Abstract
It was well accepted that only B-lymphocytes and plasma cells expressed immunoglobulin (Ig) gene. However, our group and others have confirmed that non-B-cells, such as epithelial cancer cells, can also express Ig. The aim of this work is to elucidate the role of non-B-cell-derived Ig by investigating the characteristics of the Ig heavy chain (IgH) gene repertoire in epithelial cancer cells. We cloned and sequenced 89 VHDJH (V-D-J recombination of the IgH variable region) transcripts by microdissecting cells from eight different types of epithelial cancers and two cancer cell lines (HT-29 and HeLa S3). The cancer-derived Ig gene repertoire showed specific restricted patterns of VHDJH recombination with seven sets of predominant VHDJH sequences. Surprisingly, within a set of VHDJH recombination, the variable (V) sequences derived from different cancer types had not only identical heavy chain variable (VH), diversity (D), and joining (JH) segments usage, but also identical junctions and mutation targets in the VH region. The VHγDJHγ (but not VHμDJHμ) in the cancer-derived sequences had a high mutation rate; however, it was shown that the mechanism of hypermutation was different from antigen selection in B-cell-derived VHγDJHγsequences. In contrast to VHμDJHμ, the VHγDJHγ sequences did not appear to originate from classical class switching. These results suggest that cancer-derived Ig genes have a distinct repertoire that may have implications for their role in carcinogenesis.
Immunoglobulins (Ig) were discovered more than a century ago, yet the understanding of these proteins continues to evolve. Until 1950, most scientists believed that cells from various types of tissues could express Ig (1). However, it was shown that B-lymphocytes from bone marrow secreted Ig, although other hematopoietic cells did not (2), and that levels of serum Ig decreased with B-cell disfigurement (2, 3). These were thought to indicate that only B-lymphocytes could express Ig; non-immunocytes could not.
In 1976, Tonegawa discovered that Ig gene recombination was the mechanism behind antibody diversity in B-lymphocyte-derived plasma cells. Ig gene recombination, as theorized previously by Dreyer and Bennett, was confirmed to exist in mouse myeloma cells using a probe against the Ig mRNA kappa chain (4, 5).
Subsequently, Cleary et al. compared the restriction enzyme map of the Ig gene in B-lymphocytes with that of the genes in cell types such as germ-line using Southern blot analysis and found that B-cell and non-B-cell restriction maps differed. These results further strengthened the hypothesis that Ig gene recombination only occurred in B-lymphocytes. Consequently, Ig gene recombination became a criterion for identifying B-cells (6, 7). Some tumor cells expressing both epithelial cell markers and Ig gene recombination were thus believed to originate from B-cells (6, 8).
Immunoglobulin gene recombination has been detected in T-cell lymphomas and acute non-lymphocytic leukemias (9, 10). However, there is no substantial evidence that Ig gene recombination, transcription, and production could occur in non-immunocytes.
Patients with non-hematopoietic tumors, including carcinomas of the brain, breast, colon, and liver, may have elevated levels of serum IgG, IgA, and/or IgM (11–13). Additionally, many patients with malignant tumors of epithelial origin have been shown to have monoclonal or oligoclonal gamma globulinemia (14–16). These antibodies had been presumed to be produced by B-lymphocytes and plasma cells. However, recent studies from our group and others have demonstrated that both malignant and normal epithelial cells could express Ig.
In 1996, we first reported the detection of IgG-like molecules in breast and colon carcinoma cells and showed that these molecules were not present in their normal epithelial cell counterparts by immunohistochemical staining and Western blot analysis (17). In studies of human cancer cell lines, IgG-like proteins were detected in both the tumor cells and the culture supernatant (18). Kimoto (19) identified transcripts of the Ig constant region and the T-cell receptor (TCR) gene in five epithelial-derived cancer cell lines (SW1116, HEp2, MCF-7, MDA-MB-231, and HC48) using nested reverse transcription-PCR (RT-PCR).3 In 2003, we demonstrated that tumor cells isolated from epithelial cancers and cell lines could secrete IgG using Western blot analysis and N terminus sequencing, and we detected both cytoplasmic and secreted IgG in cells from carcinomas of the lung, breast, liver, and colon, as well as epithelial cell lines (20). IgG transcription was also detected by in situ hybridization, Northern blot analysis, and single cell RT-PCR (20). In 2004, it was reported that human cervical cancer cells could express Ig mRNA and protein (21). Recent studies have also confirmed the expression of Ig and activation-induced cytidine deaminase (AID) in six breast cancer cell lines (BT474, MDA-MB-231, MCF-7, SKBR3, T47D, and ZR75-1) (22). Furthermore, we recently reported that IgA and IgG were expressed in numerous oral epithelial tumor cells (23). Despite the detection of Ig in numerous cancer cell types, Ig specificity and variable region repertoire are poorly characterized.
B-cells are known to generate Ig diversity by several mechanisms. During the formation of Ig in B-cells from bone marrow, two recombinant events bring different VH, DH, and JH exons together to form heavy chains. Additionally, short sequences are inserted between VH and DH and between DH and JH to generate further diversity. Subsequent encounters with antigens in the germinal centers drive B-cell to undergo somatic hypermutation (SHM) and class switching, thus generating even greater diversity.
In the present study, we analyzed the V region transcripts in rearranged IgH in eight cancer cell samples from microdissected epithelial cancer tissue and in two cell lines (HT-29 and HeLaS3) using RT-PCR and sequence analysis. We found that cancer-derived Ig genes show classic VHDJH recombination. Reminiscent of classical recombination, additional short sequences were inserted between the VH and DH segments and the DH and JH segments, Rearranged Ig μ- and γ-chain gene sequences were expressed in cells, and SHM was observed in the VH segment of the γ-chain genes. However, the cancer-derived Ig gene repertoire also displayed several distinct characteristics.
EXPERIMENTAL PROCEDURES
Sample Assays—For laser capture microdissection (LCM) and RT-PCR analysis of the cancer-derived Ig gene, eight tumor samples from therapeutic excisions of breast invasive ductal carcinoma (n = 3), colon carcinoma (n = 2), squamous cell carcinoma of the lung (n = 1, from the tissue bank of Peking University School of Oncology), squamous cell carcinoma of the oral cavity (n = 1), and basal cell carcinoma of the oral cavity (n = 1) from the Department of Pathology at Peking University School of Stomatology were included with informed consent from the patients. Ethical approval of the study was granted by the Peking University Health Service Trust Research Ethics Committee. Carcinoma samples were embedded in Tissue-Tek OCT Compound (Sakura, IMEB International Medical Equipment, Inc., San Marcos, CA) and snap-frozen in liquid nitrogen immediately after surgery. Serial frozen sections (8 μm) were cut with a cryostat and mounted on slides treated with 0.1% DEPC for sterilization. Sections were air-dried, fixed in 70% ethanol, and evaluated with hematoxylin and eosin stain or immunohistochemical staining for LCM.
Immunohistochemistry—The slides were then incubated with 0.3% hydrogen peroxide for 5 min, washed with PBS, and blocked in PBS with 10% normal goat serum for 10 min. After removal of excess blocking buffer, indirect immunohistochemical staining was performed with monoclonal antibodies against human epithelial cell adhesion molecules (EpCAM) (1:100, Abcam, Cambridge, MA). Slides were incubated at 37 °C for 45 min, washed thoroughly, and then incubated with horseradish peroxidase-labeled goat anti-mouse IgG (1:100, Dako, Carpinteria, CA) at 37 °C for 45 min. Slides were washed again and bound antibodies were detected using 3,3′-diaminobenzidine (DAB, Sigma Aldrich). The mouse IgG was used as an isotype control.
