Abstract
Glycolipid transfer proteins (GLTPs) are small, soluble proteins that selectively accelerate the intermembrane transfer of glycolipids. The GLTP fold is conformationally unique among lipid binding/transfer proteins and serves as the prototype and founding member of the new GLTP superfamily. In the present study, changes in human GLTP tryptophan fluorescence, induced by membrane vesicles containing glycolipid, are shown to reflect glycolipid binding when vesicle concentrations are low. Characterization of the glycolipid-induced “signature response,” i.e. ∼40% decrease in Trp intensity and ∼12-nm blue shift in emission wavelength maximum, involved various modes of glycolipid presentation, i.e. microinjection/dilution of lipid-ethanol solutions or phosphatidylcholine vesicles, prepared by sonication or extrusion and containing embedded glycolipids. High resolution x-ray structures of apo- and holo-GLTP indicate that major conformational alterations are not responsible for the glycolipid-induced GLTP signature response. Instead, glycolipid binding alters the local environment of Trp-96, which accounts for ∼70% of total emission intensity of three Trp residues in GLTP and provides a stacking platform that aids formation of a hydrogen bond network with the ceramide-linked sugar of the glycolipid headgroup. The changes in Trp signal were used to quantitatively assess human GLTP binding affinity for various lipids including glycolipids containing different sugar headgroups and homogenous acyl chains. The presence of the glycolipid acyl chain and at least one sugar were essential for achieving a low-to-submicromolar dissociation constant that was only slightly altered by increased sugar headgroup complexity.
Glycolipid transfer protein (GLTP)4 is a soluble (∼24-kDa) protein that selectively transfers glycosphingolipids (GSLs) between membranes. GSLs play key roles in cell recognition, adhesion, differentiation, proliferation, and programmed death in normal and disease states (1–8). Phylogenetic/evolutionary analyses show GLTP to be highly conserved among vertebrates (9–11). The conformational uniqueness of the GLTP fold when compared with other lipid binding/transfer proteins (12–14) has resulted in GLTP being designated the prototype and founding member of the GLTP superfamily (15, 16). GLTP employs a novel two-layer “sandwich motif,” dominated by α-helices and achieved without intramolecular disulfide bridges, to accommodate glycolipid within a single lipid binding site and to form a membrane-interaction domain that differs from other known membrane targeting/translocation domains, i.e. C1, C2, PH, PX, and FYVE (9, 13, 17–21). The glycolipid binding site of GLTP consists of a sugar headgroup recognition center that anchors the ceramide-linked sugar to the protein surface via multiple hydrogen bonds and a hydrophobic tunnel that accommodates the hydrocarbon chains of ceramide. The crystal structures of glycolipid-free GLTP and of GLTP complexed with a half-dozen glycolipids differing in sugar headgroup and/or lipid acyl composition reveal the basis for specific recognition and adaptive accommodation of various GSLs. A conserved, concerted sequence of events, initiated by anchoring of the GSL headgroup to the sugar headgroup recognition center, seems to facilitate entry and exit of the lipid chains in the membrane-associated state (13). Glycolipid uptake occurs via a cleft-like gating mechanism involving conformational changes to one α-helix and two interhelical loops (12). The selectivity of GLTP for glycolipids makes this protein a prime candidate for molecular manipulation of GSL-enriched microdomains in membranes as well as a potential vehicle for selectively delivering glycolipids to cells. However, the binding affinity of various glycolipids for GLTP and the time frame of GSL uptake by GLTP remain unclear. In the present study, these issues are investigated using fluorescence approaches.
GLTP is intrinsically fluorescent by virtue of having 3 Trp and 10 Tyr residues among its 209 amino acids. All 3 Trp residues reside on or near the surface of GLTP (12–14, 17, 22, 23), where they could help form a membrane-interaction site. Only one, Trp-96, is directly involved in glycolipid binding (12–14). Given the likely roles in membrane interaction and GSL binding, our goal was to define the relative contributions of the Trp fluorescence changes caused by membrane interaction versus glycolipid binding. A signature Trp emission response, indicative of GSL binding by WT-GLTP, has been identified and characterized using select GLTP point mutants and different modes of glycolipid presentation, i.e. ethanol injection of pure GSLs and titration with membrane vesicles (LUVs and SUVs) containing GSLs as minor components. The signature Trp emission response has been used to comprehensively assess the glycolipid binding affinity of the novel GLTP fold for the first time, focusing on the impact of compositional variation of the sugar headgroup and nonpolar acyl chain moieties of the glycolipid.
EXPERIMENTAL PROCEDURES
Most GalCers, LacCers, and sphingomyelins with homogeneous acyl chains were synthesized by reacylating GalSph, Lac-Sph, or lysosphingomyelin with the desired fatty acyl residue (24, 25) (see supplemental information for details). Other glycolipids and all phosphoglycerides were purchased from Avanti Polar Lipids (Alabaster, AL). n-Hexyl-β-d-glucoside was obtained from Anatrace Inc. (Maumee OH). Stock phospholipid concentrations were quantitated by the Bartlett method (26), and glycolipids were quantitated by gravimetric analyses. Lipid vesicles were produced by extrusion or sonication (see supplemental information).
Protein Expression and Purification—Our GLTP purifications have been described previously in detail (22, 27). Briefly, the open reading frame encoding human GLTP (National Center for Biotechnology Information (NCBI) GenBank™ accession number AF209704) was subcloned into pET-30 Xa/LIC expression vector (Novagen) by ligation-independent cloning, enabling cleavage of the N-terminal His6-S tag to yield protein identical in sequence to native GLTP (12, 27). His6-GLTP also was prepared because structural analyses indicate that glycolipid binding might be subtly affected by the presence of the N-terminal fusion tag (13). Site-directed mutants were produced by QuikChange® site-directed mutagenesis (Stratagene) and verified by sequencing. Near UV CD analysis confirmed the global folding similarity of WT-GLTP and W96F-GLTP (12). Protein purity and concentration were determined by SDS-PAGE and bicinchoninic acid (22). Glycolipid intervesicular transfer activity of purified GLTP was monitored using established assays involving fluorophore- or radiolabeled glycolipid (see supplemental information).
