either via Western blot analysis (C) or counted via DAPI staining (D). The data in each set represent the mean ± S.D. of two independent experiments.
TRAIL and TZD-induced Apoptosis Is Associated with Reduced β-Catenin Expression in Different Cancer Cells—To determine any correlation of decreased β-catenin expression and TRAIL-TZD-induced apoptosis in other cancer cells, similar studies were performed with TRAIL-resistant prostate cancer (LNCaP), ovarian cancer (SKOV3), and colon cancer (RKO) cells. TRAIL-TZD combinatorial treatment resulted in a significant induction of apoptosis in LNCaP cells, as indicated by the apoptosis assays (Fig. 3A and supplemental Fig. S3A) and Western blot analysis showing PARP and caspase-3 cleavage (Fig. 3B). Increased apoptosis was also observed in SKOV3 (Fig. 3C and supplemental Fig. S3B), and RKO cells (Fig. 3D and supplemental Fig. S3C). This drug combination thus was effective in sensitizing various TRAIL-resistant cancer cells toward apoptosis. Western blot analysis carried out following a similar combinatorial drug treatment also showed a corresponding decrease of β-catenin expression following co-incubation with TRAIL and TZD (Fig. 3, E and F). These results indicated a correlation between reduced β-catenin expression and increased apoptosis, and suggested the possibility that decreasing β-catenin expression might increase drug-induced apoptosis in drug-resistant cancer cells.
FIGURE 3.
Effect of TRAIL-TZD combinatorial treatment on the apoptotic potential andβ-catenin expression in various cancer cells. LNCaP prostate cancer cells (A and B), SKOV3 ovarian cancer cells (C), and RKO colon cancer cells (D) were treated with 50 μm TZD or 100 ng/ml TRAIL either alone or in combination for 24 h. Apoptosis was estimated via the cell death detection ELISAPLUS kit (A, C, and D) and Western blot analysis with the indicated antibodies (B). The data in each set represent the mean ± S.D. of at least two independent experiments. LNCaP (E) and SKOV3 (F) cells were treated as in A, total cell extracts were prepared and Western blot analysis was performed with antibodies against β-catenin and GAPDH (as control).
In an attempt to identify candidate downstream targets of β-catenin mediating this apoptotic response, we focused on the following β-catenin target proteins: cyclin D1 (which mediates cell growth) and Bcl-xL and AKT (which mediate cell survival) (45–47). However, Western blot analysis of total protein extracts from Huh-7 and LNCaP cells following treatment with these drugs showed a significant decrease in the expression of cyclin D1, AKT, and Bcl-xL with TZD alone (supplemental Fig. S4, A and B, second lane), and did not seem to correlate with the apoptotic response (Figs. 1A and 3B).
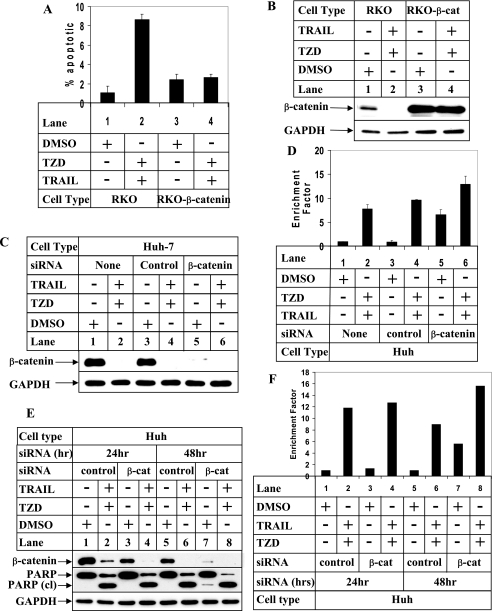
TRAIL and TZD-induced Apoptosis Involves β-Catenin—To determine whether β-catenin was involved in providing resistance to apoptosis, we utilized the RKO and RKO-β-catenin cells, the latter stably overexpressing a mutant stable form of β-catenin (39). Apoptosis assays showed an increase in the apoptotic potential of the RKO cells following treatment with TRAIL-TZD (Fig. 4A and supplemental Fig. S5, RKO panels), which was attenuated in the RKO-β-catenin cells (RKO-β-catenin panels). Western blot analysis showed higher levels of β-catenin expression in the RKO-β-catenin cells even after TRAIL-TZD treatment (Fig. 4B, RKO-β-catenin panel) compared with lower levels in the RKO cells (RKO panel). These results suggested that β-catenin overexpression can antagonize TRAIL-TZD-mediated apoptosis and might contribute to TRAIL resistance. To establish the involvement of β-catenin more conclusively, we designed siRNA studies to knockdown endogenous β-catenin expression in the Huh-7 hepatocellular carcinoma cells. Treatment with β-catenin siRNA resulted in a significant decrease in endogenous β-catenin expression (Fig. 4C, lane 5), which was unaffected by control siRNA (lane 3). Apoptosis assays designed following β-catenin knockdown showed an increase in basal apoptotic response in the absence of either TRAIL or TZD (Fig. 4D, lane 5), which was increased further following treatment with the TRAIL and TZD combination (lane 6). In addition, a time course analysis carried out following β-catenin knockdown showed that at 24 h where β-catenin knockdown is partial (Fig. 4E, compare lanes 1 and 3), there is no increase in spontaneous apoptosis in the absence of TRAIL and TZD (Fig. 4, E, compare lanes 1 and 3, PARP cleaved panel, and F, lanes 1 and 3). However, at 48 h when β-catenin knockdown is significant (Fig. 4E, compare lanes 5 and 7, β-catenin panel), there is an increase in spontaneous apoptosis in the absence of drugs as evident from PARP cleavage (Fig. 4E, lanes 5 and 7, PARP cleaved panel), and apoptosis assays (Fig. 4F, compare lanes 5 and 7). Addition of TRAIL and TZD at 48 h produces a synergistic effect on apoptosis (Fig. 4F, lane 8). These studies showed that overexpression of β-catenin provides a survival advantage to the cancer cells and knocking down β-catenin expression sensitizes them toward apoptosis.