HT-29 and HeLa S3 Cell Cultures—HT-29 (from colon cancer) and HeLa S3 (from cervical cancer) cell lines were supplied by the Peking University Center for Human Disease Genomics. These two cancer cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone/Thermo Fisher Scientific Inc., Waltham, MA) and l-glutamine (2 mm) at 37°C in a humidified 5% CO2.
Isolation and Preparation of Mononuclear Cells from Peripheral Blood—Two samples of peripheral blood were obtained from two healthy donors. Mononuclear cells (MNC) were isolated from 5 ml of peripheral blood using two-step discontinuous Ficoll-Hypaque gradients (Second Chemistry Factory, Shanghai, China). The white gradient layer containing MNC was recovered and washed with 0.01 m PBS, and the isolated MNC used immediately for total RNA extraction.
LCM and RT-PCR of Cancer Cells—LCM was carried out as previously described (20). Briefly, to minimize contamination of infiltrating B-lymphocytes or plasma cells in cancer tissues, only large EpCAM+ cells in cancer cell nests without lymphocyte or plasma cell infiltration were dissected from fresh biopsy tissues of carcinomas of the breast, colon, oral cavity, and lung by LCM. Total RNA of microdissected cancer cells was extracted using RNeasy Micro kit (Qiagen) according to the manufacturer's instructions. Reverse transcription was carried out with the Sensiscript RT kit (Qiagen) according to the manufacturer's instructions. Touchdown PCR was then performed using 1 μl of each reverse transcription reaction with LA Taq polymerase (TaKaRa Bio USA, Madison, WI) as previously described (20). To amplify the human IgVH gene of the γ chain and the μ chain by nested PCR, the first round of PCR was carried out with upstream primers for VH1 (5′-GAGGTGCAGCTCGAGGAGTCTGGG-3′), VH2 (5′-CAGGTGCAGCTCGAGCAGTCTGGG-3′), VH3 (5′-CAGGTACAGCTCGAGCAGTCAGG-3′), and VH4 (5′-CAGGTGCAGCTGCTCGAGTCGGG-3′), coupled with CH1 region primer (CγCH1, 5′-ACACCGTCACCGGTTCGG-3′;CμCH1, 5′-ACGCTGCTCGTATCCGACGGG-3′). Conditions for the second round of PCR were the same as the first round, except with the JH primer (5′-GTGACCAGGGTNCCTTGGCCCCAAG-3′) replacing the CH1 primer. To confirm that there was no B-lymphocyte contamination, we studied CD19 (a B-lymphocyte marker) transcription using the same cDNA used for the amplification of the IgH V gene, and the following primer set: CD19 up, 5′-AAGGGGCCTAAGTCATTGCT-3′ (sense), and CD19 down, 5′-CACGTTCCCGTACTGGTTCT-3′ (antisense).
RT-PCR for Cell Lines and Peripheral Blood Lymphocytes— Total RNA were extracted from HT-29, HeLa S3, and peripheral blood MNC using TRIzol reagent (Invitrogen). Reverse transcription of total RNA from each of these samples was performed using a Superscript II RT kit (Invitrogen) according to the manufacturer's instructions. The human IgVH gene of the γ-or μ-chain and CD19 gene were amplified using the same PCR conditions and primers as those employed for the RT-PCR of cells obtained by LCM.
Sequencing and Analysis of Rearranged Genes—PCR products were cloned in a pGEM-T Easy Vector (Promega, Madison, WI) and sequenced with an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). The VHDJH sequences were compared with those found in the BLAST and Immunogenetics data bases (24) to identify the best matching germline gene segments and V(D)J junctions. V gene sequences belonging to a set of VHDJH recombinants were defined on the basis of identical VH, DH, and JH gene usage and V-D and D-J junction sequences. The repertoire of the cancer-derived Ig V genes was compared with that of the two healthy donors' peripheral blood lymphocytes (PBL) and published B-lymphocyte-derived Ig variable region sequences (25–28).
Determination of Taq Polymerase and Sequencing Fidelity— To analyze whether SHM was present in the cancer-derived VH genes, we first calculated both the error rate resulting from the TaKaRa LA Taq polymerase used for PCR and from the sequencing. The results showed that in a known sequence of 17 clones generated by the same RT-PCR method and cloning procedures that were used for the VH genes, the LA-Taq error rate was 0.07%, which equals to 0.02 mutations per VH clone. The sequencing induced error rate was 0.
Analysis of IgVH Gene Mutations—The pattern of mutations of each sequence was compared with that of germline sequence to identify hybrid sequences derived from recombinant VH gene segments. To analyze whether the mechanism of SHM occurring in cancer-derived Ig variable region was similar to that caused by antigen selection in B-cell-derived Ig, the mutation frequency of both the RGYW and the WRCY motifs (the mutable position is G:C, which is underlined; r = A or G, Y = C or T, and W = T or A) used as a principal hotspot for AID-induced G:U lesions was calculated (29–31). In addition, we determined the replacement-to-silent mutation (R/S) ratio in the CDRII and FWRIII regions. A VH sequence was considered to be antigen-selected when the R/S ratio was higher than 2.9 in the CDRII and lower than 1.5 in the FWRIII region (32). The error-prone polymerases mainly induced A/T mutations that were identified as a principal site, and the dinucleotide target WA (AA or TA) mutation induced by the error-prone polymerases was involved in the mismatch repair of SHM (33–35). Therefore the WA/TW ratio was analyzed using JOINSOLVER (36).
Statistical Analysis—The distribution of VH and JH gene family usage and the calculation of mutations in WA versus TW were assessed using the Chi-squared test. Values were considered statistically significant when p < 0.05.
RESULTS
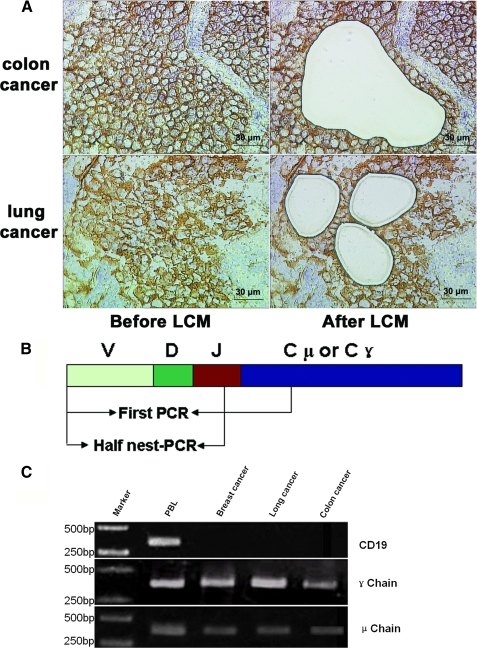
Cancer-derived IgVH Transcripts Were Amplified by RT-PCR— The Ig gene transcripts and repertoires were detected following LCM of cells from carcinomas from the colon, breast, oral, and lung (Fig. 1A) and HT-29 and HeLa S3 cell lines. Peripheral blood lymphocytes from two healthy donors served as positive controls. The rearranged V region genes of the γ and μ chains were successfully amplified and cloned from both the cancer cells and PBL (Fig. 1B). As expected, CD19 transcript was not detected in any of the cancer cell cDNA libraries, but was detected in PBL from the two donors (data not shown).