Fluorescence Measurements—Emission spectra were measured using a SPEX FluoroMax instrument (HORIBA Scientific, Edina, NJ). Excitation and emission band passes were 5 nm, and the cuvette holder was temperature-controlled to 25 ± 0.1 °C (Neslab RTE-111, (Thermo Fisher Scientific)). To eliminate contributions from residues other than Trp, the excitation wavelength was 295 nm. Emission spectra were recorded from 305 to 500 nm using GLTP concentrations at optical densities >0.1 to avoid inner filter effects. GSL addition (2-μl aliquots; 0.1 mm GSL stock in ethanol) to protein (2.5 ml; 1 μm) involved constant stirring. Rapid equilibration of the fluorescence emission signal (<3 min) was observed for glycolipids containing either short saturated or longer unsaturated acyl chains (supplemental Fig. S2). Binding isotherms were adjusted for the constant fractional contributions of Trp-85 and Trp-142 determined from the emission response of W96F-GLTP. Dissociation constants were determined by nonlinear curve fitting using Prism 4.0 (GraphPad Software, La Jolla, CA) (see supplemental information).
Mass Spectrometry—WT-GLTP and GLTP·glycolipid complex were analyzed using an Agilent 6210 LC/MS-TOF mass spectrometer (Santa Clara, CA) by preparing (10 μm protein) in 5 mm ammonium acetate plus 5% methanol and infusing directly into the electrospray source. Spectra were collected in positive mode over an m/z range of 500–5000 using parameters optimized for complex stability, e.g. capillary, 3000 V; fragmentor, 300 V; skimmer, -60 V; octopole radio frequency, -300 V; octopole direct current, -32 V. Raw spectra data were transformed into relative molecular masses using the Agilent time-of-flight Protein Confirmation software.
SHARP2 Analyses—WT-GLTP surface interaction site analysis was performed using SHARP2. The algorithm calculates parameters for overlapping patches of residues on the surface of a protein from Protein Data Bank (PDB) data (28). Six parameters are assessed: solvation potential, hydrophobicity, accessible surface area, residue interface propensity, planarity, and protrusion (SHARP2) (29). Patches are ranked according to combined parameter scores, enabling assessment as potential protein-protein interaction sites.
RESULTS
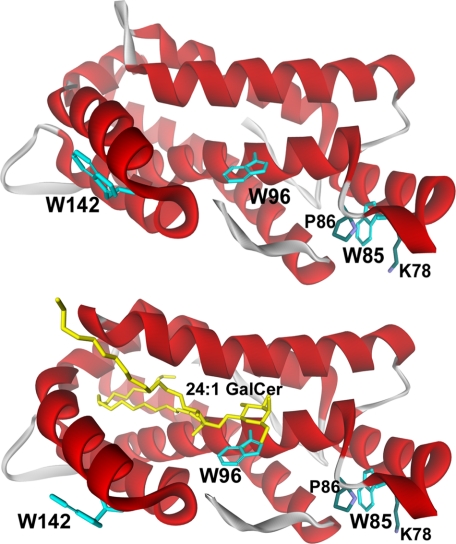
Our previous x-ray crystallographic analyses showed that the 3 Trp residues of human GLTP are located in the vicinity of the glycolipid binding site (12–14). Trp-96 and Trp-142 are surface-localized, 14–16Å apart, and reside within a putative membrane-interaction region that encompasses the glycolipid binding site (Fig. 1, upper panel, and supplemental Fig. S1, upper panel). Trp-96 also helps bind glycolipid by providing a stacking platform for the initial ceramide-linked sugar and facilitating formation of multiple hydrogen bonds between the sugar ring hydroxyls and Asp-48, Asn-52, and Lys-55 (Figs. 1B and 6). Trp-85 resides near the protein surface but is partially buried, with its indole ring sandwiched between Pro-86 (cis configuration) and Lys-78 (supplemental Fig. S1, lower panel). The topology of the 3 Trp residues in glycolipid-free WT-GLTP is reflected by the steady-state Trp fluorescence signal, which is relatively red-shifted (λmax ∼ 347 nm) and is diminished by aqueous quenchers (22, 23). When glycolipid-free GLTP encounters membranes containing glycolipid, dramatic changes in Trp emission occur (22, 23), consistent with Trp involvement in both the glycolipid binding site and the membrane-interaction region of GLTP. The changes include: (i) a large decrease in intensity and (ii) a substantial blue shift (∼12 nm) in wavelength maximum (λmax). Our initial goal was to establish whether the changes in Trp emission could be used to assess the binding affinity and uptake kinetics of various glycolipids by WT-GLTP.
FIGURE 1.
Tryptophan topology in human GLTP. The upper panel shows the crystal structure of the glycolipid-free form of human GLTP (PDB 1swx). The lower panel shows the crystal structure of human GLTP complexed with N-nervonoyl GalCer (PDB 2euk). Trp residues are turquoise, and 24:1 GalCer is yellow. Trp-142 and Trp-96 are surface-localized and completely accessible to the aqueous milieu. Trp-85 is near the surface, but its indole ring is sandwiched between Pro-86 and Lys-78. Trp-85 is located 30–32 Å from Trp-142 and 14–16 Å from Trp-96.
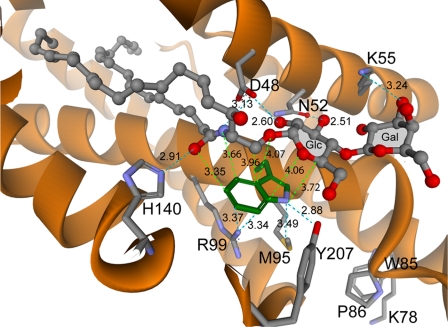
FIGURE 6.
Tryptophan 96 and neighboring residues in the GLTP·N-oleoyl LacCer complex. Depictions are based on the crystal coordinates of human GLTP liganded with 18:1 LacCer (PDB 1sx6). Interaction distances between the Trp-96 indole ring and the ceramide-linked sugar of the glycolipid headgroup are indicated by lime green dashed lines. Side chains of residues that stabilize Trp-96 position in GLTP and interactions of Asp-48, Asn-52, Lys-55, and His-140 with the glycolipid polar headgroup region are indicated by turquoise dashed lines. Numerical values indicate distances in angstroms.