FIGURE 4.
TRAIL-TZD-induced apoptosis is mediated via down-regulation ofβ-catenin expression. A, RKO or RKO-β-catenin cells were treated with either DMSO or the TRAIL (100 ng/ml) and TZD (50 μm) combination for 24 h. At the end of the incubation cells were harvested and apoptosis assays were performed using DAPI staining. The data in each set represent the mean ± S.D. of at least two independent experiments. B, RKO and RKO-β-catenin cells were treated as in A followed by Western blot analysis with antibodies against β-catenin and GAPDH (as control). C and D, Huh-7 cells were transfected with either none (lanes 1 and 2) or control siRNA (lanes 3 and 4), or β-catenin siRNA (lanes 5 and 6) followed by treatment with the TRAIL-TZD combination for 24 h. At the end of the incubation Western blot analysis was performed utilizing the antibodies indicated (C) or apoptosis assays were performed utilizing the cell death detection ELISAPLUS kit (D). The data represent the mean ± S.D. of two independent experiments. E and F, Huh-7 cells were transfected with control siRNA (lanes 1, 2 and 5, 6) or β-catenin siRNA (lanes 3, 4 and 7, 8) as in C for the indicated periods of time, followed by treatment with DMSO or a combination of 50 μm TZD and 100 ng/ml TRAIL for 24 h. At the end of the incubation Western blot analysis was performed utilizing the antibodies indicated (E) or apoptosis assays were performed utilizing the cell death detection ELISAPLUS kit (F).
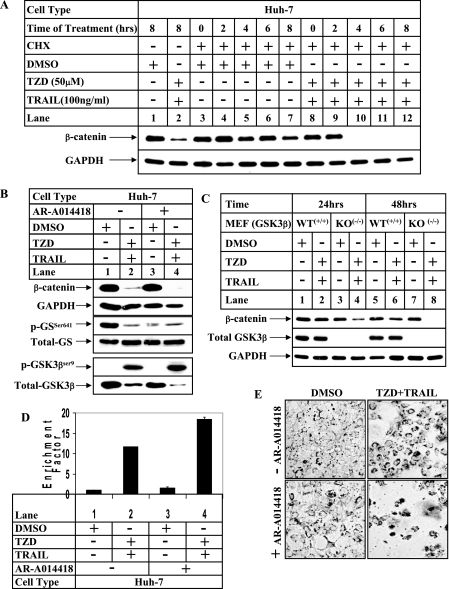
TRAIL-TZD Attenuates β-Catenin Expression at a Posttranslational Level Independent of GSK3β Activity—Because β-catenin expression can be regulated via different pathways, studies were designed next to determine the mechanism by which TRAIL-TZD stimulation might be regulating β-catenin expression. To determine whether this regulation was at a post-translational level, TRAIL-TZD studies were designed following treatment with the protein synthesis inhibitor cycloheximide. These studies showed a significant decrease in the half-life of β-catenin protein following treatment with the TRAIL-TZD combination compared with untreated controls (Fig. 5A, compare lanes 3–7 with lanes 8–12). This suggested that this drug combination regulated β-catenin expression at a post-translational level. In the conventional pathway of β-catenin degradation, GSK3β phosphorylates specific N-terminal residues of β-catenin in the presence of functional APC and Axin, which targets it toward proteasomal degradation (29). However, our earlier studies showed that TZD alone can degrade β-catenin via a novel GSK3β-APC and p53-independent pathway (34). To determine any involvement of GSK3β in the TRAIL-TZD-mediated decrease of β-catenin expression, cells were pretreated with a pharmacological inhibitor of GSK3β (AR-A014418) prior to TRAIL-TZD treatment. Pretreatment with AR-A014418 resulted in a decrease in phosphorylation of glycogen synthase (downstream target of GSK3β) as evident from the low phospho-glycogen synthaseSer641 levels in the absence of TRAIL-TZD (Fig. 5B, phospho-GSSer641 panel, compare lanes 1 and 3), indicating that AR-A014418 inhibited GSK3β activity. However, inhibition of GSK3β by AR-A014418 was unable to restore β-catenin expression following TRAIL-TZD treatment (β-catenin panel, compare lanes 2 and 4). Similar studies with other known GSK3β inhibitors including kenpaullone and lithium chloride were also unable to restore β-catenin expression in the presence of TRAIL-TZD (data not shown). In addition, treatment with TRAIL-TZD resulted in an increase in GSK3βSer9 phosphorylation (phospho-GSK3βSer9 panel) indicating an inactivation of the kinase. This is also evident from a reduction of phospho-glycogen synthaseSer641 levels following TRAIL-TZD treatment (phospho-GSSer641 panel, lane 2). To rule out the participation of GSK3β conclusively, TRAIL-TZD treatments were carried out with control (WT+/+) and GSK3β knock-out (KO-/-) MEFs (40). Time course analysis of these cells showed that TRAIL-TZD treatment was capable of reducing β-catenin expression in WT+/+ and to a greater extent in the GSK3β KO-/- MEFs (Fig. 5C, compare lanes 2 and 4, and lanes 6 and 8). In addition, pretreatment with proteasomal inhibitors MG132 or lactacystin were unable to restore the β-catenin full-length form following TRAIL-TZD treatment, indicating no involvement of proteasomes (Fig. 8C, compare lanes 2, 4, and 6, β-catenin-FL-panel). These results indicated that the TRAIL-TZD combination regulated β-catenin expression via a post-translational mechanism, independent of the conventional degradation pathway mediated by GSK3β.