FIGURE 1.
A, capture of the cancer cells by LCM. After frozen cancer tissue slices were stained with anti-human EpCAM antibody, about 500 EpCAM+ cells per cancer nest sample were identified and dissected from squamous cell carcinoma of the lung and colon cancer tissues by LCM. Scale bars, 30 μm. B, diagrams (not to scale) of the DNA segments analyzed in this study. The arrows indicate the positions of the primers used for amplification of the segments. C, results of LCM-related RT-PCR. The variable region fragments of Ig γ chain and μ chain were amplified. CD19 was used as positive control for B-lymphocytes.
Functional IgVH Transcripts Were Expressed by Cancer Cells— Sequencing results from the eight cancer samples and two cancer cell lines demonstrated that 31 of the 35 VHμDμ JHμ sequences and 47 of the 54 VHγDγJHγ sequences showed functional V region gene recombination (Table 1). Among the 89 sequences examined, 11 (12.3%) were identified as nonfunctional because of mutations that introduced stop codons into the V region. This rate of nonfunctional VHDJH recombination was similar to that in B-cell-derived Ig (25). The functional V region genes showed typical VHDJH recombination with an N or P insert, and the JH gene recombination occurred primarily at TG nucleotide sequences, as in B-cell-derived Ig (33). None of the 78 functional VHDJH recombinations assessed was identified in the two control PBL samples or in normal B-lymphocytes, B-cell chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), or tumor-infiltrating B-lymphocytes from ductal or medullary breast carcinoma and cervical carcinoma (based on data obtained from public databases, Refs. 25–28). Some of the cancer-derived VHDJH sequences identified from our study were submitted to the GenBank™ data base (GenBank™ Accession Numbers: AY270187 -AY270190, AY247234, AY286495, and AY505568).
TABLE 1.
Rate of functional rearrangement in 89 VHDJH recombinations from different cancer types
| Sample origin | Ig type | No. of sample | No. of clone | No. of functional VHDJH recombination | No. of non-functional VHDJH recombination | |
|---|---|---|---|---|---|---|
| PBL | γ chain | 2 | 6 | 5 | 1 | |
| μ chain | 2 | 4 | 4 | 0 | ||
| Total | 4 | 10 | 9 | 1 (10%)a | ||
| Breast cancer | γ chain | 3 | 12 | 10 | 2 | |
| μ chain | 3 | 10 | 10 | 0 | ||
| Lung cancer | γ chain | 1 | 6 | 6 | 0 | |
| μ chain | 1 | 2 | 2 | 0 | ||
| Colon cancer | γ chain | 2 | 11 | 10 | 1 | |
| μ chain | 1 | 5 | 4 | 1 | ||
| Oral carcinoma | γ chain | 2 | 12 | 10 | 2 | |
| μ chain | 1 | 9 | 7 | 2 | ||
| HT29 cell | γ chain | 1 | 7 | 7 | 0 | |
| μ chain | 1 | 6 | 5 | 1 | ||
| Hela S3 cell | γ chain | 1 | 6 | 4 | 2 | |
| μ chain | 1 | 3 | 3 | 0 | ||
| Total (cancer) | 18 | 89 | 78 | 11 (12%) | ||
The percentage of all sequences corresponding to each group of genes is shown in parentheses
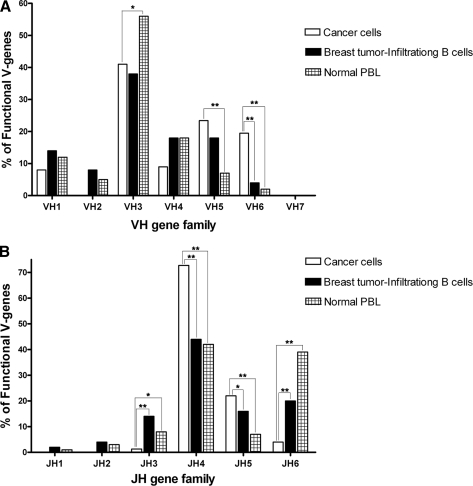
Cancer-derived IgVH Genes Showed Restricted Pattern of VHDJH Combinations—To detect usage of cancer-derived IgVH genes, we used primers that could apply for almost all VHDJH rearrangements. When comparing the cancer-derived IgVH gene distribution with the best matching functional germline IgVH genes within each family, all seven of the VH families were amplified, except VH2. The VH3 gene family was used most often, and the most frequently encountered germline sequences were VH5-51, VH6-1, VH3-33, VH3-15, VH3-30, and VH3-23, which accounted for 23.7, 18.7, 14.67, 8.0, 6.67, and 5.33% of all of the potentially possible functional cancer-derived VH genes, respectively. The VH5-51 and VH6-1 frequencies were remarkably higher than expected compared with normal B-cell-derived VH (37) and tumor-infiltrating B-cells-derived VH (25) (Fig. 2A).
FIGURE 2.
VH (A) and JH (B) gene family usage profiles of cancer-derived Ig compared with the normal PBL-derived Ig. We analyzed the usage of 78 functional VH genes from the cancer-derived Ig and collected five groups of usage profiles from healthy donors PBL-derived Ig from previous studies (22, 23, 25, 26, 33). The VH and JH gene family usage data from the combined patient group differed significantly from that of the healthy donors PBL-derived Ig (p < 0.05 for both VH and JH).
We measured the immunoglobulin heavy chain DH gene distribution, and found that the DH3, DH6, and DH5 sequences were most frequently used. Expression of the JH of the IgH showed that JH4 was expressed most often (72.7%), although all six JH genes were detected (Fig. 2B).
Several Sets of Predominant Functional VHDJH Gene Recombinations Were Detected in Human Cancer Cells—A noteworthy finding in this study was that the cancer-derived Ig gene repertoire exhibited distinct characteristics. LCM-isolated cancer cells usually expressed one to three sets of dominant VHDJH recombination patterns, as well as some individual VHDJH recombination (Table 2). Notably, seven sets of predominant VHDJH recombinations were detected in more than one cancer cell type (Table 3). Of the 26 VHμDμ JHμ sequences studied, three sets of VHDJH recombinations were predominant: VH3-15/D3-10/JH4, found in 5 of 26 (19.2%) cases of carcinomas from the breast and colon; VH6-1/D6-13/JH4, found in 5 of 26 (19.2%) cases of carcinomas from the lung and oral cavity; and VH4-30-2/D3-22/JH4, found in 4 of 26 (15.4%) cases of carcinomas from the breast and colon. Of the 41 VHγDγJHγsequences studied, the following types of VHDJH recombinations were predominant: VH5-51/D3-9/JH4 in 19 of 38 (50%) cases of carcinomas of the breast, colon, oral, and lung; and VH3-30/D6-19/JH4 in 4 of 38 (10.5%) cases of carcinomas of the breast, colon, and lung. There was no correlation between the patterns of VHDJH recombinations and the histological origins of the cancers.
TABLE 2.
