Abstract
Sox10 is a member of the group E Sox transcription factor family and plays key roles in neural crest development and subsequent cellular differentiation. Sox10 binds to regulatory sequences in target genes via its conserved high mobility group domain. In most cases, Sox10 exerts its transcriptional effects in concert with other DNA-binding factors, adaptor proteins, and nuclear import proteins. These interactions can lead to synergistic gene activation and can be cell type-specific. In earlier work, we demonstrated that Sox10 transactivates the nicotinic acetylcholine receptor α3 and β4 subunit genes and does so only in neuronal-like cell lines, raising the possibility that Sox10 mediates its effects via interactions with co-regulatory factors. Here we describe the identification of the armadillo repeat-containing protein, ARMCX3, as a Sox10-interacting protein. Biochemical analyses indicate that ARMCX3 is an integral membrane protein of the mitochondrial outer membrane. Others have shown that Sox10 is a nucleocytoplasmic shuttling protein. We extend this observation and demonstrate that, in the cytoplasm, Sox10 is peripherally associated with the mitochondrial outer membrane. Both Sox10 and ARMCX3 are expressed in mouse brain and spinal cord as well as several cell lines. Overexpression of ARMCX3 increased the amount of mitochondrially associated Sox10. In addition, although ARMCX3 does not possess intrinsic transcriptional activity, it does enhance transactivation of the nicotinic acetylcholine receptor α3 and β4 subunit gene promoters by Sox10. These results suggest that Sox10 is a membrane-associated factor whose transcriptional function is increased by direct interactions with ARMCX3 and raise the possibility of a signal transduction cascade between the nucleus and mitochondria through Sox10/ARMCX3 interactions.
Sox proteins are important transcription factors involved in a wide variety of developmental processes, including sex determination, chondrogenesis, and neurogenesis (1, 2). To date, 20 Sox proteins have been identified in mammals (2–4). The Sox family is defined by a highly conserved high mobility group (HMG)2 domain that functions as a DNA-binding and -bending domain (5, 6) as well as a protein-protein interaction domain (7). Sequences outside the HMG domain are variable among the Sox family members and include transcriptional regulatory domains and additional protein-protein interaction domains. The Sox family is phylogenetically subdivided into several groups, A–J (2). Similar to other transcriptional regulatory factors, the Sox proteins perform their functions via interactions with partner proteins. This may occur through one of a number of interactions. For example, Sox proteins may interact directly with other transcription factors, they may interact indirectly through adaptor proteins, and they may interact with nuclear import proteins (8). It is likely that this repertoire of interactions will expand as additional Sox-interacting proteins are identified.
Sox10 is a member of the E group of Sox factors. Other members of this group include Sox8 and Sox9 (8). Sox10 is a key player in neural crest development (9) as reflected in the observation that Sox10 mutations are associated with Waardenburg-Shah syndrome in humans, which is characterized by hypopigmentation of the skin, heterochromia irides, deafness, and usually aganglionic megacolon (10, 11). Similarly, mutations in the mouse Sox10 gene, as seen in the dominant megacolon strain, give rise to aganglionosis of the colon, pigmentation defects, significant losses of neurons and glia in the peripheral nervous system, and a complete loss of the enteric nervous system (12, 13). Consistent with these defects is the demonstration that Sox10 is a critical factor in terminal oligodendrocyte differentiation as well as pigment cell development (5, 14). More recently, a direct role for Sox10 in the specification of sensory neurons derived from the neural crest has been reported (15). This latter point is particularly germane to our previous work in which we demonstrated that the nicotinic acetylcholine (nACh) receptor α3 and β4 subunit gene promoters are transactivated by Sox10 in a cell type-specific manner, occurring only in neuronal-like cell lines but not in non-neuronal cell lines (16). This raised the possibility that, as discussed above, Sox10 exerts its effects through interactions with other regulatory factors. Directly related to this, we previously showed that, in addition to Sox10, the transcription factors Sp1, Sp3, Purα, and heterogeneous nuclear ribonucleoprotein K physically interact with the β4 subunit gene promoter region (17–20). Furthermore, we showed that Sox10 directly interacts with all four factors (21) and that Sox10 and the Sp factors can synergistically transactivate gene expression (22). However, the Sp factors as well as Purα and heterogeneous nuclear ribonucleoprotein K are ubiquitously expressed suggesting that other cofactors may be involved in mediating the cell type-specific transcriptional function of Sox10.
To gain further insight to the mechanism underlying the transcriptional regulatory function of Sox10, we carried out a protein-protein interaction screen using Sox10 as the bait. Here we report the identification of a heretofore functionally uncharacterized armadillo (ARM) repeat-containing protein, ARMCX3, as a Sox10-interacting protein. We show that ARMCX3 is a mitochondrial outer membrane protein that increases the level of mitochondrially associated Sox10. In addition, we demonstrate that ARMCX3 and Sox10 are co-expressed in the central nervous system as well as in several cell lines and, further, that endogenous ARMCX3 and Sox10 physically associate. Importantly, we show that ARMCX3 enhances the Sox10-mediated transactivation of the nACh receptor α3 and β4 subunit gene promoters.
EXPERIMENTAL PROCEDURES
Plasmids—A fragment of the rat Sox10 cDNA (GenBank™ number AJ001029) that encodes the N-terminal 100 amino acids of the Sox10 protein was amplified by PCR from pCMV5-Sox10 (a kind gift from Dr. Michael Wegner). This fragment was subcloned into the EcoRI and SalI sites of pGBKT7 to generate the bait plasmid pGBKT7-Sox10N100 (Fig. 1A). The prey plasmid, pACT2-ARMCX3, was used as the PCR template to amplify full-length and deletion fragments of ARMCX3. pCMV-Myc-ARMCX3 was generated by subcloning the full-length ARMCX3 cDNA into the SalI and NotI sites of pCMV-Myc (Clontech). Amplified deletion fragments were also subcloned into the SalI and NotI sites of pCMV-Myc to generate pCMV-Myc-ARMCX3Δ1 (encoding amino acids 1–272 of ARMCX3), pCMV-Myc-ARMCX3Δ2 (encoding amino acids 1–200 of ARMCX3), and pCMV-Myc-ARMCX3Δ3 (encoding amino acids 1–109 of ARMCX3). pCMV-Myc-ARMCX3TMDΔ was generated by deleting nucleotides 19–75 of the ARMCX3 coding sequence using a QuikChange® II site-directed mutagenesis kit (Stratagene). pCMV-HA-FLAG was generated by inserting a DNA fragment encoding a FLAG tag into the SfiI site of pCMV-HA (Clontech). Full-length rat Sox10 cDNA and a cDNA fragment encoding amino acids 101–466 were amplified from pCMV5-Sox10 and subcloned into the EcoRI and XhoI sites of pCMV-HA-FLAG, respectively, to generate pCMV-HA-FLAG-Sox10 and pCMV-HA-FLAG-Sox10ΔN.
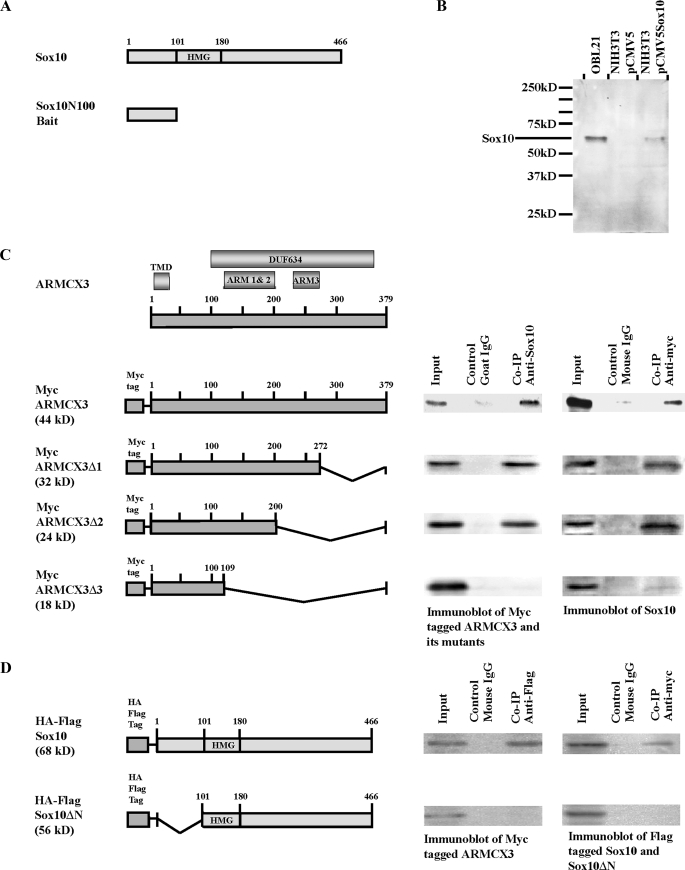
FIGURE 1.