FIGURE 5.
TRAIL-TZD regulatesβ-catenin expression via a post-translational mechanism independent of GSK3β. A, Huh-7 cells were treated with DMSO or the TRAIL-TZD combination in the presence of 20 μg/ml cycloheximide (CHX, lanes 3–12). Cells were harvested at 0, 2, 4, 6, and 8 h after treatment followed by Western blot analysis with the indicated antibodies. The Huh-7 cells treated with either DMSO or the TRAIL-TZD combination in the absence of cycloheximide were included in lanes 1 and 2, respectively, as positive controls to show TRAIL-TZD effects on β-catenin. B, Huh-7 cells were treated with the TRAIL-TZD combination for 24 h in the presence (+) or absence (-) of a pretreatment with the GSK3β inhibitor AR-A014418. At the end of the incubation total cell extracts were prepared and Western blot analysis was performed with the indicated antibodies. C, wild type (WT+/+) or GSK3β knock-out (KO-/-) MEFs were incubated with either DMSO or the TRAIL-TZD combination. Total cell extracts were prepared at the indicated time points followed by Western blot analysis with the antibodies shown. D, Huh-7 cells were treated as in B followed by apoptosis assay utilizing the cell death detection ELISAPLUS kit. The data represent the mean ± S.D. of two independent experiments. E, phase-contrast microscopic pictures showing apoptotic morphology of Huh-7 cells following treatment with either DMSO or the TRAIL-TZD combination in the presence (+) or absence (-) of AR-A014418.
FIGURE 8.
Effect of inhibition of caspase-3 and -8 on TRAIL-TZD-induced apoptosis and β-catenin cleavage. A, Huh-7 cells were treated with the TRAIL and TZD combination for 24 h following a pretreatment in the absence (-) or presence (+) of 50 μm Z-VAD-fmk. Western blot analyses of the cell extracts were performed with the indicated antibodies. B, Huh-7 cells were treated as in A, followed by apoptosis assay utilizing the cell death detection ELISAPLUS kit. The data represent the mean ± S.D. of two independent experiments. C, Huh-7 cells were treated with either DMSO or a combination of TRAIL and TZD for 10 h following pretreatment with 10 μm MG132 (lanes 3 and 4) or 5 μm lactacystin (lanes 5 and 6) for 1 h. Western blot analysis was performed with the cell extracts utilizing the antibodies indicated. D, Huh-7 cells were treated with the TRAIL and TZD combination for 24 h following pretreatment in the absence (-) or presence (+) of 50 μm Z-VAD-fmk (lanes 3 and 4), 25 μm DEVD-fmk (lanes 5 and 6), or 25 μm IETD-fmk (lanes 7 and 8). Western blot analyses of cell extracts were performed with the indicated antibodies. E, Huh-7 cells were transfected with either control-siRNA (lanes 1 and 2), caspase-3-siRNA (lanes 3 and 4), caspase-8-siRNA (lanes 5 and 6), or caspase-3 and -8 siRNA (lanes 7 and 8), followed by treatment with TRAIL and TZD for 24 h. Western blot analyses were then performed with the indicated antibodies.
The fact that GSK3β KO-/- MEFs showed a greater reduction of β-catenin when incubated with TRAIL-TZD indicated the interesting possibility that inhibition of GSK3β expression or activity might increase TRAIL-TZD-mediated apoptosis. In fact, apoptosis assays showed that AR-A014418-mediated inhibition of GSK3β activity produced synergistic effects on TRAIL-TZD-induced apoptosis (Fig. 5, D and E). These results are consistent with earlier reports that showed a profound increase in liver cell apoptosis in GSK3β knock-out mice (40).
