Abstract
Small ubiquitin-like modifier 1 (SUMO-1) modification of IκBα has been described to actively participate in NFκB regulation. Following proteosomal degradation of IκBα, an auto-regulatory loop consisting of transcriptional activation of IκBα gene and SUMO-1 modification of newly synthesized IκBα proceeds. The SUMOylated IκBα form is resistant to signal-induced degradation, consequently halting NFκB activation. We describe a mechanistic model by which adenosine (Ado) signaling results in significant accumulation of SUMO-1 modified IκBα with subsequent attenuation of NFκB activation. Using models of hypoxia followed by reoxygenation (H/R), we have documented an H/R cycle-dependent increase in extracellular Ado correlating with increases in the cytoplasmic pool of IκBα/SUMO-1. We demonstrate a dose-dependent increase in IκBα/SUMO in cells treated with the general Ado receptor agonist NECA and abolished by Ado receptor antagonists. Experiments in cells exposed to cycles of H/R followed by hypoxia demonstrated differential patterns of SUMOylation and phosphorylation of IκBα, greatly impacting its proteosomal degradation by the 26 S proteasome. Assays targeting knockdown and overexpression of SUMO-1 demonstrated significant regulation of NFκB activation and NFκB-mediated gene transcription (interleukin-6). These results were confirmed in vivo using wild type and cd73 null mouse lung tissue. In summary, we present an endogenous mechanism by which cells and tissues acquire anti-inflammatory properties by recruiting a nondegradable form of IκBα, a major control point for NFκB activation via Ado signaling.
Endogenous anti-inflammatory mechanisms exist in cells to adapt to changes in tissue metabolism (1). These changes often involve low oxygen concentrations triggering the synthesis of anti-inflammatory molecules such as adenosine (Ado)3 (2). Ado-mediated responses influence multiple levels of cell functioning and specifically aids in the complex modulation of inflammation (3). Ado signaling results in inhibition of NFκB activation by active deneddylation of Cullin-1, an integral component of the E3 SCF ubiquitin-ligase complex (SKP1, Cullin-1, and the F-box domain of β-TrCP). Under enhanced Ado signaling the deneddylated form of E3 ligase looses its activity and is unable to polyubiquitinate IκBα, preventing proteosomal degradation by the 26 S proteasome and nuclear translocation of NFκB. This effect proved of physiologic significance to different cell types and was specifically relevant to lung tissue (4).
Post-translational modifiers of IκBα degradation have been described including the small ubiquitin-like protein (SUMO-1) (5, 6). SUMO-1 is one of a family of small ubiquitin-like proteins (SUMO-1–4) described to participate in a multitude of cellular functions including transcription (7), nuclear translocation (8), and inflammation (9). SUMO-1 directly modifies IκBα using three steps similar to ubiquitination using the E1 equivalent activating complex (SAE I and II) and E2 also known as Ubc9 (10). Although Ubc9 can directly conjugate SUMO-1 to IκBα without the use of E3, SUMO-1 E3-related proteins have been described to participate in the efficient SUMO-1 modification of IκBα (11). Ubc9 utilizes the same ubiquitination site (Lys21/Lys22) of IκBα, transferring a single molecule of SUMO-1 preventing ubiquitination and proteosomal degradation (12, 13). SUMO modification of IκBα confers anti-inflammatory properties and provides a nondegradable isoform inhibiting the transcriptional activity of NFκB. The SUMO modification of targeted proteins is a dynamic process and also involves removal by specific SUMO isopeptidases returning the protein to its native state (14). Cytoplasmic levels of SUMO-1 and unmodified IκBα control NFκB activity independent of phosphorylation or ubiquitination events. As such, we sought to investigate whether SUMO-1 modification of IκBα would also be under the influence of Ado signaling and potentially contributing to the potent anti-inflammatory phenotype observed with H/R.
EXPERIMENTAL PROCEDURES
Murine Hypoxia and Reoxygenation (H/R) Model—As described previously (4), C57BL/6 (Charles River Laboratories), and CD 73 null male mice 8–10 weeks old were used in this study. The Animal Care and Use Committee of Children's Hospital approved all procedures. Briefly, the mice were placed in humidified environmental chambers and subjected to FiO2 of 8% for 10 min followed by FiO2 21% for 10 min for three cycles. Hypoxic conditions were achieved by exposure to FiO2 5% for 10 min. For each experiment (n = 3 mice), the experiments were performed in triplicate.