Identification of ARMCX3 as a Sox10-interacting protein. A, schematic representation of the rat Sox10 protein indicating the HMG box (top figure). Numbering refers to amino acids. A cDNA fragment that encodes the first 100 amino acids of Sox10 was subcloned into pGBKT7 to yield the bait plasmid pGBKT7-Sox10N100 (lower figure). B, detection of endogenous Sox10 in OBL21 whole cell extracts and exogenous Sox10 in whole cell extracts of pCMV5-Sox10-transfected NIH3T3 cells. Parallel cultures of pCMV5-transfected NIH3T3 cells were analyzed on the same Western blot. C, schematic representation of ARMCX3 indicating the three ARM repeats, the putative transmembrane domain (TMD) and the DUF634 domain (top figure). Myc-tagged ARMCX3 and its three deletion constructs are shown in the lower portion of the figure. Cell lysates were prepared from OBL21 cells transfected with pCMV-Myc-ARMCX3 and the three deletion constructs. Co-IPs were done using anti-Sox10 and anti-Myc antibodies, as indicated. Mouse IgG and goat IgG were used as negative controls. The left immunoblot presents the results from a precipitation with anti-Sox10 followed by blotting with anti-Myc while the right immunoblot presents the reciprocal experiment in which anti-Myc was used for immunoprecipitation and anti-Sox10 was used for the immunoblotting. D, pCMV-Myc-ARMCX3 was co-transfected with either pCMV-HA-FLAG-Sox10 or pCMV-HA-FLAG-Sox10ΔN into Neuro-2A cells. Cell lysates were prepared, and Co-IPs were done using anti-FLAG and anti-Myc antibodies.
Cell Lines—The mouse olfactory bulb cell line OBL21 (23) was grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Mediatech), 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Neuro-2A cells (24) were grown in minimal essential medium (Invitrogen) containing 10% fetal bovine serum.
Yeast Two-hybrid Screening—The Matchmaker™ Two-Hybrid System 3 (Clontech) was used for protein-protein interaction screening. pGBKT7-Sox10N100 was used as the bait plasmid to screen a pre-transformed mouse brain MATCHMAKER cDNA library (Clontech) according to the manufacturer's suggested protocol. 56 prey plasmids were isolated and sequenced (University of Massachusetts Medical School Nucleic Acid Facility). One of the prey plasmids contained the complete coding sequence of the ARMCX3 gene (GenBank™ number NM_027870). The ARMCX3 coding sequence was fused in-frame with the Gal4-transactivating domain sequence in the pACT2 plasmid. The resulting prey plasmid, pACT2-ARMCX3, and pGBKT7-Sox10N100, were co-transformed into AH109 yeast cells. Transformation of the bait plasmid alone and co-transformation of pACT2-ARMCX3 and pGBKT7 were used as negative controls while co-transformation of pGBKT7–53 and pTD1–1 was used as a positive control (data not shown). Single colonies were selected and streaked onto a quadruple dropout/X-α-Gal plate. Large blue colonies indicated positive interactions in yeast. Peptide sequence analysis was carried out using the NCBI blastp program.
Bioinformatics—GCG, version 11.1.3 (Wisconsin package), was used for bioinformatic analysis. The ARMCX3 transmembrane domain was predicted by the GCG TransMem program, whereas the other conserved domains were predicted by the GCG HmmerPfam program.
Test of Anti-Sox10 Antibody Specificity—Endogenously expressed Sox10 was detected using Western blot analysis of OBL21 cells, which express relatively high levels of Sox10 (see Fig. 4), with 1 μg of anti-Sox10 antibody (Santa Cruz Biotechnology, #sc-17342). As controls, NIH3T3 cells, which do not express endogenous Sox10, were transfected either with pCMV5-Sox10 or the empty expression vector, pCMV5. Transfected cells were harvested 48 h after transfection and subjected to Western blot analysis using the same anti-Sox10 antibody as above.
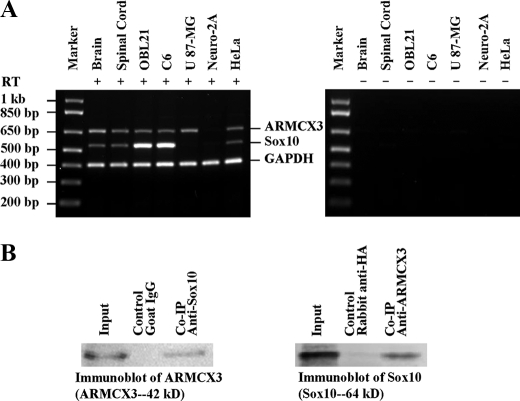
FIGURE 4.
Co-expression and direct interaction between endogenous Sox10 and ARMCX3. A, RNA was extracted from mouse brain and spinal cord, rat C6 cells, human U87-MG and HeLa cells, and mouse Neuro-2A cells. The RNA was used in multiplex RT-PCR using primers that recognize rat, mouse, and human Sox10, ARMCX3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, as endogenous control). The left panel presents results obtained from experiments in which reverse transcriptase was included in the RT-PCR, while the right panel presents results obtained when the enzyme was not included in the RT-PCR. B, mitochondria were isolated from OBL21 cells using differential centrifugation. Lysates prepared from the mitochondria were used for Co-IP experiments using anti-Sox10 and anti-ARMCX3 antibodies. Goat IgG and rabbit anti-HA antibody were used as negative controls.
Interaction Domain Mapping—The interacting domains of Sox10 and ARMCX3 were identified by co-immunoprecipitation (Co-IP) assays using deletion constructs of the two proteins. OBL21 cells, which endogenously express Sox10, were used to express Myc-tagged ARMCX3. OBL21 cells were grown to 70% confluency and transfected with pCMV-Myc-ARMCX3 using a calcium phosphate approach (CalPhos Mammalian Transfection Kit, Clontech). Forty-eight hours after transfection, cells were washed with cold PBS and lysed in Co-IP buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1× protease inhibitor mixture (Roche Applied Science) and 1 mm dithiothreitol). Two 1-ml aliquots of whole cell lysate (1 μg/ml) were precleared with 100 μl of a 50% protein G-agarose suspension (Invitrogen) for 1 h at 4 °C. After pre-clearing, lysates were centrifuged at 4,000 rpm, and the supernatants were transferred to new tubes and incubated with 8 μg of anti-Sox10 antibody (Santa Cruz Biotechnology) and 8 μg of control goat IgG (Santa Cruz Biotechnology, #sc-2028), respectively. The same procedure was performed on another two aliquots of whole cell extract using anti-Myc antibody (Clontech #631206) and control mouse IgG (Sigma #I5381), respectively. Following overnight incubation at 4 °C, 100 μl of a 50% protein G-agarose suspension were added to each Co-IP aliquot and incubated for 4 h at 4 °C to precipitate the Sox10 and ARMCX3 complexes. The protein G-agarose beads were washed with lysis buffer four times and then resuspended in 50 μl of 2× SDS loading buffer. After boiling for 5 min, the samples were subjected to Western blot analysis. Myc-tagged ARMCX3 was detected by anti-Myc antibody (Clontech) at 1 μg/ml. The secondary antibody was a goat anti-mouse horseradish peroxidase conjugate antibody (Pierce #1858413) used at 1:1,000 dilution. Sox10 was detected using an anti-Sox10 antibody (Santa Cruz Biotechnology) at 1 μg/ml. The secondary antibody was a rabbit anti-goat IgG peroxidase-conjugated antibody used at 1:10,000 dilution (Sigma #A5420). Proteins were detected by SuperSignal West Pico Chemiluminescent substrate (Pierce) according to the manufacturer's protocol. Blots were scanned using a Bio-Rad Versa-Doc scanner while quantification was done using Bio-Rad Quantity One version 4.6.5 software. Similar procedures were performed to investigate whether Sox10 can interact with the Myc-tagged mutant proteins ARMCX3Δ1, ARMCX3Δ2, and ARMCX3Δ3. Co-IP experiments were also performed to map the ARMCX3-interacting domain of Sox10: Neuro-2A cells were co-transfected with pCMV-HA-FLAG-Sox10ΔN and pCMV-Myc-ARMCX3. As a positive Co-IP control, Neuro-2A cells were transfected with pCMV-HA-FLAG-Sox10 and pCMV-Myc-ARMCX3. Co-IP procedures were done as described above except that anti-FLAG antibody (Stratagene #200472) was used instead of anti-Sox10 antibody and control mouse IgG (Sigma) was used instead of goat IgG.