TRAIL-TZD-induced Apoptosis Is Preceded by a Cleavage of β-Catenin—Earlier reports have shown that β-catenin can undergo caspase-induced cleavage during cellular apoptosis (48). Studies were designed next to determine whether the TRAIL-TZD combinatorial treatment induced any cleavage of β-catenin protein. Western blot assays were performed with an antibody created against the C-terminal 210 amino acid residues of β-catenin (from BD Biosciences) as well as with the one used in earlier studies from Zymed Laboratories Inc., created against the C-terminal 100 amino acid residues of β-catenin. The antibody from BD Biosciences detected the full-length (FL) form as well as a cleaved β-catenin fragment (<97 kDa), the latter specifically following TRAIL-TZD treatment (Fig. 6A, lane four, β-catenin-cleaved BD panel), whereas the Zymed Laboratories Inc. antibody only detected the full-length form (β-catenin, FL-Zymed panel). It was thus conceivable that TRAIL-TZD-mediated activation of caspases might lead to β-catenin cleavage, which contributes to the induction of apoptosis. The BD antibody also detected the full-length form following TRAIL-TZD treatment (Fig. 6A, lane four, β-catenin-FL-BD panel), which could be due to the presence of both apoptotic and non-apoptotic cells in this preparation. To confirm this, floating cells (fully apoptotic) and adherent cells (partially apoptotic) were separately isolated after TRAIL-TZD treatment and analyzed. Western blot analysis performed with these cells indicated the presence of both the cleaved and noncleaved forms of β-catenin in the partially apoptotic (adherent) population (Fig. 6B, lane 2), and only the cleaved form in the fully apoptotic (floating) population (lane 3). TRAIL-TZD-mediated cleavages of β-catenin were also detected in LNCaP (Fig. 6C) and RKO cells (Fig. 6D), indicating a similar pathway operating in all cancer cells. Interestingly, the degree of β-catenin cleavage correlated with the degree of apoptosis, with greater apoptosis in cells showing more cleavage (LNCaP, Figs. 3A and 6C) than those with less cleavage (Huh, Figs. 1E and 6A).
FIGURE 6.
Effect of TRAIL-TZD on β-catenin cleavage. A, Huh-7 cells were treated with 50 μm TZD or 100 ng/ml TRAIL either alone or in combination for 24 h. Western blot analysis was performed with the cell extracts utilizing antibodies against β-catenin from Zymed Laboratories Inc., recognizing the full-length (β-catenin-FL-Zymed panel), or from BD Biosciences, recognizing the full-length (β-catenin-FL-BD panel) and cleaved forms (β-catenin-cleaved-BD panel). The samples were also blotted with GAPDH antibody as control (GAPDH panel). B, Huh-7 cells were treated with either DMSO or a combination of 50 μm TZD and 100 ng/ml TRAIL for 24 h. At the end of the incubation, adherent (non-apoptotic, lane 1), adherent (partially apoptotic, lane 2), and floating (fully apoptotic, lane 3) populations of cells were separately isolated and analyzed by Western blot analysis with antibodies against β-catenin (BD Biosciences), PARP, cleaved caspase-3 or GAPDH (as control). LNCaP (C) and RKO (D) cells were treated as in A followed by Western blot analysis with antibodies against β-catenin (from BD Biosciences) and GAPDH.
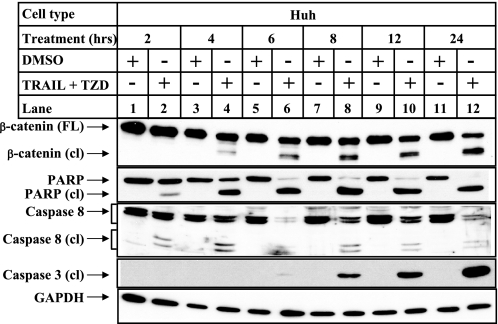
TRAIL-TZD-induced Apoptosis and Cleavage of β-Catenin Is Mediated via Caspases-3 and -8—To determine any involvement of caspases in β-catenin cleavage, and to understand whether cleavage of β-catenin occurred early or late during the apoptotic process, detailed time course studies were designed following incubation of Huh cells with the TRAIL and TZD combination. The results indicated an early activation of caspase-8 starting at 2 h (Fig. 7, lane 2, caspase-8 cleaved panel), followed by β-catenin cleavage starting at 4 h (lane 4, β-catenin cleaved panel) and caspase-3 cleavage starting at 6 h (lane 6, caspase-3 cleaved panel). Although β-catenin cleavage was initiated following caspase-8 activation and preceded caspase-3 cleavage, it was maximal at 24 h (lane 12), and correlated well with maximum caspase-3 cleavage. This suggested that β-catenin might be cleaved by both caspase-3 and -8 in this apoptotic cascade. In addition, this sequential and early cleavage pattern suggested that β-catenin cleavage might be a critical step in the progression of apoptosis.
FIGURE 7.
Time course of β-catenin and caspase cleavage during TRAIL-TZD-induced apoptosis. Huh-7 cells were treated with either DMSO or a combination of 50 μm TZD and 100 ng/ml TRAIL for the indicated periods of time. Western blot analysis was performed with the cell extracts utilizing the following antibodies: β-catenin antibody (from BD Biosciences), and PARP and caspase-8 antibodies recognizing full-length and cleaved forms of the respective proteins (panels 1, 2, and 3, respectively), caspase-3 antibody recognizing only the cleaved form (panel 4) and GAPDH antibody as control (panel 5).