Assignment of the likely matching germline variable region genes to the VHDJH recombinants from different cancer types and analysis of the V gene somatic mutation rate
| Cases | No. of clones | VHγDJHγ (no. of clone) | Mutation rate | No. of clones | VHμDJHμ (no. of clone) | Mutation rate |
|---|---|---|---|---|---|---|
| Breast cancer (case 1) | 5 | VH5-51/D3-9/JH4(3) | 15.6% (3) | 3 | VH4-30-2/D3-22/JH4(1) | 3.3% (1) |
| VH7-4-1/D2-8JH4(1) | 1% (1) | VH3-33/D3-10/JH4(1) | 2.2% (1) | |||
| VH3-9/D3-9/JH3(1) | stopa (1) | VH3-33/D5-12/JH5(1) | 0 (1) | |||
| Breast cancer (case 2) | 5 | VH5-51/D3-9/JH4(4) | 15.6% (2); 17.5% (1) | 3 | VH4-30-2/D3-22/JH4(1) | 0 (1) |
| stop (1) | VH3-33/D5-12/JH5(1) | 4.4% (1) | ||||
| VH1-16/NI/JH4(1) | stop (1) | VH3-15/D3-10/JH4(1) | 1.1% (1) | |||
| Breast cancer (case 3) | 2 | VH5-51/D3-9/JH4(1) | 15.6% (1) | 4 | VH4-30-2/D3-22/JH4(1) | 3.3% (1) |
| VH3-33/D5-12/JH5(1) | 0 (1) | |||||
| VH3-30/D6-19/JH4(1) | 13.5% (1) | VH4-15/D6-13/JH4(1) | 10% (1) | |||
| VH4-61/D6-13/JH4(1) | 0 (1) | |||||
| Lung cancer | 6 | VH5-51/D3-9/JH4(2) | 15.6% (2) | 2 | VH6-1/D6-13/JH4(1) | 3.3% (1) |
| VH3-30/D6-9/JH4(2) | 12.5% (2) | VH6-1/D1-7/JH6(1) | 2.1% (1) | |||
| VH1-3/D5-12/JH4(2) | 28.1% (2) | |||||
| Colon cancer (case 1) | 4 | VH5-51/D3-9/JH4(4) | 15.6% (4) | 5 | VH3-15/D3-10/JH4(4) | 3.5% (4) |
| VH4-30-2/D3-22/JH4(1) | stop (1) | |||||
| Colon cancer (case 2) | 7 | VH5-51/D3-9/JH4(4) | 15.6% (3), 17.5% (1) | ndb | nd | nd |
| VH3-30/D6-9/JH4(1) | 13.5% (1) | |||||
| VH1-69/D6-15/JH4(1) | stop (1) | |||||
| VH7-4-1/D3-10/JH4(1) | 4% (1) | |||||
| Oral cancer (case 1) | 3 | VH5-51/D3-9/JH4(2) | 15.6% (1), stop (1) stop (1) | 9 | VH6-1/D6-13/JH4(4) | 0 (4) |
| VH1-2/D2-8/JH4(1) | VH6-1/D1-7/JH6(3) | 7.6% (3) | ||||
| VH6-1/D2-2/JH4(2) | 2.5% (2) | |||||
| Oral cancer (case 2) | 9 | VH3-33/D6-19/JH5(6) | 11.5% (5); | nd | nd | nd |
| VH3-23/D1-26/JH4(3) | 16.3% (1) 7.3% (3) | |||||
| HT-29 | 7 | VH5-51/D2-2/JH4(1) | 15.% (1) | 6 | VH6-1/D6-13/JH4(6) | 3.3% (4), 1% (1), 7.6% (1) |
| VH3-33/D3-10/JH5(1) | 5% (1) | |||||
| VH1-8/D5-18/JH5(1) | 18% (1) | |||||
| VH1-69/D1-26/JH5(1) | stop (1) | |||||
| VH3-23/D2-15/JH4(1) | 7.1% (1) | |||||
| VH3-30/D6-13/JH4(1) | 9.2% (1) | |||||
| V1-3/D3-10/JH5(5) | 8.3% (1) | |||||
| HelaS3 | 6 | VH1-8/D1-14/JH4(1) | 8.1% (1) | 3 | VH3-30-3/D6-19/JH5(3) | 1.5% (3) |
| VH1-8/D2-2/JH4(1) | 6.2% (1) | |||||
| VH4-59/D3-10/JH4(1) | 10.4% (1) | |||||
| VH4-59/D3-22/JH4(1) | 7.5% (1) | |||||
| VH3-30/N1/JH5(1) | stop (1) | |||||
| VH3-7/D3-10/JH4(1) | 9.1% (1) | |||||
| PBL | 3 | VH3-23/D3-10/JH4(1) | 4.8% (1) | 4 | VH3-21/D1-26/JH4(1) | 0 (1) |
| VH4-59/D1-20/JH4(1) | 4.8% (1) | |||||
| (case 1) | VH1-69/D5-5/JH4(1) | 11.5% (1) | VH4-39/D3-10/JH5(1) | 2.9% (1) | ||
| VH1-8/D2-2/JH5(1) | 12.5% (1) | VH3-23/D6-19/JH5(1) | 14.4% (1) | |||
| PBL | 3 | VH4-59/D3-3/JH4(1) | 20% (1) | nd | nd | nd |
| VH4-59/D4-17/JH4(1) | 17.9% (1) | |||||
| (case 2) | VH3-43/NI/JH5 (1) | 3.1% (1) |
Stop, non-functional VHDJH sequences
nd, not done
TABLE 3.
Seven sets of predominant VHDJH recombinants found in different cancer types The clone name appears in four letters as follows: the first represents the tissue of origin (B for breast cancer, L for lung cancer, C for colon cancer, O for oral cancer, and H for HT-29 cell line); the second represents immunoglobulin isotype (G for IgG, and M for IgM); the third represents the patient number; and the last number in brackets represents the number of clones in this set of VHDJH rearrangements.