Co-immunoprecipitation of Endogenous Sox10 and ARMCX3— OBL21 cells were used to verify the interaction between endogenous Sox10 and ARMCX3. Twenty-seven 150-mm dishes of OBL21 cells were grown, harvested, and homogenized in homogenization buffer (0.25 m sucrose, 20 mm HEPES, pH 7.4, 2 mm MgCl2, 2 mm EGTA, 2 mm EDTA, 1 mm dithiothreitol, and 1× protease inhibitor mixture). Mitochondria were isolated by differential centrifugation as previously described (25). Mitochondria were lysed in Co-IP lysis buffer as described above and then divided into four 0.5-ml aliquots. One aliquot of lysate was incubated with 5 μg of anti-ARMCX3 antibody (Sigma #HPA000967), while another was incubated with 5 μg of anti-Sox10 antibody (Santa Cruz Biotechnology). Negative Co-IP controls were performed by incubating lysates with 5 μg of rabbit anti-HA antibody (Clontech #631207) and 5 μg of goat IgG (Santa Cruz Biotechnology). Co-IP experiments were done as described above except using 40 μl of a 50% protein G-agarose suspension. The Co-IP products, together with inputs, were analyzed by Western blot analysis using anti-Sox10 (1 μg/ml) and anti-ARMCX3 (0.5 μg/ml).
Immunofluorescence—For endogenous Sox10 immunostaining, OBL21 cells were grown on coverslips to 70% confluency. Medium was removed, and the cells were washed briefly with 1× PBS. Cells were fixed in 4% paraformaldehyde for 10 min and then washed 2 times for 5 min each with 1× PBS. Cells were permeabilized with 0.5% saponin (Sigma) for 10 min and washed 3 times for 5 min each with 1× PBS. Cells were blocked with 1× PBS containing 2% BSA (PBS-BSA) for 30 min and then incubated for 1 h with anti-Sox10 antibody (Santa Cruz Biotechnology) at a concentration of 1 μg/ml in PBS-BSA. After 3 washes with PBS-BSA for 5 min each, cells were incubated with Alexa Fluor 488 chicken-anti-goat IgG (Invitrogen #A21467) at 1:200 dilution for 30 min in the dark. Cells were washed in the dark 3 times with PBS for 5 min each and stained for 30 min with 4′,6-diamidino-2-phenylindole (0.2 μg/ml). Coverslips were mounted with one drop of Antifade mounting medium (Invitrogen). Double immunofluorescence was done to show the co-localization of Myc-tagged ARMCX3 and the mitochondrial marker COX IV (26): OBL21 cells transfected with pCMV-Myc-ARMCX3 were immunostained with anti-Myc antibody (2 μg/ml, Clontech). Subsequently, cells were blocked by M.O.M. Blocking Reagent (Vector Laboratories) for 1 h according to the manufacturer's instructions. Cells were then immunostained with anti-COX IV antibody (Abcam #14744) at 1:2,500 dilution. OBL21 cells transfected with pCMV-Myc-ARMCX3TMDΔ were also double immunostained to show the lack of co-localization of mutant ARMCX3 and COX IV.
Endogenous Sox10 Nuclear:Cytoplasmic Ratio Analysis—Endogenously expressed Sox10 in OBL21 cells was detected by standard Western blot analysis. Total nuclear and cytoplasmic proteins were isolated from OBL21 cells (∼2 × 106 cells) using NE-PER nuclear and cytoplasmic extraction reagents according to the manufacturer's protocol (Pierce). Nuclear and cytoplasmic fractions were run in SDS-PAGE gels. Three independent protein isolations were done for each experimental condition. Western blot analysis was carried out using standard approaches. Successful isolation of nuclear and cytoplasmic proteins was demonstrated by exclusive detection of TATA-binding protein (TBP) in the nuclear fraction and α-tubulin in the cytoplasmic fraction. The working concentrations of primary antibodies were 1 μg/ml anti-Sox10 (Santa Cruz Biotechnology, #sc-173432), 1:10,000 dilution of anti-α-tubulin (Abcam #ab7291) and 1:2,000 dilution of anti-TBP antibody (Abcam #ab818). The secondary antibody for Sox10 detection was rabbit anti-goat IgG peroxidase (Sigma #A5420) at 1:10,000 dilution while the secondary antibody for TBP and α-tubulin detection was goat anti-mouse horseradish peroxidase conjugate (Pierce #1858413) at 1:1,000 dilution. Proteins were detected by SuperSignal West Pico Chemiluminescent Substrate (Pierce) according to the manufacturer's protocol. Blots were scanned using a Bio-Rad VersaDoc scanner and quantified by Bio-Rad Quantity One 4.6.5 software.
Subcellular Localization of ARMCX3 and Sox10—The fractionation protocol described below is shown schematically in Fig. 3C. Eight 150-mm dishes of OBL21 cells were transfected with pCMV-Myc-ARMCX3. Forty-eight hours after transfection, cells were harvested and homogenized in 4 times the cell pellet volume of homogenization buffer (0.25 m sucrose, 10 mm HEPES, pH 7.4, 2 mm MgCl2, 2 mm EGTA, 2 mm EDTA, 1 mm dithiothreitol, and 1× protease inhibitor mixture) according to the published protocol (25). The homogenate was centrifuged at 800 × g for 10 min, and the supernatant (post-nuclear supernatant, PNS) was saved. One milliliter of the PNS was layered onto a discontinuous sucrose gradient containing 1 ml each of 12.56%, 18.30%, 26.30%, 33.81%, 38.98%, 47.00%, 55.50%, 64.51%, 73.90%, and 83.80% sucrose (w/v) in 10 mm HEPES, pH 7.4, 2 mm MgCl2, 2 mm EGTA, 0.1% BSA, and 1 mm dithiothreitol. The gradient was centrifuged at 4 °C at 15,000 × g for 3 h as previously described (27). Eleven fractions of 1 ml each were harvested from the top to the bottom (numbered 1–11). Western blot analysis was done to detect the distribution of Sox10, ARMCX3, and the mitochondrial marker COX IV (Fig. 3D). Another 1 ml of the PNS was centrifuged at 100,000 × g for 30 min, and the supernatant was saved as the S100 fraction. The remaining PNS was divided into seven aliquots. Mitochondria were isolated from these aliquots by differential centrifugation as described earlier. Pellets from four aliquots of the mitochondrial preparations were resuspended in 2 mm HEPES, pH 7.4, 220 mm mannitol, 70 mm sucrose and then digested with varying concentrations of proteinase K for 90 min at room temperature. After proteinase K digestion, mitochondria were pelleted and washed as previously described (28). Western blot analysis was done to detect the distribution of Sox10, ARMCX3, and COX IV (Fig. 3E). Pellets from the remaining three aliquots of the mitochondrial preparations were resuspended in either 50 mm Tris-HCl, pH 7.2, 0.2 m Na2CO3, pH 11.4, or 1% Triton X-100, respectively, as previously described (29). After incubation on ice for 30 min, samples were centrifuged at 100,000 × g for 30 min. Supernatants and pellets, together with the PNS and S100 fractions, were subjected to Western blot analysis to detect Sox10 and Myc-tagged ARMCX3 (Fig. 3F).
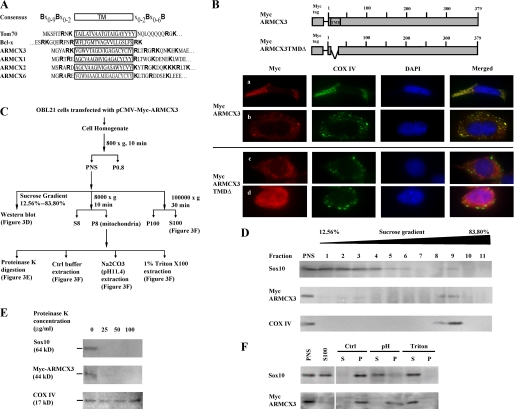
FIGURE 3.