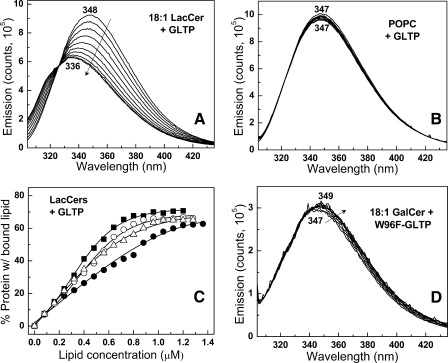
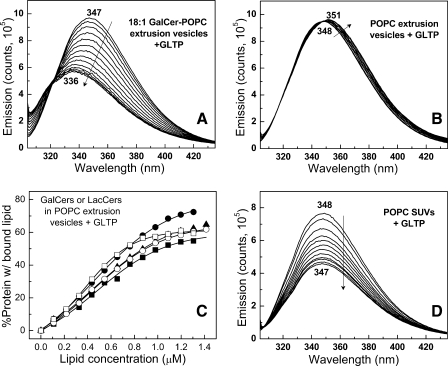
Glycolipid Binding to WT-GLTP Induces Dramatic Changes in Tryptophan Fluorescence—To determine whether the emission changes reflect uptake of glycolipid or protein partitioning to the membrane surface, WT-GLTP was presented with limited amounts of glycolipids, dissolved in ethanol, in titration-like fashion. The strategy of using ethanol injection was based on our previous findings showing that ethanolic solutions of glycolipids can facilitate cocrystallization of GLTP complexed with various glycolipid ligands (12, 13) and that GLTP is highly adaptable in its ability to acquire glycolipids presented in various forms (27). Fig. 2 illustrates a typical Trp emission response upon stepwise injections of LacCer ethanolic solution. The Trp emission λmax becomes progressively blue-shifted, i.e. 347–336 nm, whereas the intensity systematically decreases. Eventually, a saturation response is achieved as the λmax reaches ∼336 nm, and the total Trp emission intensity declines by ∼40%. It is important to note that each glycolipid injection corresponds to <10 mol % of total WT-GLTP and that the changes in Trp fluorescence occurred rapidly (<3–5 min) for glycolipids with short saturated or long monounsaturated acyl chains (supplemental Fig. S2), permitting fast equilibration. The emission response was similar for LacCers with oleoyl (18:1), nervonoyl (24:1), or octanoyl (8:0) acyl chains (e.g. Fig. 2A) and resulted in similar binding isotherms (Fig. 2C). In contrast, almost no changes in Trp emission were observed when POPC (Fig. 2B), l-α-dimyristoylphosphatidylcholine, 1-myristoyl,2-palmitoyl-sn-glycero 3-phosphocholine, or 1-myristoyl-2-stearoyl-sn-glycero 3-phosphocholine were presented with WT-GLTP.
FIGURE 2.
Changes in Trp emission of WT-GLTP and W96F-GLTP induced by incubation with glycolipid. A, 18:1 LacCer + WT-GLTP. B, POPC + WT-GLTP. C, binding Isotherms for WT-GLTP + various LacCer species. •, 24:1 LacCer; ▵, 18:1 LacCer; ▪, 8:0 LacCer; ○, 12:0 LacCer. Binding isotherms were adjusted for the constant fractional contributions of Trp-85 and Trp-142 determined from the emission response of W96F-GLTP. D, 18:1 GalCer + W96F-GLTP. WT-GLTP Trp signal (first scan acquired at 1 min) remains stable for >30 min in the absence of lipid (not shown). The Trp emission scans correspond to lipid concentrations of 0, 0.08, 0.16, 0.24, 0.32, 0.40, 0.48, 0.56, 0.64, 0.72, 0.80, 0.88, 0.96, 1.04, 1.12, and 1.20 μm with respect to decreasing emission intensity (see “Experimental Procedures” for details). The stepwise addition of glycolipid to free Trp produced no reduction in emission intensity (data not shown).
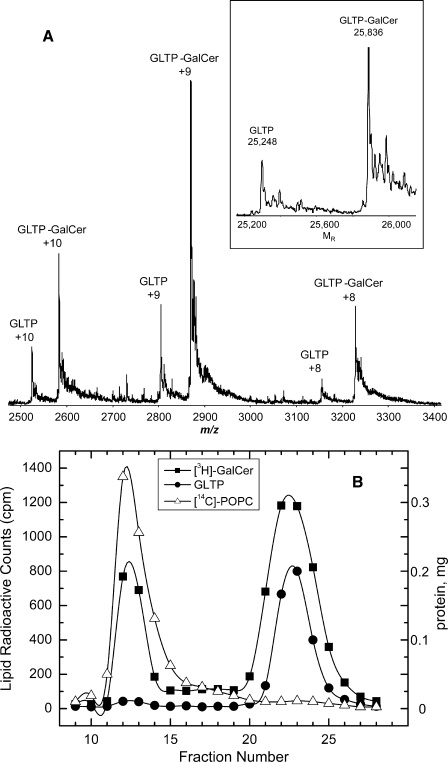
Formation of a WT-GLTP·glycolipid complex by GSL ethanol injection was verified by electrospray ionization mass spectrometry (ESI-MS). Fig. 3A shows spectra obtained by direct infusion of glycolipid-free GLTP plus GLTP·GalCer complex under nondenaturing conditions. Ions corresponding to monomeric, glycolipid-free rGLTP (25,248 Da) and monomeric complex (rGLTP + N-octanoyl GalCer (25,836 Da)) dominate the spectra.
FIGURE 3.
Formation of GLTP·glycolipid complex. A, ESI-MS analysis of the GLTP·glycolipid complex. Three main charge states are observed from the direct infusion of the GLTP·N-octanoyl GalCer complex (2:1 molar ratio) under nondenaturing conditions. The transformed spectra (see inset) results in molecular masses of 25,248 Da for the GLTP and 25,836 Da for the GLTP·N-octanoyl GalCer complex. The N-terminal His6 tag accounted for the molecular mass being 5% higher than human WT-GLTP (23,850 Da). B, SEC analysis of GLTP after interaction with vesicles containing GalCer. Elution of [14C]POPC vesicles (▵) containing 20 mol % [3H]GalCer (▪) from Sephacryl S-300 (1.5 × 30 cm) after incubation at room temperature for 3 h with WT-GLTP (•) is shown.
To further confirm that the observed changes in the Trp emission reflected glycolipid binding by WT-GLTP, alternate possibilities were evaluated. Adsorption of GLTP·glycolipid complex to the quartz cuvette walls, resulting in diminished protein solution concentration, was ruled out because applying a film of polyethyleneimine, which prevents protein adsorption to silica (30), did not alter the outcome (data not shown). Another possibility was Trp environmental polarity change caused by nonspecific partitioning of WT-GLTP to lipid aggregates/vesicles formed in solution after ethanol injection. However, adjustment of the WT-GLTP concentration from 1 μm to either 0.5 μm or 2 μm proportionally lowered or raised the glycolipid amount needed to saturate the spectral emission changes. Also, SEC analyses using Sephacryl S-300 showed that overnight incubations of WT-GLTP, with POPC vesicles containing glycolipid, resulted in vesicles eluting in the void volume (with almost no adsorbed WT-GLTP when vesicle concentrations were kept relatively low), whereas WT-GLTP (>95%) remained soluble and eluted in the included volume (Fig. 3B). The Trp emission signal of the soluble WT-GLTP exhibited a blue-shifted λmax (∼335 nm), a finding consistent with complexation of glycolipid.