| No. of sets | Clone | V gene name | V region | N1 | P | D region | N1 | P | J region | J gene name | D gene name |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | BG1 (3) | IGHV5-51*01 | tgtgcgaga | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | ||
| BG2 (4) | IGHV5-51*01 | tgtgcgaga | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| BG3 (1) | IGHV5-51*01 | tgtgcgaga | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| CG1 (4) | IGHV5-51*01 | tgtgcgaga | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| CG2 (4) | IGHV5-51*01 | tgtgcgaga | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| LG1 (1) | IGHV5-51*01 | tgtgcgaga | tggaatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| OG1 (2) | IGHV5-51*01 | tgtgcgaga. | tgggatg | ...........ttatgattggattttata.. | cagc | .......tgactactgg | IGHJ4*02 | IGHD3-9*01 | |||
| 2 | BG3 (1) | IGHV3-30*01 | tgtgccagaga | ......aacaatggctg.... | cctc | ....ctttgacaattgg | IGHJ4*03 | IGHD6-19*01 | |||
| LG1 (2) | IGHV3-30*01 | tgtgccagaga | ......aacaatggctg.... | cctc | ....ctttgacaattgg | IGHJ4*03 | IGHD6-19*01 | ||||
| CG2 (1) | IGHV3-30*01 | tgtgccagaga | ......aacaatggctg.... | cctc | ....ctttgacaattgg | IGHJ4*02 | IGHD6-19*01 | ||||
| 3 | HM (3) | IGHV6-1*01 | tgtgcaagaga | at | .....tagcagcagct..... | .....tttgactactgg | IGHJ4*03 | IGHD6-13*01 | |||
| OM1 (2) | IGHV6-1*01 | tgtgcaagaga | at | .....tagcagcagct..... | .....tttgactactgg | IGHJ4*02 | IGHD6-13*01 | ||||
| LM (1) | IGHV6-1*01 | tgtgcaagaga | at | .....tagcagcagct..... | .....tttgactactgg | IGHJ4*02 | IGHD6-13*01 | ||||
| OM2 (2) | IGHV6-1*01 | tgtgcaagaga | at | .....tagcagcagct..... | .....tttgactactgg | IGHJ4*02 | IGHD6-13*01 | ||||
| 4 | BM1 (1) | IGHV4-30-2*01 | tgtgccgg... | cc | ..............gaagtggttattact | c | .cccctttgactactgg | IGHJ4*02 | IGHD3-22*01 | ||
| BM2 (1) | IGHV4-30-2*01 | tgtgccgg... | cc | ..............gaagtggttattact | c | .cccctttgactactgg | IGHJ4*02 | IGHD3-22*01 | |||
| BM3 (1) | IGHV4-30-2*01 | tgtgccgg... | cc | ..............gaagtggttattact | c | .cccctttgactactgg | IGHJ4*02 | IGHD3-22*01 | |||
| 5 | BM21 (1) | IGHV3-15*01 | tgtaccacaaa | cctga | ac | gtattactatggttcggggaccga... | accccc | ........gactactgg | IGHJ4*02 | IGHD3-10*01 | |
| CM1 (4) | IGHV3-15*01 | tgtaccacaaa | cctga | ac | gtattactatggttcggggaccga... | accccc | ........gactactgg | IGHJ4*02 | IGHD3-10*01 | ||
| 6 | LM1 (1) | IGHV6-1*02 | tgtgcaagag. | ggcgtacgtgg...... | ccagc | ..........ggacgtctgg | IGHJ6*02 | IGHD1-7*01 | |||
| OM1 (3) | IGHV6-1*01 | tgtgcaagag. | ggcgtacgtgg...... | ccagc | ..........ggacgtctgg | IGHJ6*02 | IGHD1-7*01 | ||||
| 7 | BM2 (1) | IGHV3-33*01 | tgtgcgagaga | .....tggggtggctacgattac | aaggaggtg | ....ctggttcgacccctgg | IGHJ5*02 | IGHD5-12*01 | |||
| BM3 (1) | IGHV3-33*01 | tgtgcgagaga | .....tggggtggctacgattac | aaggaggtg | ....ctggttcgacccctgg | IGHJ5*02 | IGHD5-12*01 |
In contrast, all of the VHγDγJHγ sequences obtained, either from the HT-29 cell line (7 clones) or from the HeLa S3 cell line (6 clones), showed distinct diversity. None of the γ chain clones from the cell lines was detected in any of the LCM-isolated cancer cells. All six VHμDμJHμ sequences from HT-29 cells showed identical VH6-1/D6-13/JH4 recombination, which was also detected in LCM-isolated carcinomas of the lung and oral cavity. All three VHμDμJHμsequences from HeLa S3 cells showed identical VH3-30-3/D6-19/JH5 recombination, which was not identified in the LCM-isolated cancer cells. Ten VHDJH recombination sequences in two control PBL samples exhibited distinct diversity (Table 2).
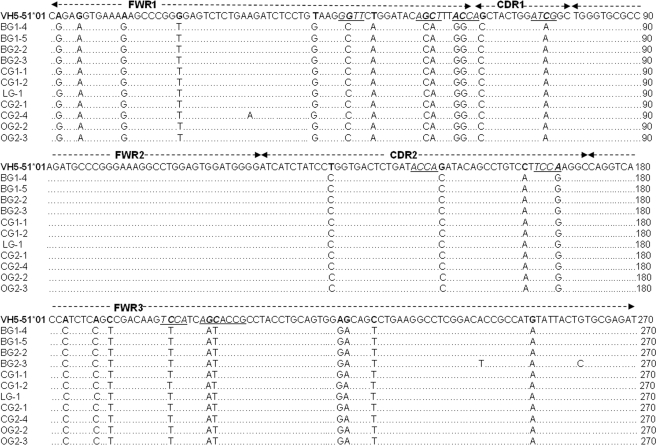
Mechanism of SHM of Functional V Genes in Cancer Cells Was Different from Antigen Selection in B-cell-derived VHDJH Sequences—Somatic hypermutation of the VH region is known to be an important event in B-cell-derived Ig following antigen stimulation. Mutational analysis demonstrated that all of the Vμ gene sequences showed fewer than 5% mutations, and 73.9% of Vμ gene sequences had fewer than 2% mutations, which should be considered as “unmutated” by definition (38). In contrast, most functional VHγ sequences were highly mutated. Approximately 90.6% of VHγ sequences had greater than 5% mutation, which should be considered as “mutated” by definition (38) (Table 2). Unexpectedly, within a set of VHDJH recombinant, the V regions from different cancer cell types showed identical VHDJH recombination and junction. The recombination was either identical or differed only by several mutation targets over the VH region. For example, within the VH5-51/D3-9/JH4 recombination set, 19 of the VH5-51 sequences showed almost identical mutation targets, which were identical to our previously published VH5-51/D3-9/JH4 recombination sequence described for lung caner cells (Fig. 3) (20). Despite the high mutation rate, the rate of homology within the set of VHDJH recombinants ranged from 92.7 to 100%.
FIGURE 3.
The analysis of VH5-51/D3-9/JH4 recombinations derived from different cancer types. These VHγ Dγ JHγ sequences showed almost identical mutation targets over the V segment. Somatic mutations were identified by comparison with the published germline VH5-51 gene in NCBI. The RGYW/WRCY motif was underlined. The first 25 nucleotides were excluded from the analysis because they were encoded by the primers. The clone name appears in four characters: the first character represents the tissue origin (B for breast cancer, L for lung cancer, C for colon cancer, and O for oral cancer); the second represents the immunoglobulin isotype (G for IgG); the third represents the patient number, and the last represents the clone number.
In B-cell-derived Ig, the mutations may either be silent or missense that changes the affinity of the Ig for the antigen; the latter may occasionally give rise to cells expressing higher affinity antibodies, usually with mutations clustered in the CDRs and as a result of antigen selection. Moreover, mutations introduced by AID activity under antigen selection typically target the known SHM hotspot, WRCY, and its complement, RGYW. Furthermore, the mutation frequency is expected to be higher in the CDRs than in the framework regions (FWRs). However, no AID transcript was detected by RT-PCR in the LCM-isolated cancer cells. In contrast, we detected an AID transcript in HT-29 cell line at a low level (Genbank™ Accession Number, AY748364), as well as in Raji cell line (B-lymphocyte-derived, Burkitt's lymphoma) which was used as a positive control (data not shown). In addition, in only 17 of 43 (39.5%) VHγ sequences were there more frequent mutations in the RGYW hot spot. In 3 of 43 VHγ sequences, the mutation frequency was higher in the CDRs than in the FWRs. In only 7 of 43 (16.3%) VHγ sequences were the R/S ratios in the CDRII and FWRIII >2.9 and <1.5, respectively. In the VH5-51/D3-9/JH4 recombination set, a higher mutation frequency, ∼40%, occurred in the RGYW motif (if the ratio was under 25.6%, it was not considered classical SHM) (Fig. 3). In contrast, the mutations in cancer-derived Ig mainly occurred in the FWRIII and not in the CDRs. This finding suggests that a large number of the mutations introduced into the cancer-derived VHγ sequences were not typical of B-cell-derived VH mutations caused by antigen selection.