Mitochondrial localization of ARMCX3 and Sox10. A, schematic representation of the consensus MOM-targeting sequence (42) and the corresponding sequences of the murine ARMCX3 proteins, the yeast translocase of the outer membrane of mitochondria70 (Tom70 (43)) and the human Bcl-2-like survival factor Bcl-x (42). Basic amino acids are indicated by bold typeface. The TMDs are enclosed by a rectangle. B, the upper portion of the figure presents schematic representations of Myc-tagged ARMCX3 and Myc-tagged ARMCX3 in which the TMD is deleted (Myc-tagged ARMCX3TMDΔ). OBL21 cells were transfected with one of the two constructs as indicated and subjected to double immunofluorescence using anti-Myc and anti-COX IV antibodies. The first set of images (row a) shows almost complete overlap in ARMCX3 and COX IV expression, while the second set (row b) shows partial overlap in expression. The lower panels (rows c and d) show the loss of co-localization when the TMD of ARMCX3 is deleted. C, flow diagram of the subcellular fractionation protocol (27) used for the studies shown in panels D–F. D, Western blot analysis of fractions from sucrose gradient fractionation of the PNS derived from OBL21 cells transfected with pCMV-Myc-ARMCX3. E, mitochondrial aliquots were digested with three concentrations of proteinase K, as indicated. The samples were then subjected to Western blot analysis using anti-Sox10, anti-Myc, and anti-COX IV antibodies. F, three mitochondrial aliquots were extracted with either 50 mm Tris, pH 7.2 (Ctrl), 0.2 m Na2CO3, pH 11.4 (pH), or 1% Triton X-100 (Triton). Following extraction, the samples were centrifuged at 100,000 × g to yield supernatants (S) and pellets (P), which were subjected to Western blot analysis using anti-Sox10 and anti-Myc antibodies. The PNS and S100 fractions were included in the analysis for comparison.
Multiplex RT-PCR—Total RNA was isolated from the indicated samples (see Fig. 4B) using an RNeasy Mini Kit (Qiagen). Genomic DNA was removed by digestion with an RNase-Free DNase Set (Qiagen). 2 μg of total RNA were reversed transcribed to cDNA using a RETROscript kit (Ambion). A reaction without reverse transcriptase was used as a negative control for each sample. 2 μl of each reverse transcription product as well as the corresponding negative control were used as templates for 50-μl multiplex PCR. The primers were designed to recognize cDNA of human, mouse, and rat. The sequences and working concentrations of the primers were as follows: ALXRTF, 5′-CAGGTATAATGACTGGTCTGATG-3′ (1 μm); ALXRTR, 5′-CTAGTCATGGCTGGATTTTCAGC-3′ (1 μm); SOXRTF, 5′-ACCCCTCAGGCCAGAGCCATG-3′ (0.5 μm); SOXRTR, 5′-GTCCTGAGGGCTGATGGTCAG-3′ (0.5 μm); GAPDHF, 5′-CCCTTCATTGACCTCAACTAC-3′ (0.25 μm); and GAPDHR, 5′-CCAAAGTTGTCATGGATGACC-3′ (0.25 μm). Thirty cycles of PCR were performed with the annealing temperature at 59 °C. Reaction products were analyzed using agarose gel electrophoresis.
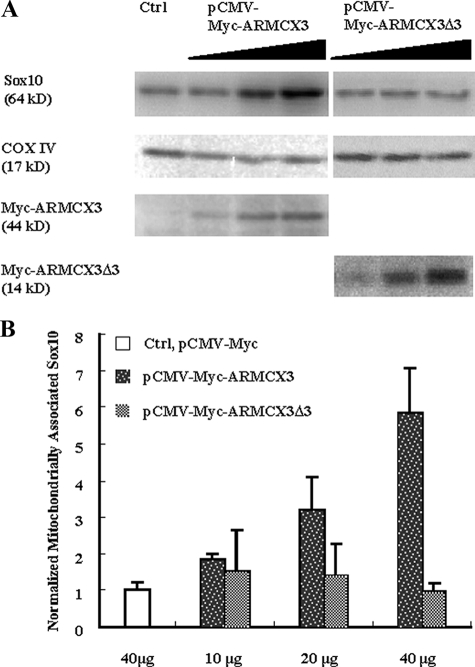
Dose Effect of ARMCX3 on the Mitochondrial Localization of Sox10—Neuro-2A cells were grown on 150-mm dishes to 70% confluency. Three dishes of cells were transfected with 10, 20, and 40 μg of pCMV-Myc-ARMCX3, respectively. Each dish was co-transfected with 20 μg of pCMV5-Sox10. Similar procedures were performed for pCMV-Myc-ARMCX3Δ3. One dish of cells was transfected with 20 μg of pCMV5-Sox10 and 40 μg of pCMV-Myc as the background control. Forty-eight hours after transfection, cells were harvested and mitochondria were isolated by differential centrifugation as described above. Sox10, COX IV, and Myc-tagged ARMCX3 and ARMCX3Δ3 were detected and quantified as described earlier. The amount of Sox10 in each sample was normalized to its loading control (COX IV).
Reporter Gene Assays—Activation of the α3 and β4 nACh receptor subunit gene promoters was measured using luciferase assays. The method used is a modification of our published approach (16, 30). pX1B4FH contains the β4 promoter driving expression of the luciferase reporter gene, whereas pX1A3HS contains the α3 promoter driving expression of the luciferase reporter gene. Neuro-2A cells grown in 6-well plates to 60% confluency were co-transfected with the relevant plasmids using Lipofectamine. To test the effects of Sox10 and ARMCX3 on the receptor gene promoters, 0.5 μg of pCMV-Sox10, 1.5 μg of pCMV-Myc-ARMCX3, and 1.25 μg of pCMV-Myc-ARMCX3Δ3 (equimolar with pCMV-Myc-ARMCX3) alone and combined, together with 0.75 μg of either pX1B4FH or pX1A3HS, were co-transfected into Neuro-2A cells. In addition, 0.125 μg of a β-galactosidase expression plasmid, pRSV-βGal, was included in each transfection as a control for normalizing the luciferase results. Prior to using pRSV-βGal for normalization, the potential effects of pCMV-Sox10 and pCMV-Myc-ARMCX3, both together and independently, on RSV promoter activity were carefully checked to ensure that normalized luciferase results were not artifacts caused by regulation of the RSV promoter. In previous work, we used a similar approach in studying the effects of growth factor regulation of nACh receptor gene promoter activity (16, 31). As a transfection background control, cells were transfected with 2.0 μg of pBluescript II SK(+) (Stratagene). Cells were also transfected with 0.5 μg of pCMV5 or 1.15 μg of pCMV-Myc as expression vector background controls. pBluescript II SK(+) was included in some cases to equalize the amount of DNA in each transfection. Forty-eight hours after transfection, cells were harvested and assayed for luciferase activity (Luciferase Assay System, Promega, Madison, WI) using a Lumimark Microplate Luminometer (Bio-Rad). All transfections were repeated at least four times with independent preparations of DNA. The Student's t test was used for statistical analysis.
RESULTS
Identification of ARMCX3 as a Sox10-interacting Protein—A yeast two-hybrid protein-protein interaction screen was done to identify Sox10-interacting proteins. Several Sox10 bait constructs were tested in preliminary experiments to determine if they possessed intrinsic transcriptional activity. One of these, Sox10N100, which contains just the 100 amino acid residues at the N-terminal of Sox10 (Fig. 1A), exhibited little intrinsic transcriptional activity (data not shown) and was therefore chosen for the library screen. A mouse brain cDNA library was screened with Sox10N100 leading to the isolation of 56 candidates. Sequence analysis revealed, not surprisingly, that there were a number of false positives that contained either portions of the 3′-untranslated regions of genes or genomic DNA or had open reading frames of less than 20 amino acids. These candidates were excluded from further analyses. Of the remaining candidates, one was particularly interesting because it encodes a protein containing several armadillo repeat domains (Fig. 1C). This gene, ARMCX3, is a member of the ARMCX gene family about which little is known functionally (32–34). Direct interaction between Sox10 and ARMCX3 was confirmed by co-transfection of pACT2-ARMCX3 and pGBKT7-Sox10N100 in the yeast strain AH109 and subsequent growth on strict selection plates (data not shown). Bioinformatic analysis suggests that ARMCX3 contains an N-terminal transmembrane domain (amino acids 7–25), 3 armadillo domains (ARM1: amino acids 110–151, ARM2: amino acids 155–192, and ARM3: amino acids 234–272) and a DUF634 domain (domain of unknown function, amino acids 100–363 (Fig. 1C)).
As a further test of the direct interaction between Sox10 and ARMCX3 and to map the Sox10-interaction domain of ARMCX3, Co-IPs were done. Before initiating these experiments, however, we tested the specificity of the anti-Sox10 antibody. Western blot analysis of whole cell extracts of OBL21 cells, which express relatively high levels of Sox10 (see Fig. 4), revealed the presence of a single band at ∼64 kDa, the predicted molecular mass of Sox10 (Fig. 1B). In parallel experiments, NIH3T3 cells, which do not express endogenous Sox10, were transfected with pCMV5-Sox10 or pCMV5. Western blot analysis was carried out using whole cell extracts of the transfected cells. As shown in Fig. 1B, a single band of ∼64 kDa was detected in the pCMV5-Sox10-transfected sample, whereas no bands were detected in the vector-transfected sample. These data indicate the anti-Sox10 antibody is specific and is suitable for the experiments described below.