Collectively, the experimental results indicated that the changes in Trp emission reflected glycolipid binding by WT-GLTP. This conclusion was further supported by experiments involving a Trp-96 point mutant (W96F), which emits via Trp-85 and Trp-142 while maintaining ∼70% or more activity (12). As shown in Fig. 2D and supplemental Fig. S3, W96F-GLTP emits at a λmax of ∼347 nm (like glycolipid-free WT-GLTP) but with only ∼30% intensity. Stepwise ethanol microinjection of 18:1 or 8:0 GalCer with W96F-GLTP resulted in only slight changes in the Trp emission response, i.e. 2–3-nm red shift in the λmax and 4–5% intensity decrease. This finding was consistent with Trp-96 accounting for the blue shift in the λmax and the vast majority of the Trp emission intensity decrease observed when WT-GLTP acquires glycolipid.
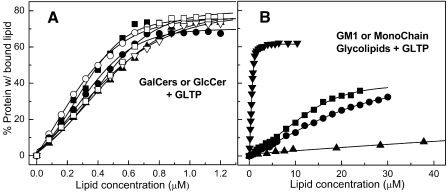
Glycolipid Structural Parameters Affecting Binding by WT-GLTP—To determine whether the changes in WT-GLTP Trp fluorescence were glycolipid-specific, the responses elicited by ceramide and sphingomyelin were investigated. As expected, removal of the glycolipid sugar headgroup (supplemental Fig. S4A) or replacement with a nonsugar headgroup (supplemental Fig. S4B) had little effect on the intrinsic Trp emission. Next, various glycolipid structural parameters were evaluated. When the sugar headgroup was changed from lactose to either glucose or galactose, the dramatic blue shift in the λmax and decrease in Trp emission intensity persisted (supplemental Fig. S5) and resulted in similar binding isotherms (Fig. 4A). Ethanol injection of ganglioside GM1, which contains five sugars including negatively charged N-acetylneuraminic acid, also elicited a λmax blue shift and decrease in Trp emission intensity, albeit reduced in magnitude and affinity when compared with glycolipids with simpler headgroups (Fig. 4A). In contrast, removal of the amide-linked acyl chain of GlcCer or LacCer elicited very modest changes, i.e. 3–6-nm λmax blue shifts and 15–25% Trp intensity decreases, but only when added in high amounts (supplemental Fig. S4, C and D). Fig. 4B shows that very weak binding occurred with hexyl-β-d-glucoside, which also can occupy the GLTP binding site (13).
FIGURE 4.
Binding isotherms for WT-GLTP and various glycolipids. A, GalCer or GlcCer with different acyl chains. ○, 18:2 GalCer; ▪, 8:0 GalCer; □, 18:1 GalCer; ▴, 12:0 GalCer; ▵, 24:1 GalCer; •, 8:0 GlcCer. B, ganglioside GM1 and monochain glycolipids. ▾, GM1; ▪, Lac-Sph; •, GlcSph; ▴, hexyl glucoside. Experimental conditions and binding isotherm calculations were the same as described in the legend for Fig. 2.
Binding Affinity of WT-GLTP for Different Glycolipids—Table 1 shows the binding constants for the spectral responses produced by titration of WT-GLTP with various glycolipids. KD values between ∼0.6 and 2.1 μm were determined for glycolipids with one or two sugars that contained either short saturated acyl chains or long acyl chains with mono- or diunsaturation. Slow equilibration was observed when these simple glycolipids contained long saturated acyl chains (>16 carbons), thus interfering with accurate assessment of KD values. With the multiglycosylated and more soluble ganglioside GM1, fast equilibration was observed despite the predominance of stearoyl acyl chains (supplemental Fig. S2). The KD for GM1 was ∼1.7 μm. The very modest spectral changes elicited by high amounts of lyso-GlcCer and lyso-LacCer translated into KD values of ∼80 and ∼60 μm, respectively. The lack of change in Trp fluorescence signal elicited by nonglycosylated lipids (e.g. ceramide, sphingomyelin, and PC) was consistent with lack of binding.
TABLE 1.
KD values of WT-GLTP for various glycolipids Dissociation constants were determined by nonlinear fitting analyses of glycolipid-induced changes in GLTP Trp emission intensity as detailed in the supplemental information. Values were calculated at fixed wavelength (353 nm) in the emission peak (50). Extrusion vesicles contained 10 mol % glycolipid. Glycolipid pool size in the POPC vesicle outer leaflet and available for interaction with GLTP was estimated from previously determined transbilayer distributions (51–53).
| KD | R2 | |
|---|---|---|
| μM | ||
| GalCer | ||
| 8:0 GalCer | 0.60 ± 0.13 | 0.962 |
| 12:0 GalCer | 1.60 ± 0.30 | 0.987 |
| 18:0 GalCera | 4.20 ± 0.39 | 0.979 |
| 18:1 GalCer | 1.41 ± 0.35 | 0.975 |
| 18:2 GalCer | 0.52 ± 0.06 | 0.985 |
| 24:0 GalCera | 9.30 ± 1.52 | 0.980 |
| 24:1 GalCer | 1.29 ± 0.16 | 0.993 |
| LacCer | ||
| 8:0 LacCer | 0.90 ± 0.18 | 0.976 |
| 12:0 LacCer | 1.23 ± 0.27 | 0.978 |
| 16:0 LacCera | 4.56 ± 0.33 | 0.988 |
| 18:0 LacCera | 6.39 ± 0.27 | 0.996 |
| 18:1 LacCer | 1.16 ± 0.11 | 0.996 |
| 24:0 LacCera | 15.88 ± 0.78 | 0.995 |
| 24:1 LacCer | 2.06 ± 0.29 | 0.992 |
| Other glycolipids | ||
| 8:0 GlcCer | 0.93 ± 0.19 | 0.980 |
| 12:0 GlcCer | 1.31 ± 0.19 | 0.982 |
| 18:1 GlcCer | 1.41 ± 0.31 | 0.993 |
| GM1 | 1.69 ± 0.30 | 0.988 |
| Monochain glycolipids | ||
| GlcSph | 79.46 ± 10.58 | 0.997 |
| LacSph | 62.46 ± 13.12 | 0.993 |
| Hexyl glucoside | 158.80 ± 21.59 | 0.992 |
| Extrusion vesicles | ||
| 8:0 GalCer | 6.47 ± 2.59 | 0.993 |
| 18:1 GalCer | 3.98 ± 1.43 | 0.986 |
| 18:2 GalCer | 1.99 ± 0.65 | 0.976 |
| 8:0 LacCer | 1.90 ± 0.42 | 0.983 |
| 18:1 LacCer | 3.45 ± 1.06 | 0.990 |
Slow equilibration (supplemental Fig. S2) for lipids containing long saturated acyl chains (e.g. 18:0, 24:0, and 16:0) increased uncertainty of these values
WT-GLTP Acquisition of Glycolipids from POPC Vesicles—To determine whether the mode of glycolipid presentation affects the binding affinity by WT-GLTP and whether WT-GLTP partitioning to the membrane surface significantly contributes to the changes in Trp emission, GSLs were presented as minor components in PC bilayer vesicles. Fig. 5A shows the changes in intrinsic Trp emission of WT-GLTP elicited by the incremental addition of POPC extrusion vesicles (0–7 μm) containing 10 mol % 18:1-GalCer. Readily apparent are the dramatic blue shift in the λmax, i.e. ∼347–336 nm, and substantially decreased (∼40%) emission intensity, eventually attaining saturation. In contrast, when the POPC extrusion vesicles lacked glycolipid (Fig. 5B), the Trp emission intensity remained nearly invariant and underwent only a slight red shift (2–3 nm). The WT-GLTP uptake kinetics for glycolipid from the POPC vesicles were similar to glycolipid presented by ethanol microinjection, reaching equilibrium after <3 min (supplemental Fig. S2).