Cell Culture and in Vitro Hypoxia/Reoxygenation Model— HeLa cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. The cells were placed in a hypoxia chamber (Coy Laboratory Products Inc.) in pre-equilibrated hypoxic media at 1% O2 for 40 min, then removed from the hypoxia chamber, and allowed to be fully reoxygenated in normoxic conditions (21% O2) for 20 min. This protocol was followed for three cycles for a total time of 3 h. Hypoxia protocol was achieved by exposure of cells to 1% O2 for 3 h.
Cell Transfection and NFκB Reporter Assays—Transfection of HeLa cells was carried out using FuGENE 6 (Roche Applied Science) as directed by the manufacturer. The plasmids used in the transfections were full-length cDNA SUMO-1 plasmid, small hairpin RNA anti-SUMO-1, and control plasmids (Origene). To measure the activity of NFκB, on the second day of the transfection, the cells were transfected for the second time with pNFκB-Luc (pNRE; Clontech). The cell lysates were assayed using the Dual-Glo Luciferase assay system (Promega) with the use of a Luminometer (Turner Biosystems).
Cell Treatment—Where indicated, the cells were treated with 8-phenyltheophylline in double distilled H2O at concentration of 100 nm; N-ethylcarbamido-adenosine (NECA) in Me2SO at a range from 100 nm to 100 μm and PSB1115 at concentration of 10 μm in double distilled H2O. All of the chemicals were purchased from Sigma-Aldrich, unless otherwise noted.
Transcriptional Analysis of IL-6—Total RNA was extracted from cells using an isolation kit (RNAqueous, Ambion Inc). RNA from lung tissue was purified using TRIzol reagent according to the manufacturer's specification (Invitrogen). PCRs for IL-6 expression were carried out using PCR Master Mix (Eurogentec) on a Thermocycler (ABI 7300). Human and mouse IL-6 primers were obtained from Applied Biosystems. All of the samples were controlled for β-actin using the following primers: sense, 5′-GGTGGCTTTTAGGATGGCAAG-3′, and antisense, 5′-ACTGGAACGGTGAAGGTGACAG-3′ (162 bp).
Western Blot Analysis—Total protein was isolated from HeLa cells using cell lysis buffer (Cell Signaling Technology) or extracted from whole lung tissue from mice with radioimmune precipitation assay buffer (Cell Signaling Technology), both containing protease inhibitor mixture (Roche Applied Science), phenylmethylsulfonyl fluoride (1 mm), and sodium orthovanadate (1 mm). The protein concentrations were measured by DC protein assay (Bio-Rad). An equal amount of protein was boiled in 2% SDS loading buffer (Bio-Rad), then resolved on 12% polyacrylamide denaturing gels, and transferred to nitrocellulose membrane (Bio-Rad). After transfer, the membranes were stained with Ponceau S stain to verify equal loading. Antibodies used for Western blotting included rabbit anti-SUMO-1 (1:1000), rabbit anti-I κBα (1:1000), and rabbit anti-phospho-IκBα (1:1000), all of which were from Cell Signaling Technology, Inc., and anti-actin antibody from Santa Cruz Biotechnology. The blots were washed, and species-matched peroxidase-conjugated secondary antibody was added. Labeled bands from washed blots were detected by enhanced chemiluminescence (Thermol Scientific).
Immunoprecipitation—Total protein was isolated from HeLa cells and measured as described above. After preclearing with 60 μl of immobilized protein A/G (Thermol Scientific) for 2 h, rabbit anti-SUMO-1 antibody or rabbit anti-IκBα antibody and normal rabbit serum (Sigma-Aldrich) were added to the protein gently shaken overnight. 60 μl of immobilized protein A/G were added into each tube, incubated for 2 h, and centrifuged at 7500 rpm for 4 min, and complex-bound gel was then collected and washed with cell lysis buffer three times. Finally, 100 μl of 2% SDS loading buffer was added into each tube, boiled at 95 °C for 5 min, centrifuged at 14,000 rpm for 30 s, loaded to 12% gel, and subjected to the Western blot steps mentioned above.