Co-IPs were carried out using OBL21 cells that were transfected with pCMV-Myc-ARMCX3 and its deletion plasmids, pCMV-Myc-ARMCX3Δ1, pCMV-Myc-ARMCX3Δ2, and pCMV-Myc-ARMCX3Δ3. Fig. 1C shows that anti-Sox10 antibody precipitates Myc-tagged ARMCX3 and in the reciprocal experiment, anti-Myc precipitates Sox10. Deletion of the 127-amino acid C terminus and ARM3 of ARMCX3 had no effect on the Co-IP results (Fig. 1C). Further deletion of another 91 amino acids, containing ARM1 and ARM2, led to a complete lack of Sox10/ARMCX3 interaction (Fig. 1C), suggesting that the Sox10-interaction domain of ARMCX3 is located within the ARM1/ARM2 region.
The bait used for the protein-protein interaction screen, pGBKT7-Sox10N100, contains just the N-terminal 100 amino acids of Sox10, strongly suggesting that this region contains the ARMCX3-interaction domain of Sox10. To confirm this idea, Neuro-2A cells were co-transfected with pCMV-Myc-ARMCX3 and pCMV-HA-FLAG-Sox10 or pCMV-HA-FLAG-Sox10ΔN (Fig. 1D). Neuro-2A cells were chosen for these experiments, because they do not express Sox10 (see Fig. 4B), thus eliminating potential competition between endogenous Sox10 and transfected tagged Sox10 for ARMCX3 binding. As shown in Fig. 1C, HA-FLAG-Sox10 and Myc-tagged ARMCX3 were co-precipitated when either anti-FLAG antibody or anti-Myc antibody was used for precipitation. However, when the N-terminal deletion of Sox10 was used, HA-FLAG-Sox10 and Myc-ARMCX3 were not co-precipitated (Fig. 1D), confirming that the ARMCX3 interaction domain is located at the N terminus of Sox10.
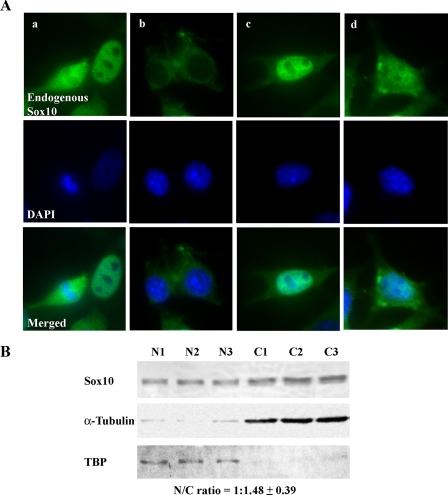
Sox10 Is a Nucleocytoplasmic Protein in OBL21 Cells—To determine the subcellular localization of Sox10, immunofluorescence of endogenous Sox10 in the olfactory bulb-derived cell line OBL21 (23) was carried out. As shown in Fig. 2A, Sox10 can be localized in the nucleus (a, right cell) or cytoplasm (b) or both with differing nuclear/cytoplasmic ratios (compare c and d and see below). In some M-phase cells, the distribution of Sox10 is diffuse (a, left cell). These observations are consistent with Sox10's function as a nucleocytoplasmic shuttling transcriptional regulatory factor (35). To estimate the nuclear:cytoplasmic ratio of Sox10, nuclear and cytoplasmic extracts were isolated from OBL21 cells and subjected to semi-quantitative Western blot analysis. TBP and α-tubulin were used as nuclear and cytoplasmic markers, respectively (Fig. 2B). Analysis of three independent experiments indicated a 1:1.48 nuclear:cytoplasmic ratio for Sox10 (Fig. 2B).
FIGURE 2.
Subcellular localization of Sox10. A, endogenous Sox10 was detected in OBL21 cells by immunofluorescence. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The four sets of images show nuclear (a), cytoplasmic (b), and nucleocytoplasmic (c and d) localization of Sox10. B, nuclear and cytoplasmic fractions were isolated from OBL21 cells. Successful isolation of nuclear protein was verified by immunoblot analysis of TBP while successful isolation of cytoplasmic protein was confirmed by immunoblot analysis of α-tubulin. Sox10 was detected by immunoblot analysis using anti-Sox10 antibody. The Sox10 nuclear:cytoplasmic (N/C) ratio was determined by semi-quantitative immunoblot analysis (see “Experimental Procedures”). The results of three independent experiments are shown.
ARMCX3 Is an Integral Membrane Protein of the Mitochondrial Outer Membrane—The armadillo gene was originally identified as a segment polarity gene in Drosophila (36–38). A family of armadillo-related proteins has since been identified and its members are characterized by a conserved 42-amino acid motif referred to as the arm repeat (39, 40). The members of the armadillo-related proteins have various functions that correspond to their varying subcellular localizations (41). As mentioned above, sequence analysis revealed the existence of a potential transmembrane domain at the N terminus of ARMCX3 (Fig. 1C). This is similar to the other ARMCX proteins, ARMCX1, ARMCX2, and ARMCX6 (Fig. 3A). Interestingly, our bioinformatic analysis also revealed substantial sequence conservation between the four ARMCX proteins and proteins anchored in the mitochondrial outer membrane (MOM, Fig. 3A). Rather strikingly, this sequence conservation falls in a region containing the putative MOM-targeting consensus sequence (shown in Fig. 3A). A key feature of this sequence is the presence of basic amino acids on both the amino and carboxyl termini of the transmembrane domain (42, 43). To determine the subcellular localization of ARMCX3, OBL21 cells were transfected with an expression construct for Myc-tagged ARMCX3. Immunofluorescence microscopy indicates that ARMCX3 is a cytoplasmic protein with a punctate staining pattern (Fig. 3B). As might be expected by the conservation of the MOM-targeting consensus sequence, ARMCX3 expression overlaps that of the mitochondrial protein COX IV (Fig. 3B). Deletion of the TMD from ARMCX3 led to the loss of overlap with COX IV, consistent with the sequence analysis and suggesting that the TMD is critical for appropriate targeting of ARMCX3 (Fig. 3B).
To further investigate the localization of ARMCX3, subcellular fractionation (27) was carried out as depicted in Fig. 3C. OBL21 cells transfected with pCMV5-Myc-ARMCX3 were used for this analysis. Given that ARMCX3 is a cytoplasmic protein, only the PNS was pursued following the initial fractionation. Sucrose density fractionation revealed, as expected, that ARMCX3 co-sedimented with COX IV to the mitochondrial fractions with very small amounts of ARMCX3 appearing in the microsomal fractions (Fig. 3D). To confirm this observation and to further biochemically characterize ARMCX3, mitochondria were isolated and subjected to both proteinase K digestion as well as protein extraction under increasingly harsh conditions (Fig. 3C). When intact mitochondria were treated with proteinase K, ARMCX3 was readily digested while COX IV, a mitochondrial inner membrane protein, was unaffected (Fig. 3E). These data indicate that ARMCX3 is exposed on the outer surface of the mitochondria while COX IV is protected from proteinase K by virtue of its mitochondrial inner membrane localization. These data are consistent with the existence of ARMCX3 in the PNS but not in the soluble S100 fraction (Fig. 3F). Furthermore, ARMCX3 was not extracted under weak buffer conditions or by high pH (Fig. 3F). It was, however, easily extracted by Triton X-100, indicating it is an integral membrane protein (Fig. 3F).
Sox10 Is Peripherally Associated with the Mitochondrial Outer Membrane—Concomitant with the biochemical characterization of ARMCX3, similar analyses of Sox10 were done. Sucrose gradient fractionation indicated that, in addition to its presence in the microsomal fractions, Sox10 is also present in the mitochondrial fractions (Fig. 3D). The presence of Sox10 in the mitochondria is consistent with its interaction with ARMCX3. The presence of Sox10 in the microsomal fractions was unexpected and may be due to technical issues (e.g. incorporation of Sox10 into microsomes during cell homogenization) or the presence of Sox10 in other cellular organelles such as the endoplasmic reticulum or the lysosomal compartment. As with ARMCX3, Sox10 was readily digested by proteinase K, consistent with localization to the exterior of the mitochondria (Fig. 3E). However, in contrast to ARMCX3, Sox10 is present in both the PNS and the S100 fractions (Fig. 3F). In addition, mild buffer conditions did not extract Sox10 from mitochondria while high pH and detergent extraction did release Sox10 from the mitochondria (Fig. 3F). These data suggest that Sox10 is, under these conditions, peripherally associated with the mitochondrial outer membrane.