To confirm whether TRAIL-TZD-induced cleavage of β-catenin involved caspase activity, Western blot analysis was performed with cell extracts obtained following pretreatment of the cells with the pan-caspase inhibitor Z-VAD-fmk. Pretreatment with caspase inhibitor abolished TRAIL-TZD-induced cleavage of β-catenin as shown in Fig. 8A (β-catenin-Cl panel, compare lanes 4 and 8), suggesting the involvement of caspases in mediating this. Z-VAD-fmk pretreatment also abolished TRAIL-TZD-induced cleavage of caspase-3 (caspase 3-Cl panel), confirming the efficacy of this inhibitor. Apoptosis assays were designed next following pretreatment with the pan-caspase inhibitor Z-VAD-fmk, to determine the role of caspases in mediating this apoptotic pathway. The results indicated a complete inhibition of TRAIL-TZD-induced apoptosis (Fig. 8B, compare lanes 2 and 4) following inhibition of caspase activity, suggesting that caspases are involved in both β-catenin cleavage and induction of apoptosis. In addition, pretreatment with two proteasomal inhibitors MG132 and lactacystin resulted in a synergistic increase in caspase-3 cleavage (Fig. 8C, compare lane 2 with 4 and 6, caspase-3 cleaved panel) and β-catenin cleavage (β-catenin cleaved panel), indicating again a correlation between increased caspase activity and β-catenin cleavage.
Because our time course analysis indicated an activation of both caspase-3 and -8 following incubation with TRAIL and TZD (Fig. 7), studies were designed next to identify the caspase(s) responsible for mediatingβ-catenin cleavage in this pathway. Western blot analysis performed with cell extracts following pretreatment separately with inhibitors of caspase-8 (IETD-fmk) or caspase-3 (DEVD-fmk) resulted in an inhibition of β-catenin cleavage (Fig. 8D, compare lanes 2, 6, and 8) as well as PARP and caspase-3 cleavage, suggesting the involvement of both of these caspases in regulating β-catenin cleavage. To further confirm the participation of these two caspases in this, siRNA studies were designed following knockdown of caspase-3 or -8 with their respective siRNA sequences. Western blot analysis showed a reduction in the expression of the full-length forms of caspase-3 (Fig. 8E, compare lane 1 with 3 and 7, caspase-3FL panel) and caspase-8 (compare lane 1 with 5 and 7, caspase-8 FL panel) following knockdown with their respective siRNA oligonucleotides. TRAIL and TZD studies designed following siRNA treatment showed a decrease in β-catenin cleavage following knockdown of both caspase-3 and caspase-8 (compare lane 2 with 4, 6, and 8, β-catenin-cleaved panel). This suggested that TRAIL-TZD-induced cleavage of β-catenin involves caspase-3 as well as 8. Cleavage of PARP and caspase-3 were also reduced by caspase-8 siRNA (PARP-cleaved and caspase-3-cleaved panels), suggesting both of them to be downstream of caspase-8 activation.
DISCUSSION
The fact that TRAIL can activate apoptosis specifically in tumor cells with little or no affect on the normal cells raises a great deal of enthusiasm toward utilizing TRAIL as a target therapeutic agent for treating cancer. However, this enthusiasm is seriously challenged by increasing reports that the majority of tumor cells develop resistance via different mechanisms that evade TRAIL-induced apoptosis (9, 10). Intense research is currently underway to develop drugs or drug combinations that can overcome TRAIL resistance and sensitize cancer cells toward TRAIL-induced apoptosis, without jeopardizing the tumor-specific pro-apoptotic properties of TRAIL. This requires a thorough understanding of the events and identification of molecular targets that contribute to tumor cell resistance. Development of an effective drug combination currently imposes the biggest challenge in designing the next generation of cancer drugs. Recent studies with TRAIL in combination with various nontoxic chemopreventive agents (49, 50), or PPARγ ligands (13, 20–24), have provided promising results in promoting TRAIL-induced apoptosis. Despite this progress, the mechanism by which this combination therapy overcomes TRAIL resistance is largely unclear and needs to be addressed to further increase the potency of this drug combination. This will also help in determining which cancer cells might respond to this combinatorial treatment.
The studies described here indicated that combinatorial treatment with TRAIL and TZD can sensitize various TRAIL-resistant cancer cells toward apoptosis, and identified β-catenin as a novel target of this apoptosis pathway. This is evident from an extensive decrease in full-length β-catenin expression, coinciding well with maximal apoptosis. This also suggested that antagonizing β-catenin expression or activity might be a critical event in promoting TRAIL sensitivity. This is confirmed by our studies showing that ectopic overexpression of β-catenin can antagonize TRAIL-TZD-mediated apoptosis (Figs. 4A and supplemental Fig. S5). Similarly, siRNA-mediated knockdown of endogenous β-catenin increased the basal level of apoptosis in the absence of TRAIL or TZD (Fig. 4, D and F). In fact β-catenin overexpression has been shown to be linked with pro-survival pathways in various cell types (27, 28, 39).