To define the role of the error-prone polymerase in cancer-derived VHγ hypermutation, we analyzed the mutation frequency in the WA and TW motifs, and found that mutation of the WA motif (36/43 VHγ sequences) was significantly more frequent than that of the TW motif (11/43 VHγ sequences, p < 0.001). These data reveal a strand bias and suggest that error-prone polymerases are involved in the mutation of cancer-derived VHγ sequences. The mutational frequency in the WRCY/RGYW region, the WA/TW motifs, and calculations of the R/S mutation ratio in CDRII and FWRIII are summarized in Table 4.
TABLE 4.
Summary of the mutation frequency in WRCY/RGYW and WA/TW motifs and calculation of the R/S mutation ratios occurring in CDRII and in FWRIII
|
Clone
|
Mutation frequency in RGYW/WRCY
|
Mutation in WA/TW
|
R/S ratio in CDRII and FWRIII
|
||||
|---|---|---|---|---|---|---|---|
| WA | TW | CDRII | FWRIII | ||||
| % | |||||||
| BG1-1 | 62.5 (≥25) | 3 | 0 | 0/1 | 9/4 | ||
| BG1-2 | 16.7 | 3 | 0 | 2/0 (>2.9) | 3/0 | ||
| BG1-3 | 0.0 | 0 | 1 | 0/0 | 0/1 (<1.5) | ||
| BG1-4 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| BG1-5 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| BG2-1 | 62.5 (≥25) | 3 | 1 | 0/1 | 13/6 | ||
| BG2-2 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| BG2-4 | 62.5 (≥25) | 2 | 0 | 0/1 | 11/4 | ||
| BG2-5 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| BG3-2 | 18.2 | 3 | 2 | 4/0 (>2.9) | 4/7 (<1.5) | ||
| CG1-1 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| CG1-2 | 62.5 (≥25) | 2 | 0 | 0/1 | 8/4 | ||
| CG1-3 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| CG1-4 | 62.5 (≥25) | 2 | 0 | 0/1 | 9/4 | ||
| CG2-2 | 0.0 | 0 | 0 | 0/0 | 0/0 | ||
| CG2-3 | 18.2 | 3 | 2 | 4/0 (>2.9) | 4/7 (<1.5) | ||
| CG2-4 | 62.5 (≥25) | 2 | 1 | 0/1 | 9/4 | ||
| LG1-1 | 18.2 | 3 | 2 | 4/0 (>2.9) | 4/7 (<1.5) | ||
| LG1-2 | 18.2 | 3 | 2 | 4/0 (>2.9) | 4/7 (<1.5) | ||
| LG1-3 | 14.3 | 4 | 0 | 2/1 | 8/5 | ||
| LG1-4 | 14.3 | 5 | 1 | 2/1 | 8/5 | ||
| LG1-5 | 62.5 (≥25) | 22 | 0 | 0/1 | 9/4 | ||
| LG1-6 | 62.5 (≥25) | 0 | 2 | 0/1 | 9/4 | ||
| OG1-2 | 62.5 (≥25) | 0 | 2 | 0/1 | 9/4 | ||
| OG2-1 | 9.1 (≥25) | 1 | 2 | 5/2 | 5/3 | ||
| OG2-2 | 23.1 | 0 | 1 | 3/0 (>2.9) | 1/3 (<1.5) | ||
| OG2-3 | 23.1 | 0 | 1 | 3/0 (>2.9) | 1/3 (<1.5) | ||
| OG2-4 | 16.7 | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| OG2-5 | 16.7 | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| OG2-6 | 16.7% | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| OG2-7 | 16.7 | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| OG2-8 | 16.7 | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| OG2-9 | 16.7 | 1 | 2 | 6/1 (>2.9) | 5/2 | ||
| SG1-1 | 18.2 | 5 | 3 | 3/1 (>2.9) | 13/5 | ||
| SG1-2 | 45.5 (≥25) | 6 | 3 | 3/0 (>2.9) | 10/6 | ||
| SG1-3 | 10.0 | 0 | 0 | 0/0 | 0/2 (<1.5) | ||
| SG1-6 | 0.0 | 2 | 1 | 1/0 (>2.9) | 7/4 | ||
| HG1-1 | 12.5 | 0 | 1 | 1/1 | 8/0 | ||
| HG1-2 | 7.7 | 0 | 0 | 0/0 | 0/0 | ||
| HG1-3 | 25.0 (≥25) | 3 | 4 | 3/3 | 10/3 | ||
| HG1-4 | 33.3 (≥25) | 5 | 3 | 7/3 | 7/8 (<1.5) | ||
| HG1-5 | 15.4 | 2 | 1 | 2/1 | 4/0 | ||
| HG1-7 | 21.4 | 2 | 2 | 3/0 (>2.9) | 7/3 | ||
| 95 | 35 | ||||||
| 20/43 of Vγ rearrangements showed ≥25% mutation frequency in RGYW/WRCY motifs | 36/43 of Vγ rearrangements showed excess mutations in WA | 8/43 of Vγ rearrangements showed both R/S >2.9 in CDRII and R/S <1.5 in FWRIII | |||||
Cancer-derived VHγDγ JHγ Sequences Did Not Appear to Originate from the Classical Class Switching—In general, the B-cell-derived VHDJH recombination pattern of the γ chain is similar to that of the μ chain (the precursor of γ chain), because class switching changed only the constant region sequence from the μ chain to the γ chain. However, there was no identical pattern between VHγDγ JHγ and VHμDμ JHμ in any of the cases studied, although each sample expressed a restricted VHγDγ JHγ or VHμDμ JHμ recombination pattern (Table 2). This suggests that cancer-derived VHγDγ JHγ sequences do not originate from the classical class switching.
DISCUSSION
B-lymphocytes have been considered the primary source of serum Ig. However, we have found that cells from epithelial cancer and hyperplasia could also express Ig (17–18, 21). In this study, we confirmed that functional Ig gene recombination and transcription occurred in a variety of cancer types. Our analysis of 78 functional cancer-derived VHDJH sequences showed that cancer-derived Ig shared some features with B-cell-derived Ig, such as VHDJH recombination, insertion of the N region into junctions, and JH gene recombination at TG nucleotide sequences. In addition, there were either low levels or a complete absence of mutations in the VHμDμ JHμ sequences and high levels of mutations in the functional VHγDγ JHγ sequences.
On the other hand, cancer-derived VHDJH recombinations also exhibited distinct features that differed from B-cell-derived Ig recombination. For example, cancer-derived Ig VH, D, and JH usage showed distinct preferences, such as that VH5-51 and VH6-1 frequencies were higher and that JH4 was expressed most often. The primers used in this experiment were designed to be applicable to almost all VHDJH recombinations; however, we found that VH5-51 and VH6-1 recombination frequency was significantly higher than expected compared with normal B-cell-derived VH (22, 26–28).