Co-expression and Co-immunoprecipitation of Endogenous Sox10 and ARMCX3—In order for the Sox10/ARMCX3 interaction to have physiological meaning, they must be co-expressed and interact in vivo. This was investigated at both the mRNA and protein levels. Using multiplex RT-PCR, ARMCX3 mRNA was detected in mouse brain RNA and spinal cord as well as in the OBL21, C6 (rat glioma), U87-MG (human glioblastoma-astrocytoma, epithelial-like), and HeLa (human cervical cancer) cell lines (Fig. 4A). Sox10 gene expression was also detected in the central nervous system and the OBL21, C6, and HeLa cell lines but not in the U-87MG line (Fig. 4A). No expression of either gene was detected in the mouse Neuro-2A neuroblastoma cell line (Fig. 4A). These results are consistent with a direct interaction between the encoded proteins in vivo but fall short of actually demonstrating such an interaction. To address this issue, Co-IPs were carried out. The experiments described above utilized Myc-tagged ARMCX3 as endogenous levels of the protein are very low in whole cell lysates (data not shown). To circumvent this problem, mitochondrial lysates, which are enriched in ARMCX3, were isolated as described earlier (Fig. 3C) and used for the Co-IP studies. As shown in Fig. 4B, immunoprecipitation of the mitochondrial lysates with anti-Sox10 followed by Western blotting with anti-ARMCX3 indicates that the two endogenous proteins do in fact directly interact. The reciprocal experiment, in which anti-ARMCX3 was used for immunoprecipitation and anti-Sox10 was used for Western blotting, confirmed this result (Fig. 4B). These data indicate that endogenous Sox10 and ARMCX3 have overlapping expression patterns and directly interact in vivo.
ARMCX3 Increases the Mitochondrial Localization of Sox10— To further investigate the relationship between Sox10 and ARMCX3, the two proteins were overexpressed in Neuro-2A cells. Neuro-2A cells, which do not express Sox10 or ARMCX3 (Fig. 4A), were used to eliminate potential interference from endogenous protein interactions. Mitochondria were isolated as described above, and the levels of mitochondrially associated Sox10 protein were quantified using Western blot analysis with COX IV as the mitochondrial loading control. As shown in Fig. 5, increasing amounts of Myc-tagged ARMCX3 increased the amount of Sox10 in the mitochondrial fraction without altering the levels of COX IV. Importantly, pCMV-Myc-ARMCX3Δ3, which lacks the Sox10-interacting domain (Fig. 1C), did not alter the levels of mitochondrially associated Sox10 (Fig. 5). These data further reinforce the conclusion that the mitochondrial protein ARMCX3 is a Sox10-interacting protein and functions, at least in part, to recruit Sox10 to mitochondria.
FIGURE 5.
ARMCX3 increases the mitochondrial localization of Sox10. A, Neuro-2A cells were co-transfected with either pCMV5-Sox10 (10 μg) and pCMV-Myc-ARMCX3 (10, 20, and 40 μg) or pCMV5-Sox10 (10 μg) and pCMV-Myc-ARMCX3Δ3 (10, 20, and 40 μg). Neuro-2A cells transfected with pCMV5-Myc (40 μg) were used as a control. Forty-eight hours following transfection, mitochondria were isolated and analyzed by immunoblotting with anti-Sox10, anti-COX IV, and anti-Myc antibodies. B, quantification of three independent experiments in which the amount of Sox10 in each sample was normalized to the loading control (COX IV). Error bars represent ± S.D.
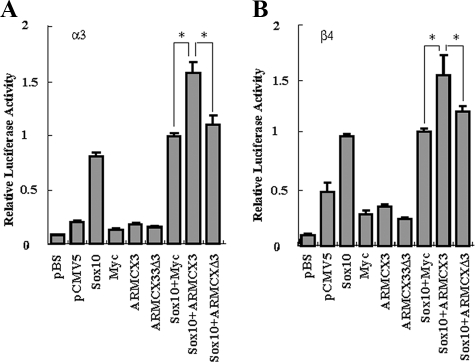
Transactivation of the nACh Receptor α3 and β4 Subunit Gene Promoters by Sox10 Is Enhanced by ARMCX3—As discussed earlier, we previously showed that Sox10 transactivates the nACh receptor α3 and β4 subunit gene promoters in a dose-dependent and cell type-specific manner (16). Given that ARMCX3 directly interacts with Sox10 and alters its subcellular localization, we tested the hypothesis that ARMCX3 also alters the transcriptional activity of Sox10. Neuro-2A cells were transiently transfected with pCMV5-Sox10, pCMV-Myc-ARMCX3, and luciferase reporter constructs for the α3 and β4 subunit gene promoters. As negative controls, the vectors pCMV5 and pCMV-Myc were included in some of the transfections. As expected from our previous work (16), Sox10 alone significantly transactivated both promoters (Fig. 6). Interestingly, although ARMCX3 by itself did not affect either promoter, ARMCX3 in combination with Sox10 led to greater promoter activity than with either protein alone (Fig. 6). The ability of ARMCX3 to increase the transcriptional effect of Sox10 on the nACh receptor gene promoters was lost when the Sox10-interacting domain of ARMCX3 was deleted (Fig. 6) suggesting that a direct interaction between the two proteins is necessary to generate a transcriptional response.
FIGURE 6.
Sox10-mediated transactivation of the nACh receptor α3 and β4 gene promoters is enhanced by ARMCX3. Neuro-2A cells were transiently transfected with either an α3/luciferase (panel A) or a β4/luciferase (panel B) reporter construct either alone (pBS) or with pCMV5 (pCMV5), pCMV-Myc (Myc) or pCMV5-Sox10 (Sox10), pCMV-Myc-ARMCX3 (ARMCX3), and pCMV-Myc-ARMCX3Δ3 (ARMCX3Δ3) individually or together as indicated. Luciferase activity was calculated relative to the normalized luciferase activity obtained by co-transfecting the β4/luciferase or the α3/luciferase reporter construct with pCMV5-Sox10 and pCMV-Myc (arbitrarily set at 1). Error bars represent ± S.D. (*, Student's t test, p < 0.01).
DISCUSSION
Sox10 was originally identified as a transcriptional regulatory factor in glial cells based on its sequence homology with other SRY-type transcription factors (44). It is now clear that Sox10 is involved in a plethora of regulatory cascades particularly in cell survival and migration of oligodendrocyte precursors (45) and cells within the neural crest and its derivatives (46–48). Furthermore, in addition to its key role in regulating glial cell development and differentiation (12–14), Sox10 is also critical for specification of neural crest-derived sensory and enteric neurons and melanophores (15, 49, 50). A key challenge is to elucidate the molecular mechanisms underlying the many roles of Sox10 in development.
Clearly, both intrinsic factors and extrinsic signals are required for proper Sox10 function. Although these factors and signals remain to be completely identified, it has been well established that Sox10 interacts with a variety of transcription factors (7, 8, 21). The majority of these interactions occur through the HMG domain of Sox10. For example, Lang and Epstein showed that Sox10 and Pax3 interact to regulate the c-RET enhancer and that the HMG domain of Sox10 is critical for the interaction (51). Wissmüller et al. performed yeast two-hybrid screens using baits containing the Sox10 HMG domain to identify a number of transcriptional regulatory factors as being Sox10 interactors (7). Interestingly, this group of proteins includes homeodomain, POU domain, and basic helix-loop-helix regulatory factors (7). These results are consistent with our own work demonstrating that Sox10 interacts, through its HMG domain, with the regulatory factors Purα, heterogeneous nuclear ribonucleoprotein K, Sp1, and Sp3 (21). Overall, these studies coupled with extensive previous work (5, 6) reveal that the HMG domain appears to have at least three distinct functions: 1) DNA binding, 2) DNA bending, and 3) protein-protein interaction. However, the HMG domain is not the whole story (2, 8).