To gain a mechanistic insight toward the pathway involved in TRAIL-TZD-mediated modulation of β-catenin expression, we designed studies with the protein synthesis inhibitor cycloheximide. These studies indicated a reduction of β-catenin protein half-life following treatment with TRAIL-TZD, suggesting regulation at a post-translational level. In the conventional degradation pathway, β-catenin is phosphorylated at specific N-terminal residues by GSK3β in the presence of functional APC and Axin, and is then targeted toward ubiquitination-mediated proteasomal degradation (29). However, pretreatment of the cells with AR-A014418, a pharmacological inhibitor of GSK3β, was unable to restore β-catenin levels following TRAIL-TZD treatment (Fig. 5B), whereas it inhibited glycogen synthase Ser-641 phosphorylation, indicating inhibition of GSK3β activity (Fig. 5B, phospho-GSSer641 panel). Studies with other known GSK3β inhibitors (kenpaullone and lithium chloride) produced similar results (data not shown). In addition, studies with GSK3β KO-/- MEFs showed a greater decrease of β-catenin expression following TRAIL-TZD stimulation compared with the WT+/+ MEFs. These results, combined with the fact that TRAIL-TZD stimulation resulted in an increase in GSK3βSer9 phosphorylation (inhibition) suggested that TRAIL-TZD-mediated degradation of β-catenin was independent of GSK3β activity. In an attempt to identify the regulator(s) modulating β-catenin expression during the onset of apoptosis, we examined any potential role of caspases in cleaving β-catenin. In fact, earlier studies have shown caspase (48, 51, 52) and Calpain (53) induced cleavage of β-catenin. Our studies detected a TRAIL-TZD-induced cleavage of β-catenin (Fig. 6, A, C, and D), and a complete loss of the full-length form in the fully apoptotic population (Fig. 6B, lane 3). Inhibition of caspase activation by a pan-caspase inhibitor (Z-VAD-fmk) inhibited TRAIL-TZD-induced cleavage of β-catenin (Fig. 8A) as well as apoptosis (Fig. 8B), suggesting the involvement of caspases in mediating both events. This also indicated that cleavage of β-catenin is an important part of this apoptosis pathway, and might be a mechanism explaining how caspases induce apoptosis. Additional studies designed to identify the caspases mediating this indicated the involvement of caspases-3 and -8 (Fig. 8, D and E) in mediating β-catenin cleavage.
The functional significance of β-catenin cleavage in these studies is still unknown, and is unlikely to be to decrease the transactivation potential of β-catenin as suggested earlier (48). This is supported by two lines of evidence: (i) expression of β-catenin target proteins showed a significant decrease with TZD alone in both Huh-7 (supplemental Fig. S4A) and LNCaP cells (supplemental Fig. S4B), which was not associated with apoptosis (Figs. 1, E, lanes 2, 6, and 10, and A, lane 2); (ii) TZD treatment alone resulted in a significant decrease in β-catenin/TCF responsive reporter activity in our earlier studies (17), but was unable to induce any apoptosis in those cells (data not shown). It thus seemed that there is no correlation between the decrease in β-catenin/TCF transcriptional activity and the induction of apoptosis. Importantly, treatment with TZD alone also reduces full-length β-catenin expression as shown in Figs. 1, B, and 3, E and F (lane 2), which might contribute to the reduced β-catenin/TCF transcriptional activity as reported earlier (17). However, this decrease is not associated with increased apoptosis, indicating the possibility that TRAIL-TZD-induced cleavage of β-catenin is important for this apoptosis. It is likely that TRAIL-TZD-mediated cleavage of β-catenin results in the disassembly of adherens junctions, as was reported earlier with NO-donating aspirin (36), which contributes toward apoptosis.
Results described here indicate the possibility that targeting β-catenin in TRAIL-resistant cancer cells might be an effective means of increasing TRAIL sensitivity. Combination of TRAIL with drugs that can antagonize the β-catenin pathway might thus be effective in ameliorating TRAIL resistance in cancer cells. Abnormal activation of the β-catenin pathway has been linked with various tumorigenic pathways including those in liver and colon (30, 31). It is important to note that this activation of the β-catenin pathway is mostly due to either inactivating mutations of APC or Axin, or stabilizing mutations of β-catenin itself, all of which result in evasion of GSK3β-mediated degradation. Because TRAIL-TZD-mediated degradation is independent of GSK3β pathway, this combinatorial therapy might be effective in treating those tumors that have GSK3β degradation-resistant β-catenin activation. Identification of the effectors and signaling pathways involved will provide important insight toward increasing the potency of this novel therapeutic approach.
Supplementary Material
Acknowledgments
We are grateful to Dr. Robert E. Lanford for providing the Huh-7 cells, Drs. Wallace K. MacNaughton and Hongying Wang for the RKO and RKO-β-catenin cells, and Dr. James R. Woodgett for the WT and GSK3β knock-out (-/-) MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grants CA121221 (to B. R.) and GM55835 (to A. R.). This work was also supported by Veterans Affairs Merit and VISN17 awards (to B. R.), the Susan Komen Breast Cancer Foundation, and a Veterans Affairs Merit award (to A. R.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: TNFα, tumor necrosis factor α; APC, adenomatous polyposis coli; DAPI, 4,6-diamidino-2-phenylindole dihydrochloride; FasL, Fas ligand; FL, full-length; GSK3β, glycogen synthase kinase 3β; KO, knockout; MEFs, mouse embryonic fibroblasts; PARP, poly(ADP-ribose) polymerase; PPARγ, peroxisome proliferator-activated receptor γ; siRNA, small interference RNA; TCF/LEF, T cell factor/lymphoid enhancer factor; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; TZD, troglitazone; WT, wild type; ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone.