It is known that each B-cell expresses a unique VHDJH recombination, including a random N region sequence. Therefore, the likelihood of identical junction sequences from two independent B-cell clones occurring in an individual should be lower than 1 in 4 million. Unexpectedly, several restricted VHDJH sequences were identified in each cancer sample. More interestingly, different cancer samples showed identical VHDJH recombination patterns, with identical junctions and VH region mutation targets. Several dominant VHDJH recombination sets were frequently expressed in different cancer types. These results suggest that there is an unknown mechanism allowing epithelial cancer cells to express several repeated sets of dominant VHDJH sequences. We eliminated the possibility of cross-contamination among cancer samples in a number of ways. First, using RT-PCR, the control tube (containing no cDNA) did not show a positive band when the VHDJH sequences were amplified. Second, there were no samples with identical Ig heavy chain gene repertoires among the eight cancer samples, suggesting that there was no cross contamination among different samples. In contrast, both HT-29 and HeLa S3 cells showed monoclonal characteristics in the VHμDμ JHμ. The VHγ Dγ JHγ derived from these two cancer cell lines showed distinct diversity, which was similar to that of B-cells, but different from primary cancer cells. Identical VHDJH sequences were not detected between the two cancer cell lines, which implied that the genetic characteristics of the cancer cell lines had changed under long-term culture in vitro.
The mechanism of SHM is another distinct feature in cancer-derived VHγ Dγ JHγ sequences as opposed to B-cell-derived VHγ Dγ JHγ sequences. In B-cell-derived Ig V regions, mutations induced by antigen selection occurred more frequently in the CDRs than in the FWRs. Moreover, there is a higher R/S mutation ratio in the CDRs. However, only a few cancer-derived VHγ Dγ JHγ sequences matched the pattern of antigen selection. In addition, AID is thought to be necessary for SHM of the Ig gene, since it can deaminate C to U on both DNA strands, resulting in the symmetrical mutation of C on both strands. The AID enzyme site preference results in hypermutation of the RGYW hotspot motif. In this study, only the VH5-51/D3-9/JH4 pattern matched the AID-induced RGYW hypermutation pattern. Moreover, mutations in VH5-51 frequently occurred in the FWRIII, but not in the CDRs. Additionally, no AID transcript was detected by RT-PCR in the LCM-isolated cancer cells. In contrast, we detected AID transcripts in the HT-29 cell line at a low level, as well as in Raji cell line. It is possible that AID expression is unnecessary for Vγ SHM in cancer cells and that other mechanisms may be involved in SHM of the cancer-derived Ig V region. Babbage et al. recently demonstrated constitutive expression of AID in six breast cancer cell lines and Matsumoto et al. demonstrated expression of AID in stomach cancer cells (22, 39). The mutational bias of A versus TinIg genes results from DNA pol η activity (40), which functions as a secondary mutator (41, 42). Thus, the excess of mutations in the WA motif suggests that DNA pol η may be involved in mutating A:T base pairs in the Ig V gene in cancer cells.
The third distinct feature that differentiates cancer-derived from B-cell-derived VHγ Dγ JHγ sequences is that the cancer-derived VHγ Dγ JHγ sequences do not seem to originate from classical class switching. Class switch recombination and SHM are two important events in Ig production. In the classical Ig class switching, the IgM-producing cells are precursors of the IgG producers, and the Vγ gene assembly mode should be similar to that of VHμDμ JHμ. Although VHμDμ JHμand VHγDγ JHγ could be synchronously detected in the same cancer cells in this study, we did not detect any identical patterns of VHγ Dγ JHγ and VHμDμ JHμ in the cases studied. This unexpected result suggested that the IgGs were completely different from the IgMs in the same cancer cells, and that IgG production in these cells did not follow the classical class switching mechanism. These results raised the possibility that IgGs might be coded by an allele on another chromosome. Our finding (by Southern blot analysis using a JH DNA segment probe) that two Ig alleles in HT-29 had been rearranged (data not shown) supported the presence of such a mechanism. Alternatively, there may be another class switching mechanism. Jhagvaral et al. (36) recently described that naive B-lymphocytes could develop into IgG-secreting cells through successive cell divisions.
In this study, a pivotal precaution was to avoid B-cell contamination. B-lymphocytes and plasma cells are capable of infiltrating cancer tissues and are mainly located in the stroma. By using the LCM method, we specifically captured the EpCAM+ cancer cells in the cancer nest regions and avoided capturing tumor-infiltrating B-lymphocytes and plasma cells. We did not detect B-cell contamination in any of the cDNA libraries from the LCM-isolated cancer cells. Importantly, none of the cancer-derived VHDJH sequences were homologous to the VHDJH recombination sequences of the two control PBL samples or published recombination sequences present in B-lymphocytes and B-cell CLL/SLL. Additionally, there was no sequence homology with published sequences for tumor-infiltrating B-lymphocytes in breast cancer tissues (25). This lack of recombinant sequence homology with B-lymphocytes indicates that the detected Ig sequences are specific to epithelial cancer cells.
To be reactive to multiple antigens, B-cells generate Ig diversity through several mechanisms. However, the biological significance of non-B-cell-derived Ig is not yet clear. The cancer cell-derived Ig repertoire displayed distinct features suggesting that non-B-cell-derived Ig may have important undiscovered activities because of their diverse origins. We previously noted that Ig expression and activity in cancer cell lines could be blocked by specific antisense DNAs and antibodies, causing the cancer cells to undergo apoptosis (20). These data suggest that non-B-cell-derived Ig is involved in the growth and survival of cells.
In summary, many nonhematopoietic tumor cells express Ig. The cancer-derived Ig gene repertoire displays several distinct characteristics, suggesting that there is an idiosyncratic mechanism for cancer-derived Ig gene expression. The gene expression patterns of Ig in different cancer cells may prove useful in understanding the structure and function of nonhematopoietic-derived Ig. In addition, these findings may have important implications for the diagnosis, targeted therapy, as well as monitoring of residual disease of cancers.
Acknowledgments
We thank Dr. Dalong Ma, Dr. Wenling Han, and Shuang Shi from the Peking University Center for Human Disease Genomics for help with experiments; we thank Dr. Lieping Chen from the Dept. of Dermatology/Oncology, Johns Hopkins University School of Medicine, Dr. Yongguang Yang from Surgery, Harvard Medical School, Dr. Jiang Gu, and Dr. Michael A. McNutt from the Dept. of Pathology, Peking University, Dr. Yu Zhang and Dr. Yanhui Yin from the Dept. of Immunology for contributions to manuscript revision. We also thank I. B. Rogozin from the National Institutes of Health/NLM/NCBI for contributions to SHM analysis.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY270187-AY270190, AY247234, AY286495, and AY505568.
This work was supported by Fundamental Research Grants 30371609 and 30572094 from the Natural Sciences Foundation, China.
Footnotes
The abbreviations used are: RT-PCR, reverse transcription-PCR; AID, activation-induced cytidine deaminase; PBS, phosphate-buffered saline; SHM, somatic hypermutation; LCM, laser capture microdissection; EpCAM, epithelial cell adhesion molecule; MNC, mononuclear cell; PBL, peripheral blood lymphocytes; CDR, complementary determining region; FWR, framework regions.