FIGURE 5.
Changes in Trp emission of WT-GLTP by incubation with curvature stress-free and curvature-stressed bilayer vesicles. A, 10 mol % 18:1 GalCer in POPC vesicles (curvature stress-free by extrusion). B, POPC vesicles (curvature stress-free by extrusion). C, binding isotherms for WT-GLTP + POPC extrusion vesicles containing GalCer or LacCer. D, POPC vesicles (curvature-stressed by probe sonication, i.e. SUVs). In A and B, the Trp emission scans correspond to extrusion vesicle POPC concentrations of 0, 1.6, 2.4, 3.2, 4.8, 6.4, 8, 9.6, 11.2, 12.8, 14.4, and 16 μm with respect to decreasing emission intensity. Glycolipid pool size in the POPC vesicle outer leaflet and available for interaction with GLTP was estimated from previously reported transbilayer distributions (51–53). In D, the resulting Trp emission scans correspond to probe-sonicated vesicle POPC concentrations of 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 22, and 24 μm with respect to decreasing emission intensity. Experimental conditions and binding isotherm calculations were the same as described in the legend for Fig. 2. No reduction in emission intensity was observed when free Trp was incubated with either probe-sonicated or extruded POPC vesicles produced.
It is noteworthy that extrusion vesicles were used in the preceding experiments. Different results have been reported with probe-sonicated vesicles (SUVs) by West et al. (23), who observed significant Trp emission intensity decreases and a moderately blue-shifted λmax (3–5 nm) upon stepwise additions of glycolipid-free POPC SUVs to WT-GLTP. Because their results indicated that GLTP partitioning to vesicles might substantially contribute to the Trp emission changes, we revisited the issue. We confirmed that stepwise additions of glycolipid-free POPC SUVs (∼25-nm diameter) significantly decreased the Trp emission intensity of WT-GLTP (Fig. 5D). However, we found little or no reduction in Trp emission intensity if the glycolipid-free POPC vesicles were produced by extrusion and were large enough to be free of curvature stress (Fig. 5B).
The KD values for GalCers in POPC extrusion vesicles were slightly larger than KD values for ethanol-injected GalCers (Table 1). For LacCers in POPC extrusion vesicles, the reliability of the KD values was decreased due to a somewhat anomalous spectral response (supplemental Fig. S6). The Trp emission response showed the expected initial changes, i.e. blue shift in the λmax and decrease in emission intensity. However, at vesicle concentrations >3–4 μm, the blue shift began to reverse, whereas the emission intensity increased slightly, consistent with “blue wing” emission attenuation known to occur when vesicle concentrations are high enough to induce light-scattering contributions to the Trp emission response (31). The phenomenon was especially evident for extrusion vesicles containing glycolipids with uncharged sugar head-groups that could extend beyond the membrane surface delineated by the phosphatidylcholine headgroup and promote stable trans-vesicle contacts (32, 33).
Does Glycolipid Binding Promote WT-GLTP Self-Association?—The Trp emission changes observed during WT-GLTP acquisition of glycolipid seemed to approach apparent saturation at lipid-to-protein molar ratios below the 1:1 stoichiometry anticipated for the single glycolipid binding site in GLTP. Similar outcomes were observed using various glycolipids and different WT-GLTP starting concentrations, i.e. 0.5 or 2 μm instead of 1.0 μm (∼25 μg/ml). The findings raised the issue of whether glycolipid acquisition by WT-GLTP could promote protein self-interaction in ways that limit access to Trp-96. The SHARP2 algorithm was used to locate such surface residue patches on WT-GLTP (28, 29). The six parameter analysis (S, solvation potential; H, hydrophobicity; A, accessible surface area; R, residue interface propensity; P, protrusion; and P, planarity) identified surface patches near Trp-96 as potential self-interaction sites in WT-GLTP. Binding of glycolipid was found to slightly enhance the interaction potential of patches located near Trp-96. However, nondenaturing gel electrophoresis performed on WT-GLTP, before and after titration with N-octanoyl GalCer, showed bands migrating only as monomers with no evidence of protein oligomerization (data not shown) under conditions similar to the fluorescence experiments.
When evaluated by ESI-MS under nondenaturing conditions (Fig. 3A), the protein·lipid complexes were found to be almost exclusively monomeric (99%). Ions corresponding to glycolipid-free, monomeric rGLTP (25,248 Da) and to monomeric complex (rGLTP + N-octanoyl GalCer (25,836 Da)) dominated the spectra, whereas no clear evidence of dimer was observed. By fast protein liquid chromatography SEC (Superdex 75 HR 10/30), there was also no evidence of WT-GLTP eluting as dimers or oligomers after binding with 24:1 GalCer or 8:0 LacCer by FPLC SEC (supplemental Fig. S7) at 10–20-fold higher protein concentrations than in the fluorescence experiments. Only at GLTP concentrations ∼500-fold higher than in the fluorescence experiments could minor amounts (<10%) of protein oligomer be detected. Thus, we concluded that glycolipid-induced formation of soluble, stable GLTP dimer did not contribute to the observed fluorescence emission response.