There is a growing body of evidence indicating that domains other than the HMG sequence are critical for protein-protein interactions with Sox proteins. Furthermore, some of these non-HMG domains play a role in Sox protein interactions with other armadillo repeat-containing proteins. For example, Sox9, a group E Sox protein with extensive homology to Sox10, interacts through its C-terminal transactivation domain with the ARM repeats of β-catenin to regulate chondrocyte differentiation (52). Similarly, another Sox protein, Sox6, interacts with the ARM repeats of β-catenin via its N-terminal LZ/Q domain to regulate pancreatic β-cell proliferation (53). A third Sox protein, Sox17, also interacts with the β-catenin ARM repeats through a C-terminal domain leading to regulation of Wnt signaling (54). These studies and others indicate that sequences outside the HMG domain of Sox proteins are critical for protein-protein interactions and the downstream functional consequences of the interactions. Consistent with these previous studies, we used the N-terminal 100 amino acids of Sox10 as the bait in a yeast two-hybrid screen; this bait is devoid of any HMG domain sequences (Fig. 1A). A number of Sox10-interacting proteins were isolated. The focus of this report is on the arm repeat-containing protein ARMCX3 (Fig. 1C). Little is known regarding the biological functions of the ARMCX proteins. Indeed, the only reports on ARMCX3 indicate that it has high sequence homology with ARMCX1 and ARMCX2 (32) and that it is expressed in the initial segment of mouse epididymis as determined by DNA microarray analysis (34). In addition, a UniGene data base search revealed matching expressed sequence tags in cDNA from heart, brain, placenta, lung, muscle, kidney, spleen, prostate, ovary, and colon (32). Our biochemical characterization of ARMCX3 demonstrates that it is an integral membrane protein localized to the MOM (Fig. 3). A similar set of experiments done with Sox10 suggests that it is a nucleocytoplasmic protein peripherally associated with the MOM (Fig. 3). Endogenous ARMCX3 and Sox10 have overlapping expression patterns and can be co-immunoprecipitated (Fig. 4). The Sox10-interacting domain was mapped to the ARM1/2 region of ARMCX3, whereas the ARMCX3-interaction domain is located in the N terminus of Sox10 (Fig. 1). Interestingly, overexpression of ARMCX3 led to a dose-dependent increase in mitochondrially associated Sox10 protein levels (Fig. 5). Importantly, in transient transfections, ARMCX3 enhanced the Sox10-mediated transactivation of the nACh receptor α3 and β4 subunit gene promoters (Fig. 6). These experiments represent the first biochemical and functional characterizations of an ARMCX family member and suggest a role for ARMCX3 in the biological function of an important developmental regulatory factor, Sox10.
A key issue raised by the present results is how does a cytoplasmic protein, ARMCX3, increase the transcriptional regulatory function of Sox10? It has been very nicely demonstrated that Sox10 is a nucleocytoplasmic shuttling protein and, further, that the translocation is critical for its transcriptional effects (35). This is similar to the previously mentioned group E Sox protein, Sox9 (56). A straightforward hypothesis to explain the shuttling feature is that the Sox proteins are post-translationally modified in the cytoplasm and then transported to the nucleus to carry out their transcriptional role (35). Shuttling back to the cytoplasm likely reflects the removal of the modification and the need for another cycle of modification (35). Consistent with this idea is the demonstration that Sox9 is phosphorylated by cAMP-dependent protein kinase A and is also SUMOylated, with both modifications having a dramatic impact of the transcriptional function of Sox9 (57–60). Similarly, SUMOylation of Sox10 has a profound effect on the transcriptional activity of Sox10 (61). It is likely that these post-translational modifications of the Sox factors occur in response to extracellular cues. The signal transduction cascade underlying the modifications and subsequent translocation to the nucleus remains unknown. One possibility is that the signaling proteins that make up the cascade are physically organized into complexes by scaffold, anchoring, and adaptor proteins as has been well described for the mitogen-activated protein kinase pathway (62–65). It is tempting to speculate that one potential role ARMCX3 may play is that of an anchoring protein in such a signaling complex. Given the MOM localization of ARMCX3, this would bring Sox10 into the proximity of the vast mitochondrial-nuclear signal transduction machinery, which includes GTPases, kinases, and phosphatases, all of which could have a profound effect on Sox10 function (66–68). Obviously, much more work needs to be done to test this idea, but the co-localization of ARMCX3 and Sox10 and the effect of ARMCX3 on Sox10 function are consistent with such a role. Similarly, the potential interaction of Sox10 with ARMCX1 and/or ARMCX2 requires further investigation given the high sequence similarity of the ARMCX proteins. Finally, as mentioned above, little is known regarding the biological functions of the ARMCX proteins. Whether ARMCX1 and ARMCX2 are mitochondrial proteins similar to ARMCX3 remains an open question. In addition, whether any of the ARMCX proteins functionally interact with other Sox family members warrants further investigation given that nucleocytoplasmic shuttling of Sox proteins is critical both in normal development as well as in a number of pathological conditions (55).
Acknowledgments
We thank Michael Wegner and Kirsten Kuhlbrodt for Sox10 plasmids and Haley Melikian for help with the fluorescence microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant R01NS030243 (to P. D. G.) from NINDS. This work was also supported by a grant from Philip Morris USA Inc. and Philip Morris International (to P. D. G.).
Footnotes
The abbreviations used are: HMG, high mobility group; nACh, nicotinic acetylcholine; CMV, cytomegalovirus; Co-IP, co-immunoprecipitation; PBS, phosphate-buffered saline; HA, hemagglutinin; BSA, bovine serum albumin; TBP, TATA-binding protein; PNS, post-nuclear supernatant; RT, reverse transcription; RSV, Rous sarcoma virus; MOM, mitochondrial outer membrane; TMD, transmembrane domain; SUMO, small ubiquitin-like modifier; ARM, armadillo.
References
- 1.Wegner, M. (1999) Nucleic Acids Res. 27 1409-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowles, J., Schepers, G., and Koopman, P. (2000) Dev. Biol. 227 239-255 [DOI] [PubMed] [Google Scholar]
- 3.Schepers, G. E., Teasdale, R. D., and Koopman, P. (2002) Dev. Cell 3 167-170 [DOI] [PubMed] [Google Scholar]
- 4.Wegner, M., and Stolt, C. C. (2005) Trends Neurosci. 28 583-588 [DOI] [PubMed] [Google Scholar]
- 5.Wegner, M. (2005) Pigment Cell Res. 18 74-85 [DOI] [PubMed] [Google Scholar]
- 6.Prior, H. M., and Walter, M. A. (1996) Mol. Med. 2 405-412 [PMC free article] [PubMed] [Google Scholar]
- 7.Wissmüller, S., Kosian, T., Wolf, M., Finzsch, M., and Wegner, M. (2006) Nucleic Acids Res. 34 1735-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson, M., and Koopman, P. (2002) Curr. Opin. Genet. Dev. 12 441-446 [DOI] [PubMed] [Google Scholar]
- 9.Kelsh, R. N. (2006) BioEssays 28 788-798 [DOI] [PubMed] [Google Scholar]
- 10.Pingault, V., Bondurand, N., Kuhlbrodt, K., Goerich, D. E., Prehu, M. O., Puliti, A., Herbarth, B., Hermans-Borgmeyer, I., Legius, E., Matthijs, G., Amiel, J., Lyonnet, S., Ceccherini, I., Romeo, G., Smith, J. C., Read, A. P., Wegner, M., and Goossens, M. (1998) Nat. Genet. 18 171-173 [DOI] [PubMed] [Google Scholar]