References
- 1.Ashkenazi, A., Pai, R. C., Fong, S., Leung, S., Lawrence, D. A., Marsters, S. A., Blackie, C., Chang, L., McMurtrey, A. E., Hebert, A., DeForge, L., Koumenis, I. L., Lewis, D., Harris, L., Bussiere, J., Koeppen, H., Shahrokh, Z., and Schwall, R. H. (1999) J. Clin. Investig. 104 155-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., Smith, C., Smolak, P., Goodwin, R. G., Rauch, C. T., Schuh, J. C., and Lynch, D. H. (1999) Nat. Med. 5 157-163 [DOI] [PubMed] [Google Scholar]
- 3.Fesik, S. W. (2005) Nat. Rev. Cancer 5 876-885 [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi, A. (2002) Nat. Rev. Cancer 2 420-430 [DOI] [PubMed] [Google Scholar]
- 5.Sprick, M. R., Weigand, M. A., Rieser, E., Rauch, C. T., Juo, P., Blenis, J., Krammer, P. H., and Walczak, H. (2000) Immunity 12 599-609 [DOI] [PubMed] [Google Scholar]
- 6.Wang, J., Chun, H. J., Wong, W., Spencer, D. M., and Lenardo, M. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 13884-13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley, S. K., and Ashkenazi, A. (2004) Curr. Opin. Pharmacol. 4 333-339 [DOI] [PubMed] [Google Scholar]
- 8.Bonavida, B., Ng, C. P., Jazirehi, A., Schiller, G., and Mizutani, Y. (1999) Int. J. Oncol. 15 793-802 [DOI] [PubMed] [Google Scholar]
- 9.Chinnaiyan, A. M., Prasad, U., Shankar, S., Hamstra, D. A., Shanaiah, M., Chenevert, T. L., Ross, B. D., and Rehemtulla, A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1754-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D., and Ashkenazi, A. (2002) Nat. Med. 8 274-281 [DOI] [PubMed] [Google Scholar]
- 11.Ashkenazi, A., and Dixit, V. M. (1998) Science 281 1305-1308 [DOI] [PubMed] [Google Scholar]
- 12.Emery, J. G., McDonnell, P., Burke, M. B., Deen, K. C., Lyn, S., Silverman, C., Dul, E., Appelbaum, E. R., Eichman, C., DiPrinzio, R., Dodds, R. A., James, I. E., Rosenberg, M., Lee, J. C., and Young, P. R. (1998) J. Biol. Chem. 273 14363-14367 [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y., Suh, N., Sporn, M., and Reed, J. C. (2002) J. Biol. Chem. 277 22320-22329 [DOI] [PubMed] [Google Scholar]
- 14.Teitz, T., Wei, T., Valentine, M. B., Vanin, E. F., Grenet, J., Valentine, V. A., Behm, F. G., Look, A. T., Lahti, J. M., and Kidd, V. J. (2000) Nat. Med. 6 529-535 [DOI] [PubMed] [Google Scholar]
- 15.Keane, M. M., Ettenberg, S. A., Nau, M. M., Russell, E. K., and Lipkowitz, S. (1999) Cancer Res. 59 734-741 [PubMed] [Google Scholar]
- 16.Kliewer, S. A., Sundseth, S. S., Jones, S. A., Brown, P. J., Wisely, G. B., Koble, C. S., Devchand, P., Wahli, W., Willson, T. M., Lenhard, J. M., and Lehmann, J. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 4318-4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma, C., Pradeep, A., Pestell, R. G., and Rana, B. (2004) J. Biol. Chem. 279 16927-16938 [DOI] [PubMed] [Google Scholar]
- 18.Sarraf, P., Mueller, E., Jones, D., King, F. J., DeAngelo, D. J., Partridge, J. B., Holden, S. A., Chen, L. B., Singer, S., Fletcher, C., and Spiegelman, B. M. (1998) Nat. Med. 4 1046-1052 [DOI] [PubMed] [Google Scholar]
- 19.Yu, J., Qiao, L., Zimmermann, L., Ebert, M. P., Zhang, H., Lin, W., Rocken, C., Malfertheiner, P., and Farrell, G. C. (2006) Hepatology 43 134-143 [DOI] [PubMed] [Google Scholar]
- 20.Goke, R., Goke, A., Goke, B., and Chen, Y. (2000) Cell Immunol. 201 77-82 [DOI] [PubMed] [Google Scholar]
- 21.Lu, M., Kwan, T., Yu, C., Chen, F., Freedman, B., Schafer, J. M., Lee, E. J., Jameson, J. L., Jordan, V. C., and Cryns, V. L. (2005) J. Biol. Chem. 280 6742-6751 [DOI] [PubMed] [Google Scholar]
- 22.Hyer, M. L., Croxton, R., Krajewska, M., Krajewski, S., Kress, C. L., Lu, M., Suh, N., Sporn, M. B., Cryns, V. L., Zapata, J. M., and Reed, J. C. (2005) Cancer Res. 65 4799-4808 [DOI] [PubMed] [Google Scholar]
- 23.Zou, W., Liu, X., Yue, P., Khuri, F. R., and Sun, S. Y. (2007) Cancer Biol. Ther. 6 99-106 [DOI] [PubMed] [Google Scholar]
- 24.Schultze, K., Bock, B., Eckert, A., Oevermann, L., Ramacher, D., Wiestler, O., and Roth, W. (2006) Apoptosis 11 1503-1512 [DOI] [PubMed] [Google Scholar]