DISCUSSION
The present study establishes that acquisition of glycolipid by GLTP is primarily responsible for the dramatic changes in intrinsic Trp fluorescence that occur when the protein encounters membranes containing glycolipids. The glycolipid-induced GLTP “signature response” includes a 40% decrease in emission intensity and a ∼12-nm blue shift in the emission in the λmax. Deciphering the molecular basis for the glycolipid-induced changes in intrinsic Trp emission of GLTP has enabled comparison of the binding affinities of glycolipids containing various structural alterations, whereas avoiding potential artifacts associated with labeling using extraneous fluorophores. Although the ability of GLTP to selectively transfer different glycolipids between membrane vesicles was recognized many years ago (9), the uniqueness of the GLTP fold among lipid binding/transfer proteins has only recently been revealed by high resolution x-ray diffraction studies (12–14). Remarkably, the GLTP·glycolipid complex does not show global conformational changes when compared with glycolipid-free GLTP, but only localized and limited expansion of a hydrophobic pocket that encapsulates the nonpolar lipid region in concert with sugar headgroup anchoring to an invariant, surface-localized, sugar-amide recognition region. These findings helped elucidate the workings of GLTP (9, 13, 17, 34). Still, many important aspects have remained uncharacterized including the physicochemical parameters of glycolipids that regulate their uptake and binding affinity by GLTP. To achieve such insights, different modes of presentation of poorly soluble GSLs, i.e. dispersed in fluid PC vesicles or as pure GSL aggregates via ethanol microinjection, were adopted because of the benefits for use with low solubility substrates/ligands that often deviate from classical enzymatic and binding behavior (e.g. Ref. 35).
The data indicate that GLTP binds glycolipids with dissociation constants in the low-to-submicromolar range in aqueous solution. The moderate strength of the binding is consistent with the ability of the protein to transfer glycolipid to membranes or other proteins and could favor the “sphingosine-out” conformation for glycolipid (with respect to localization in or out of the GLTP hydrophobic pocket/tunnel) as being the predominant complexation mode in aqueous solution. Previously, both sphingosine-out and “sphingosine-in” glycolipid conformations were observed in GLTP·glycolipid crystal complexes (12–14). Among the glycolipids, the lowest KD values are observed for GalCers and LacCers with flexible acyl chains that can probably aid rapid encapsulation by the hydrophobic pocket of GLTP. These two glycolipids are tethered primarily through the initial ceramide-linked sugar to the GLTP sugar headgroup recognition site via an identical number of multiple hydrogen bonds (12, 13). GlcCer species showed similar KD values despite having one less hydrogen bond involved in tethering the sugar to the GLTP surface (13). KD values for ganglioside GM1 were 2–3-fold higher than those of various mono- and diglycosylated species, consistent with increased GM1 solubility provided by the pentaglycosylated headgroup including negatively charged N-acetylneuraminic acid. The key role played by the glycolipid amide-linked acyl chain during binding was evident by the much weaker affinities of GlcSph and Lac-Sph when compared with the various acylated species, i.e. 30–40-fold higher KD values.
Differing Trp Emission Response of WT-GLTP with LUVs and SUVs—Unexpectedly, partitioning of GLTP from the aqueous milieu to membrane vesicles free of curvature stress induced little change in Trp emission. It had been suggested previously that protein partitioning to the membrane is a major contributor to the alterations in Trp emission of GLTP based on experiments with relatively high concentrations (0–500 μm) of curvature-stressed bilayer vesicles, i.e. POPC SUVs (23). This finding seemed reasonable given the known behavior of other monotopic and amphitropic peripheral proteins and the established localization of Trp-96 and Trp-142 on the GLTP surface within a potential membrane-interaction region enriched in lysines and nonpolar residues (9, 12–14). The data (Fig. 5D) show that when POPC SUV concentrations are kept low (≤24 μm PC), decreases in Trp fluorescence intensity do occur but with little change in emission λmax. The decrease in Trp emission intensity observed with POPC SUVs is consistent with shallow penetration by GLTP and confinement of the Trp residues to the SUV interfacial region of the membrane outer surface (17) where curvature stress alters lipid packing. The altered lipid packing does not increase interfacial hydration but rather increases the mobility of hydrated functional groups in the vicinity of the glyceride backbone of the outer surface phospholipids (36), possibly enhancing their quenching potential for Trp. It is noteworthy that examples of decreases in Trp emission intensity have been reported for other proteins interacting with high curvature SUVs produced by probe sonication (37–39).
When the glycolipid-free POPC vesicles lack curvature stress, the Trp emission intensity of GLTP remains nearly unchanged, whereas displaying a slightly red-shifted λmax. The minimal fluorescence changes are consistent with tighter lipid interfacial packing in planar membranes and diminished partitioning of GLTP to LUVs when compared with SUVs (17, 40). Even so, the POPC LUV effectiveness for glycolipid presentation to GLTP is restricted to monoglycosylated lipids with sugar headgroups not prone to promote trans-membrane contacts/aggregation. Fortunately, microinjection of ethanol-solubilized glycolipids in small, limited amounts with GLTP elicits changes in Trp emission with various glycolipids but not phosphatidylcholine or other non glycolipids. Isolation of the GLTP·glycolipid complex by FPLC SEC reveals that the soluble GLTP·glycolipid complex has a blue-shifted λmax (∼335 nm) consistent with the changes in GLTP Trp emission reflecting glycolipid binding and formation of a GLTP·glycolipid complex.
Trp-96 Is Chiefly Responsible for Glycolipid-induced Emission Changes of GLTP—Among the 3 Trp residues of GLTP, Trp-96 dominates the emission changes observed during glycolipid binding. A W96F-GLTP mutant capable of transferring glycolipid between vesicles shows almost no changes in Trp emission signal when incubated with ethanol-injected glycolipid. Moreover, with WT-GLTP, where glycolipid binding of various glycolipids induces little change in global architecture (12–14), local conformational mobility, indicated by B-factor analysis, is restricted in the vicinity of the sugar headgroup recognition center containing Trp-96, which resides near the bottom of a shallow, exposed depression on the GLTP surface (12). Nearby residues (e.g. Arg-99, Met-95, His-140, Tyr-207) that could potentially quench Trp-96 emission signal remain unaltered by glycolipid binding with respect to their distance and orientation to Trp-96 (Fig. 6). Stacking of the ceramide-linked sugar of the glycolipid above Trp-96 positions the ceramide amide nitrogen within ∼3.7 Å of the Trp-96 benzene ring, the glycosidic oxygen linking ceramide to the sugar within ∼4.1 Å of the Trp-96 heterocyclic junction and the sugar ring itself within ∼3.7–4.3 Å of the nitrogen-containing pyrrole ring of Trp-96 (Fig. 6). The coplanar stacking of the b-face of the ceramide-linked sugar-amide entity close to Trp-96 is expected to affect the local polarity associated with the π electrons of the indole aromatic ring. An inductive effect of the O and OH groups of sugars on the C–H-bond hydrogen atoms would render them more positively charged and potentially promote interaction with the electron-rich aromatic ring (41). The close proximity of the amide nitrogen of the glycolipid ceramide could exacerbate the effect, possibly accounting for the simultaneous large magnitude quenching and blue-shifting of the Trp emission signal, which reflects the environmental polarity change brought to Trp-96 by the juxtapositioning of glycolipid. Indeed, stacking interactions between free sugar ligands and Trps of sugar binding and transport proteins reportedly alter Trp fluorescence responses, albeit those responses are vastly reduced in magnitude (42–48).