References
- 1.Jerne, N. K. (1955) Proc. Natl. Acad. Sci. U. S. A. 41 849-857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glick, B., Chang, T. S., and Jaap, G. (1956) Poultry Sci. 35 224-225 [Google Scholar]
- 3.Mitchell, G. F., and Miller, J. F. (1968) J. Exp. Med. 128 821-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tonegawa, S. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 203-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hozumi, N., and Tonegawa, S. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 3628-3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neri, A., Jakobiec, F. A., Pelicci, P. G., Dalla-Favera, R., and Knowles, D. M., 2nd (1987) Blood 70 1519-1529 [PubMed] [Google Scholar]
- 7.Cleary, M. L., Chao, J., Warnke, R., and Sklar, J. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 593-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraternali-Orcioni, G., Falini, B., Quaini, F., Campo, E., Piccioli, M., Gamberi, B., Pasquinelli, G., Poggi, S., Ascani, S., Sabattini, E., and Pileri, S. A. (1999) Am. J. Surg. Pathol. 23 717-721 [DOI] [PubMed] [Google Scholar]
- 9.Bartram, C. R., Raghavachar, A., and Heimpel, H. (1986) Blut. 52 203-210 [DOI] [PubMed] [Google Scholar]
- 10.Zuniga, M. C., D'Eustachio, P., and Ruddle, N. H. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 3015-3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olubuyide, I. O., Salimonu, L. S., and Adeniran, S. O. (1993) Afr. J. Med. Med. Sci. 22 57-62 [PubMed] [Google Scholar]
- 12.Manjula, S., Aroor, A. R., Raja, A., Rao, S. N., and Rao, A. (1992) Acta Neurochir. (Wien) 115 103-105 [DOI] [PubMed] [Google Scholar]
- 13.Taylor, D. D., and Gercel-Taylor, C. (1998) Oncol. Rep. 5 1519-1524 [DOI] [PubMed] [Google Scholar]
- 14.Tenenbaum, N., Meignan, S., and Vincent, J. P. (1992) Ann. Med. Interne (Paris) 143 89-93 [PubMed] [Google Scholar]
- 15.Hori, H., Kihara, Y., Nagashio, Y., Hirohata, Y., Abe, S., Murata, I., and Otsuki, M. (2000) Nippon Shokakibyo Gakkai Zasshi 97 1373-1377 [PubMed] [Google Scholar]
- 16.Gregersen, H., Mellemkjaer, L., Ibsen, J. S., Dahlerup, J. F., Thomassen, L., and Sorensen, H. T. (2001) Haematologica 86 1172-1179 [PubMed] [Google Scholar]
- 17.Qiu, X., and Yang, G. (1996) Chinese J. Immunol. 5 296 [Google Scholar]
- 18.Wang, D., Qiu, X., Zhu, X., Lv, P., and Gao, X. (2000) J. Beijing Med. Univ. 32 310 [Google Scholar]
- 19.Kimoto, Y. (1998) Genes Chromosomes Cancer 22 83-86 [DOI] [PubMed] [Google Scholar]
- 20.Qiu, X., Zhu, X., Zhang, L., Mao, Y., Zhang, J., Hao, P., Li, G., Lv, P., Li, Z., Sun, X., Wu, L., Zheng, J., Deng, Y., Hou, C., Tang, P., Zhang, S., and Zhang, Y. (2003) Cancer Res. 63 6488-6495 [PubMed] [Google Scholar]
- 21.Li, M., Feng, D. Y., Ren, W., Zheng, L., Zheng, H., Tang, M., and Cao, Y. (2004) Int. J. Biochem. Cell Biol. 36 2250-2257 [DOI] [PubMed] [Google Scholar]
- 22.Babbage, G., Ottensmeier, C. H., Blaydes, J., Stevenson, F. K., and Sahota, S. S. (2006) Cancer Res. 66 3996-4000 [DOI] [PubMed] [Google Scholar]
- 23.Zhu, X., Li, C., Sun, X., Mao, Y., Li, G., Liu, X., Zhang, Y., and Qiu, X. (2008) Appl. Immunohistochem. Mol. Morphol. 16 232-238 [DOI] [PubMed] [Google Scholar]
- 24.Lefranc, M. P., Giudicelli, V., Ginestoux, C., Bodmer, J., Muller, W., Bontrop, R., Lemaitre, M., Malik, A., Barbie, V., and Chaume, D. (1999) Nucleic Acids Res. 27 209-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nzula, S., Going, J. J., and Stott, D. I. (2003) Cancer Res. 63 3275-3280 [PubMed] [Google Scholar]
- 26.Messmer, B. T., Albesiano, E., Messmer, D., and Chiorazzi, N. (2004) Blood 103 3490-3495 [DOI] [PubMed] [Google Scholar]
- 27.Hansen, M. H., Nielsen, H., and Ditzel, H. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12659-12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien, P. M., Tsirimonaki, E., Coomber, D. W., Millan, D. W., Davis, J. A., and Campo, M. S. (2001) Cancer Immunol. Immunother. 50 523-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogozin, I. B., and Kolchanov, N. A. (1992) Biochim. Biophys. Acta 1171 11-18 [DOI] [PubMed] [Google Scholar]
- 30.Yoshikawa, K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H., and Honjo, T. (2002) Science 296 2033-2036 [DOI] [PubMed] [Google Scholar]
- 31.Martin, A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J., and Scharff, M. D. (2002) Nature 415 802-806 [DOI] [PubMed] [Google Scholar]
- 32.Tamaru, J., Hummel, M., Marafioti, T., Kalvelage, B., Leoncini, L., Minacci, C., Tosi, P., Wright, D., and Stein, H. (1995) Am. J. Pathol. 147 1398-1407 [PMC free article] [PubMed] [Google Scholar]
- 33.Dorner, T., Brezinschek, H. P., Brezinschek, R. I., Foster, S. J., Domiati-Saad, R., and Lipsky, P. E. (1997) J. Immunol. 158 2779-2789 [PubMed] [Google Scholar]
- 34.Spencer, J., Dunn, M., and Dunn-Walters, D. K. (1999) J. Immunol. 162 6596-6601 [PubMed] [Google Scholar]
- 35.Oprea, M., Cowell, L. G., and Kepler, T. B. (2001) J. Immunol. 166 892-899 [DOI] [PubMed] [Google Scholar]
- 36.Souto-Carneiro, M. M., Longo, N. S., Russ, D. E., Sun, H. W., and Lipsky, P. E. (2004) J. Immunol. 172 6790-6802 [DOI] [PubMed] [Google Scholar]
- 37.Brezinschek, H. P., Brezinschek, R. I., and Lipsky, P. E. (1995) J. Immunol. 155 190-202 [PubMed] [Google Scholar]
- 38.Yamada, M., Wasserman, R., Reichard, B. A., Shane, S., Caton, A. J., and Rovera, G. (1991) J. Exp. Med. 173 395-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto, Y., Marusawa, H., Kinoshita, K., Endo, Y., Kou, T., Morisawa, T., Azuma, T., Okazaki, I. M., Honjo, T., and Chiba, T. (2007) Nat. Med. 13 470-476 [DOI] [PubMed] [Google Scholar]
- 40.Mayorov, V. I., Rogozin, I. B., Adkison, L. R., and Gearhart, P. J. (2005) J. Immunol. 174 7781-7786 [DOI] [PubMed] [Google Scholar]
- 41.Neuberger, M. S., Di Noia, J. M., Beale, R. C., Williams, G. T., Yang, Z., and Rada, C. (2005) Nat. Rev. Immunol. 5 171-178 [DOI] [PubMed] [Google Scholar]
- 42.Delbos, F., De Smet, A., Faili, A., Aoufouchi, S., Weill, J. C., and Reynaud, C. A. (2005) J. Exp. Med. 201 1191-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]