- 25.Bienz, M., and Clevers, H. (2000) Cell 103 311-320 [DOI] [PubMed] [Google Scholar]
- 26.Nhieu, J. T., Renard, C. A., Wei, Y., Cherqui, D., Zafrani, E. S., and Buendia, M. A. (1999) Am. J. Pathol. 155 703-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng, G., Apte, U., Cieply, B., Singh, S., and Monga, S. P. (2007) Neoplasia 9 951-959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, X., Yuan, Y., Zeng, G., Apte, U., Thompson, M. D., Cieply, B., Stolz, D. B., Michalopoulos, G. K., Kaestner, K. H., and Monga, S. P. (2008) Hepatology 47 1667-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubinfeld, B., Albert, I., Porfiri, E., Fiol, C., Munemitsu, S., and Polakis, P. (1996) Science 272 1023-1026 [DOI] [PubMed] [Google Scholar]
- 30.de La Coste, A., Romagnolo, B., Billuart, P., Renard, C. A., Buendia, M. A., Soubrane, O., Fabre, M., Chelly, J., Beldjord, C., Kahn, A., and Perret, C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 8847-8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korinek, V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B., and Clevers, H. (1997) Science 275 1784-1787 [DOI] [PubMed] [Google Scholar]
- 32.Matsuzawa, S. I., and Reed, J. C. (2001) Mol. Cell 7 915-926 [DOI] [PubMed] [Google Scholar]
- 33.Xiao, J. H., Ghosn, C., Hinchman, C., Forbes, C., Wang, J., Snider, N., Cordrey, A., Zhao, Y., and Chandraratna, R. A. (2003) J. Biol. Chem. 278 29954-29962 [DOI] [PubMed] [Google Scholar]
- 34.Sharma, C., Pradeep, A., Wong, L., Rana, A., and Rana, B. (2004) J. Biol. Chem. 279 35583-35594 [DOI] [PubMed] [Google Scholar]
- 35.Miyoshi, Y., Iwao, K., Nagasawa, Y., Aihara, T., Sasaki, Y., Imaoka, S., Murata, M., Shimano, T., and Nakamura, Y. (1998) Cancer Res. 58 2524-2527 [PubMed] [Google Scholar]
- 36.Gao, J., Liu, X., and Rigas, B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 17207-17212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinzler, K. W., and Vogelstein, B. (1996) Cell 87 159-170 [DOI] [PubMed] [Google Scholar]
- 38.Sureau, C., Moriarty, A. M., Thornton, G. B., and Lanford, R. E. (1992) J. Virol. 66 1241-1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, H., and MacNaughton, W. K. (2005) Cancer Res. 65 8604-8607 [DOI] [PubMed] [Google Scholar]
- 40.Hoeflich, K. P., Luo, J., Rubie, E. A., Tsao, M. S., Jin, O., and Woodgett, J. R. (2000) Nature 406 86-90 [DOI] [PubMed] [Google Scholar]
- 41.Barthwal, M. K., Sathyanarayana, P., Kundu, C. N., Rana, B., Pradeep, A., Sharma, C., Woodgett, J. R., and Rana, A. (2003) J. Biol. Chem. 278 3897-3902 [DOI] [PubMed] [Google Scholar]
- 42.Nayak, T. K., Norenberg, J. P., Anderson, T. L., Prossnitz, E. R., Stabin, M. G., and Atcher, R. W. (2007) Nucl. Med. Biol. 34 185-193 [DOI] [PubMed] [Google Scholar]
- 43.Verma, U. N., Surabhi, R. M., Schmaltieg, A., Becerra, C., and Gaynor, R. B. (2003) Clin. Cancer Res. 9 1291-1300 [PubMed] [Google Scholar]
- 44.Yamanaka, T., Shiraki, K., Sugimoto, K., Ito, T., Fujikawa, K., Ito, M., Takase, K., Moriyama, M., Nakano, T., and Suzuki, A. (2000) Hepatology 32 482-490 [DOI] [PubMed] [Google Scholar]
- 45.Tetsu, O., and McCormick, F. (1999) Nature 398 422-426 [DOI] [PubMed] [Google Scholar]
- 46.Xie, H., Huang, Z., Sadim, M. S., and Sun, Z. (2005) J. Immunol. 175 7981-7988 [DOI] [PubMed] [Google Scholar]
- 47.Dihlmann, S., Kloor, M., Fallsehr, C., and von Knebel Doeberitz, M. (2005) Carcinogenesis 26 1503-1512 [DOI] [PubMed] [Google Scholar]
- 48.Steinhusen, U., Badock, V., Bauer, A., Behrens, J., Wittman-Liebold, B., Dorken, B., and Bommert, K. (2000) J. Biol. Chem. 275 16345-16353 [DOI] [PubMed] [Google Scholar]
- 49.He, Q., Luo, X., Huang, Y., and Sheikh, M. S. (2002) Oncogene 21 6032-6040 [DOI] [PubMed] [Google Scholar]
- 50.Fulda, S., and Debatin, K. M. (2004) Cancer Res. 64 337-346 [DOI] [PubMed] [Google Scholar]
- 51.Fukuda, K. (1999) Int. J. Biochem. Cell Biol. 31 519-529 [DOI] [PubMed] [Google Scholar]
- 52.Ling, Y., Zhong, Y., and Perez-Soler, R. (2001) Mol. Pharmacol. 59 593-603 [DOI] [PubMed] [Google Scholar]
- 53.Abe, K., and Takeichi, M. (2007) Neuron 53 387-397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.