- 11.Bondurand, N., Dastot-Le Moal, F., Stanchina, L., Collot, N., Baral, V., Marlin, S., Attie-Bitach, T., Giurgea, I., Skopinski, L., Reardon, W., Toutain, A., Sarda, P., Echaieb, A., Lackmy-Port-Lis, M., Touraine, R., Amiel, J., Goossens, M., and Pingault, V. (2007) Am. J. Hum. Genet. 81 1169-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbarth, B., Pingault, V., Bondurand, N., Kuhlbrodt, K., Hermans-Borgmeyer, I., Puliti, A., Lemort, N., Goossens, M., and Wegner, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5161-5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southard-Smith, E. M., Kos, L., and Pavan, W. J. (1998) Nat. Genet. 18 60-64 [DOI] [PubMed] [Google Scholar]
- 14.Britsch, S., Goerich, D. E., Reithmacher, D., Peirano, R. I., Rossner, M., Nave, K.-A., Birchmeier, C., and Wegner, M. (2001) Genes Dev. 15 66-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney, T. J., Dutton, K. A., Greenhill, E., Delfino-Machin, M., Dufourcq, P., Blader, P., and Kelsh, R. N. (2006) Develop. 133 4619-4630 [DOI] [PubMed] [Google Scholar]
- 16.Liu, Q., Melnikova, I. N., Hu, M., and Gardner, P. D. (1999) J. Neurosci. 19 9747-9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bigger, C. B., Casanova, E. A., and Gardner, P. D. (1996) J. Biol. Chem. 271 32842-32848 [DOI] [PubMed] [Google Scholar]
- 18.Bigger, C. B., Melnikova, I. N., and Gardner, P. D. (1997) J. Biol. Chem. 272 25976-25982 [DOI] [PubMed] [Google Scholar]
- 19.Du, Q., Melnikova, I. N., and Gardner, P. D. (1998) J. Biol. Chem. 273 19877-19883 [DOI] [PubMed] [Google Scholar]
- 20.Du, Q., Tomkinson, A. E., and Gardner, P. D. (1997) J. Biol. Chem. 272 14990-14995 [DOI] [PubMed] [Google Scholar]
- 21.Melnikova, I. N., Yang, Y., and Gardner, P. D. (2000) Eur. J. Pharmacol. 393 75-83 [DOI] [PubMed] [Google Scholar]
- 22.Melnikova, I. N., Lin, H. R., Blanchette, A. R., and Gardner, P. D. (2000) Neuropharm. 39 2615-2623 [DOI] [PubMed] [Google Scholar]
- 23.Ryder, E. F., Snyder, E. Y., and Cepko, C. L. (1990) J. Neurobiol. 21 356-375 [DOI] [PubMed] [Google Scholar]
- 24.Olmsted, J. B., Carlson, K., Klebe, R., Ruddle, F., and Rosenbaum, J. (1970) Proc. Natl. Acad. Sci. U. S. A. 65 129-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frezza, C., Cipolat, S., and Scorrano, L. (2007) Nat. Protoc. 2 287-295 [DOI] [PubMed] [Google Scholar]
- 26.Capaldi, R. A. (1990) Annu. Rev. Biochem. 59 569-596 [DOI] [PubMed] [Google Scholar]
- 27.Srinivas, K. S., Chandrasekar, G., Srivastava, R., and Puvanakrishnan, R. (2004) J. Biochem. Biophys. Methods 60 23-27 [DOI] [PubMed] [Google Scholar]
- 28.Armstrong, L. C., Saenz, A. J., and Bornstein, P. (1999) J. Cell. Biochem. 74 11-22 [PubMed] [Google Scholar]
- 29.Hay, J. C., Hirling, H., and Scheller, R. H. (1996) J. Biol. Chem. 271 5671-5679 [DOI] [PubMed] [Google Scholar]
- 30.Fuentes Medel, Y. F., and Gardner, P. D. (2007) J. Biol. Chem. 282 19062-19070 [DOI] [PubMed] [Google Scholar]
- 31.Hu, M., Whiting Theobald, N. L., and Gardner, P. D. (1994) J. Neurochem. 62 392-395 [DOI] [PubMed] [Google Scholar]
- 32.Kurochkin, I. V., Yonemitsu, N., Funahashi, S., and Nomura, H. (2001) Biochem. Biophys. Res. Commun. 280 340-347 [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. A., McClive, P. J., and Sinclair, A. H. (2005) Dev. Dyn. 233 188-193 [DOI] [PubMed] [Google Scholar]
- 34.Hsia, N., and Cornwall, G. A. (2004) Biol. Reprod. 70 448-457 [DOI] [PubMed] [Google Scholar]
- 35.Rehberg, S., L. P., Glaser, G., Stamminger, T., Wegner, M., and Rosorius, O. (2002) Mol. Cell. Biol. 22 5826-5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nüsslein-Volhard, C., and Wieschaus, E. (1980) Nature 287 795-801 [DOI] [PubMed] [Google Scholar]
- 37.Riggleman, B., Wieschaus, E., and Schedl, P. (1989) Genes Dev. 3 96-113 [DOI] [PubMed] [Google Scholar]
- 38.Wieschaus, E., and Riggleman, R. (1987) Cell 49 177-184 [DOI] [PubMed] [Google Scholar]
- 39.Peifer, M., Berg, S., and Reynolds, A. B. (1994) Cell 76 789-791 [DOI] [PubMed] [Google Scholar]
- 40.Hatzfeld, M. (1999) Int. Rev. Cytol. 186 179-224 [DOI] [PubMed] [Google Scholar]
- 41.Coates, J. C. (2003) Trends Cell Biol. 13 463-471 [DOI] [PubMed] [Google Scholar]
- 42.Kaufmann, T., Schlipf, S., Sanz, J., Neubert, K., Stein, R., and Borner, C. (2003) J. Cell Biol. 160 53-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rapaport, D. (2003) EMBO Rep. 4 948-952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhlbrodt, K., Herbarth, B., Sock, E., Hermans-Borgmeyer, I., and Wegner, M. (1998) J. Neurosci. 18 237-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finzsch, M., Stolt, C. C., Lommes, P., and Wegner, M. (2008) Development 135 637-646 [DOI] [PubMed] [Google Scholar]
- 46.Paratore, C., Goerich, D. E., Suter, U., Wegner, M., and Sommer, L. (2001) Development 128 3949-3961 [DOI] [PubMed] [Google Scholar]
- 47.Kim, J., Lo, L., Dormand, E., and Anderson, D. J. (2003) Cell 38 17-31 [DOI] [PubMed] [Google Scholar]
- 48.Maka, M., Stolt, C. C., and Wegner, M. (2005) Dev. Biol. 277 155-169 [DOI] [PubMed] [Google Scholar]
- 49.Elworthy, S., Lister, J. A., Carney, T. J., Raible, D. W., and Kelsh, R. N. (2003) Development 130 2809-2818 [DOI] [PubMed] [Google Scholar]
- 50.Elworthy, S., Pinto, J. P., Pettifer, A., Cancela, M. L., and Kelsh, R. N. (2005) Mech. Dev. 122 659-669 [DOI] [PubMed] [Google Scholar]
- 51.Lang, D., and Epstein, J. A. (2003) Hum. Mol. Genet. 12 937-945 [DOI] [PubMed] [Google Scholar]
- 52.Akiyama, H., Lyons, J. P., Mori-Akiyama, Y., Yang, X., Zhang, R., Zhang, Z., Deng, J. M., Taketo, M. M., Nakamura, T., Behringer, R. R., McCrea, P. D., and De Crombrugghe, B. (2004) Genes Dev. 18 1072-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iguchi, H., Urashima, Y., Inagaki, Y., Ikeda, Y., Okamura, M., Tanaka, T., Uchida, A., Yamamoto, T. T., Kodama, T., and Sakai, J. (2007) J. Biol. Chem. 282 19052-19061 [DOI] [PubMed] [Google Scholar]
- 54.Zorn, A. M., Barish, G. D., Williams, B. O., Lavender, P., Klymkowsky, M. W., and Varmus, H. E. (1999) Mol. Cell 4 487-498 [DOI] [PubMed] [Google Scholar]
- 55.Smith, J. M., and Koopman, P. A. (2004) Trends Genet. 20 4-8 [DOI] [PubMed] [Google Scholar]
- 56.Gasca, S., Cañizares, J., de Santa Barbara, P., Méjean, C., Poulat, F., Berta, P., and Boizet-Bonhoure, B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11199-11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang, W., Chung, U., Kronenberg, H. M., and De Crombrugghe, B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 160-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang, W., Zhou, X., Lefebvre, V., and De Crombrugghe, B. (2000) Mol. Cell. Biol. 20 4149-4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, K. M., and LaBonne, C. (2005) Dev. Cell 9 593-603 [DOI] [PubMed] [Google Scholar]
- 60.Hattori, T., Eberspaecher, H., Lu, J., Zhang, R., Nishida, T., Kahyo, T., Yasuda, H., and De Crombrugghe, B. (2006) J. Biol. Chem. 281 14417-14428 [DOI] [PubMed] [Google Scholar]
- 61.Girard, M., and Goossens, M. (2006) FEBS Lett. 580 1635-1641 [DOI] [PubMed] [Google Scholar]
- 62.Pawson, T., and Scott, J. D. (1997) Science 278 2075-2080 [DOI] [PubMed] [Google Scholar]
- 63.Burack, W. R., and Shaw, A. S. (2000) Curr. Opin. Cell Biol. 12 211-216 [DOI] [PubMed] [Google Scholar]
- 64.Bhattacharyya, R. P., Reményi, A., Yeh, B. J., and Lim, W. A. (2006) Annu. Rev. Biochem. 75 655-680 [DOI] [PubMed] [Google Scholar]
- 65.Bashor, C. J., Helman, N. C., Yan, S., and Lim, W. A. (2008) Science 319 1539-1543 [DOI] [PubMed] [Google Scholar]
- 66.McBride, H. M., Neuspiel, M., and Wasiak, S. (2006) Curr. Biol. 16 R551-R560 [DOI] [PubMed] [Google Scholar]
- 67.Ryan, M. T., and Hoogenraad, N. J. (2007) Annu. Rev. Biochem. 76 701-722 [DOI] [PubMed] [Google Scholar]
- 68.Pagliarini, D. J., and Dixon, J. E. (2006) Trends Biochem. Sci. 31 26-34 [DOI] [PubMed] [Google Scholar]