Significance—The significance of the present findings with human WT-GLTP lies in the increased understanding of the molecular workings of the GLTP fold. Although other lipid binding/transfer proteins use conformational folds dominated by a β-sheet, i.e. β-grooves/concave cups and β-barrels, or helical bundles stabilized by multiple disulfide bridges, i.e. saposin folds (9, 12–14), GLTP relies on a two-layer fold, dominated by α-helices, with an expandable lipid binding pocket. The GLTP fold employs a membrane-interaction domain that differs from the C1, C2, PH, PX, and FYVE domains found in many monotopic and amphitropic proteins (17–21), endows other larger proteins, such as FAPP2, with key functionality (49, 50), and delineates the new GLTP superfamily, with orthologs occurring widely in eukaryotes (9–11, 15, 16). Among the mostly highly conserved residues in the active GLTP binding pocket is Trp, located at positions equivalent to Trp-96 of human GLTP. Whether the changes in Trp emission induced by glycolipid binding extend to more distant members of the GLTP superfamily is presently unknown but will be of interest to explore in future investigations.
Supplementary Material
Acknowledgments
We thank Prof. Julian G. Molotkovsky (Shemyakin Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences) for providing the fluorescent glycolipids used to verify transfer activity and Dr. Xin-Min Li for synthesizing several of the chain-pure glycolipids.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AF209704, AY372530, AY372531, AY372532. The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession numbers AAF33210 and NP_057517.
This work was supported, in whole or in part, by National Institutes of Health Grants CA121493 and GM45928 from the United States Public Health Service and NCI. This work was also supported by grants from the Abby Rockefeller Mauzé Trust and the Dewitt Wallace Fund, the Maloris Foundation, the Mayo Foundation for Medical Education and Research, and The Hormel Foundation.
The on-line version of this article (available at http://www.jbc.org) contains supplemental information and seven supplemental figures.
Footnotes
The abbreviations used are: GLTP, glycolipid transfer protein; WT-GLTP, wild-type GLTP; rGLTP, recombinant GLTP; LacCer, lactosylceramide; GalCer, galactosylceramide; GlcCer, glucosylceramide; PC, phosphatidylcholine; POPC, 1-palmitoyl-2-oleyl-phosphatidylcholine; GM1, monosialoganglioside; SUV, small unilamellar vesicle; LUV, large unilamellar vesicle; GSL, glycosphingolipid; FPLC, fast protein liquid chromatography; ESI-MS, electrospray ionization mass spectrometry; SEC, size exclusion chromatography.
References
- 1.Viard, M., Parolini, I., Rawat, S. S., Fecchi, K., Sargiacomo, M., Puri, A., and Blumenthal, R. (2004) Glycoconj. J. 20 213-222 [DOI] [PubMed] [Google Scholar]
- 2.Schnaar, R. L. (2004) Arch. Biochem. Biophys. 426 163-172 [DOI] [PubMed] [Google Scholar]
- 3.Bektas, M., and Spiegel, S. (2004) Glycoconj. J. 20 39-47 [DOI] [PubMed] [Google Scholar]
- 4.Morales, A., Colell, A., Mari, M., Garcia-Ruiz, C., and Fernandez-Checa, J. C. (2004) Glycoconj. J. 20 579-588 [DOI] [PubMed] [Google Scholar]
- 5.Gouaze-Andersson, V., and Cabot, M. C. (2006) Biochim. Biophys. Acta 1758 2096-2103 [DOI] [PubMed] [Google Scholar]
- 6.Sabourdy, F., Kedjouar, B., Sorli, S. C., Colié, S., Milhas, D., Salma, Y., and Levade, T. (2008) Biochim. Biophys. Acta 1781 145-183 [DOI] [PubMed] [Google Scholar]
- 7.Hannun, Y. A., and Obeid, L. M. (2008) Nat. Rev. Mol. Cell Biol. 9 139-150 [DOI] [PubMed] [Google Scholar]
- 8.Prinetti, A., Loberto, N., Chigorno, V., and Sonnino, S. (2009) Biochim. Biophys. Acta 1778 184-193 [DOI] [PubMed] [Google Scholar]
- 9.Brown, R. E., and Mattjus, P. (2007) Biochim. Biophys. Acta 1771 746-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou, X., Chung, T., Lin, X., Malakhova, M. L., Pike, H. M., and Brown, R. E. (2008) BMC Genomics 9 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West, G., Viitanen, L., Alm, C., Mattjus, P., Salminen, T. A., and Edqvist, J. (2008) FEBS J. 275 3421-3437 [DOI] [PubMed] [Google Scholar]
- 12.Malinina, L., Malakhova, M. L., Teplov, A., Brown, R. E., and Patel, D. J. (2004) Nature 430 1048-1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malinina, L., Malakhova, M. L., Kanak, A. T., Lu, M., Abagyan, R., Brown, R. E., and Patel, D. J. (2006) PLoS Biol. 4 e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Airenne, T. T., Kidron, H., Nymalm, Y., Nylund, M., West, G. P., Mattjus, P., and Salminen, T. A. (2006) J. Mol. Biol. 355 224-236 [DOI] [PubMed] [Google Scholar]
- 15.Murzin, A. G., Brenner, S. E., Hubbard, T., and Chothia, C. (1995) J. Mol. Biol. 247 536-540 [DOI] [PubMed] [Google Scholar]
- 16.Madera, M., Vogel, C., Kummerfeld, S. K., Chothia, C., and Gough, J. (2004) Nucleic Acids Res. 32 D235-D239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao, C. S., Chung, T., Pike, H. M., and Brown, R. E. (2005) Biophys. J. 89 4017-4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho, W., and Stahelin, R. V. (2005) Annu. Rev. Biophys. Biomol. Struct. 34 119-151 [DOI] [PubMed] [Google Scholar]
- 19.Hurley, J. H. (2006) Biochim. Biophys. Acta 1761 805-811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho, W. (ed) (2006) Biochim. Biophys. Acta (Special Issue: Lipid-Binding Domains) 1761 803-968 [Google Scholar]
- 21.Lemmon, M. A. (2008) Nat. Rev. Mol. Cell Biol. 9 99-111 [DOI] [PubMed] [Google Scholar]
- 22.Li, X.-M., Malakhova, M. L., Lin, X., Pike, H. M., Chung, T., Molotkovsky, J. G., and Brown, R. E. (2004) Biochemistry 43 10285-10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West, G., Nylund, M., Slotte, J. P., and Mattjus, P. (2006) Biochim. Biophys. Acta 1758 1732-1742 [DOI] [PubMed] [Google Scholar]
- 24.Smaby, J. M., Momsen, M., Kulkarni, V. S., and Brown, R. E. (1996) Biochemistry 35 5696-5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai, X., Li, X.-M., Momsen, M. M., Brockman, H. L., and Brown, R. E. (2006) Biophys. J. 91 2490-2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartlett, G. R. (1959) J. Biol. Chem. 234 466-468 [PubMed] [Google Scholar]
- 27.Malakhova, M. L., Malinina, L., Pike, H. M., Kanack, A. T., Patel, D. J., and Brown, R. E. (2005) J. Biol. Chem. 280 26312-26320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, S., and Thornton, J. M. (1997) J. Mol. Biol. 272 133-143 [DOI] [PubMed] [Google Scholar]
- 29.Jones, S., and Thornton, J. M. (1997) J. Mol. Biol. 272 121-132 [DOI] [PubMed] [Google Scholar]
- 30.Persson, D., Thorén, P. E. G., Lincoln, P., and Nordén, B. (2004) Biochemistry 43 11045-11055 [DOI] [PubMed] [Google Scholar]
- 31.Ladokhin, A. S., Jayasinghe, S., and White, S. H. (2000) Anal. Biochem. 285 235-245 [DOI] [PubMed] [Google Scholar]
- 32.Corti, M., Cantù, L., Brocca, P., and Del Favero, E. (2007) Curr. Opin. Colloid Interface Sci. 12 148-154 [DOI] [PubMed] [Google Scholar]
- 33.Yu, Z., Calvert, T. L., and Leckband, D. (1997) Biochemistry 37 1540-1550 [DOI] [PubMed] [Google Scholar]
- 34.Rao, C., Lin, X., Pike, H. M., Molotkovsky, J. G., and Brown, R. E. (2004) Biochemistry 43 13805-13815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg, O. G., Gelb, M. H., Tsai, M. D., and Jain, M. K. (2001) Chem. Rev. 101 2613-2653 [DOI] [PubMed] [Google Scholar]
- 36.Sy`kora, J., Jurkiewicz, P., Epand, R. M., Kraayenhof, R., Langner, M., and Hof, M. (2005) Chem. Phys. Lipids 135 213-221 [DOI] [PubMed] [Google Scholar]
- 37.Joseph, M., and Nagaraj, R. (1995) J. Biol. Chem. 270 16749-16755 [DOI] [PubMed] [Google Scholar]
- 38.Christiaens, B., Symoens, S., Verheyden, S., Engelborghs, Y., Joliot, A., Prochiantz, A., Vandekerckhove, J., Rosseneu, M., and Vanloo, B. (2002) Eur. J. Biochem. 269 2918-2926 [DOI] [PubMed] [Google Scholar]
- 39.Chowdary, T. K., Raman, B., Ramakrishna, T., and Rao, C. M. (2007) Biochem. J. 401 437-445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nylund, M., Fortelius, C., Palonen, E. K., Molotkovsky, J. G., and Mattjus, P. (2007) Langmuir 23 11726-11733 [DOI] [PubMed] [Google Scholar]
- 41.Vázquez-Ibar, J. L., Guan, L., Svrakic, M., and Kaback, H. R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12706-12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas, N. K., Vyas, M. N., and Quiocho, F. A. (1988) Science 242 1290-1295 [DOI] [PubMed] [Google Scholar]
- 43.Sixma, T. K., Pronk, S. E., Kalk, K. H., van Zanten, B. A., Berghuis, A. M., and Hol, W. G. (1992) Nature 355 561-564 [DOI] [PubMed] [Google Scholar]
- 44.Weis, W. I., and Drickamer, K. (1996) Annu. Rev. Biochem. 65 441-473 [DOI] [PubMed] [Google Scholar]
- 45.Soisson, S. M., MacDougall-Shackleton, B., Schleif, R., and Wolberger, C. (1997) J. Mol. Biol. 273 226-237 [DOI] [PubMed] [Google Scholar]
- 46.Zolotnitsky, G., Cogan, U., Adir, N., Solomon, V., Shoham, G., and Shoham, Y. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11275-11280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sujatha, M. S., and Balaji, P. V. (2004) Proteins 55 44-65 [DOI] [PubMed] [Google Scholar]
- 48.Sujatha, M. S., Sasidhar, Y. U., and Balaji, P. V. (2004) Protein Sci. 13 2502-2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Angelo, G., Polishchuk, E., Di Tullio, G., Santoro, M., Di Campli, A., Godi, A., West, G., Bielawski, J., Chuang, C.-C., van der Spoel, A. C., Platt, F. M., Hannun, Y. A., Polishchuk, R., Mattjus, P., and De Matteis, M. A. (2007) Nature 449 62-67 [DOI] [PubMed] [Google Scholar]
- 50.Halter, D., Neumann, S., van Dijk, S. M., Wolthoorn, J., de Mazière, A. M., Vieira, O. V., Mattjus, P., Klumperman, J., van Meer, G., and Sprong, H. (2007) J. Cell Biol. 179 101-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattjus, P., Malewicz, B., Valiyaveettil, J. T., Baumann, W. J., Bittman, R., and Brown, R. E. (2002) J. Biol. Chem. 277 19476-19481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malewicz, B., Valiyaveettil, J. T., Jacob, K., Byun, H.-S., Mattjus, P., Baumann, W. J., Bittman, R., and Brown, R. E. (2005) Biophys. J. 88 2670-2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sillence, D. J., Raggers, R. J., Neville, D. C. A., Harvey, D. J., and van Meer, G. (2000) J. Lipid Res. 41 1252-1260 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.