For p65 nuclear translocation studies, HeLa cells were plated on 4-well glass chamber slides (Nalgene Nunc International) and allowed to grow to ∼70% confluence. The cells were exposed to protocols of hypoxia and H/R as described earlier or coincubated with NECA (10 μm) for 24 h. The cells were fixed in 1% paraformaldehyde, phosphate-buffered saline at 4 °C for 10 min and permeabilized with prechilled 0.2% Triton X-100, phosphate-buffered saline, 2% bovine serum albumin. The cells were incubated with rabbit anti-p65 (1:200; Rockland Immunochemicals) in 1% normal goat serum in phosphate-buffered saline for 1 h followed by anti-rabbit Oregon Green 488 (1:100, Molecular Probes; Invitrogen) in the same buffer for 30 min. The cell images were captured on a fluorescence microscope.
Statistics—The values are expressed as the means ± S.D. Where appropriate, groups were compared by analysis of variance. Otherwise, comparisons between groups were conducted using two-tailed Student's t test. A p value of less than 0.05 was considered to be significant.
RESULTS
Hypoxia/Reoxygenation Attenuates NFκB Activation in a Cycle-dependent Manner—We profiled the pattern of NFκB activity in cells exposed to H/R followed by hypoxia. We addressed whether H/R would attenuate NFκB activity in a cycle-dependent fashion consistent with cumulative concentrations of Ado. To this end, we transfected a NFκB luciferase reporter plasmid and exposed cells to increasing cycles of H/R followed by 24 h of hypoxia. These experiments revealed a significant cycle-dependent attenuation NFκB activity when compared with the individual response of one or three cycles of H/R followed by hypoxia (Fig. 1A). Further, to document that, in fact, NFκB attenuation was related to enhanced Ado receptor signaling, we studied NFκB activity in the presence of increasing concentrations of the NECA in hypoxic cells. For this, we exposed cells to 24 h of hypoxia coincubated with increasing concentrations of NECA. These experiments demonstrated a dose-dependent attenuation of NFκB activity with increasing concentrations of NECA, similar to the effect observed for H/R (Fig. 1B). Given the potential association between cycles of H/R, Ado concentrations, and NFκB activity, we pursued NFκB regulatory mechanisms potentially modulated by extracellular Ado accumulation.
FIGURE 1.
H/R attenuates NFκB activation in a cycle-dependent manner. A, HeLa cells transfected with pNRE were cultured under normoxia or hypoxia (24 h) or subjected to one to three cycles of H/R followed by 24 h of hypoxia. Each cycle consisted of hypoxia (1%, O2) for 40 min followed by reoxygenation (21% O2) for 20 min. The cells exposed to three cycles demonstrated significant NFκB attenuation compared with hypoxia (*, p < 0.01) and a single cycle of H/R (**, p < 0.01). B, HeLa cells transfected with pNRE were incubated with Log concentrations of NECA and NFκB activation determined after 24 h of hypoxia. Significant (*, p < 0.001) attenuation of NFκB activity was observed at 10 μm compared with hypoxic cells incubated with vehicle (Me2SO) only.
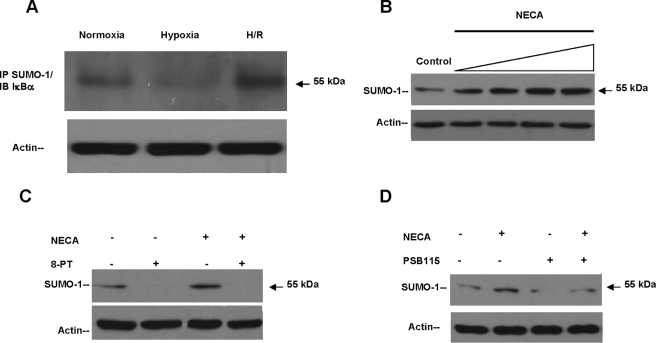
Adenosine Signaling Promotes Sumoylation of IκBα—We explored the possibility that known modulators of IκBα ubiquitination would also be regulated by Ado signaling. Activation of NFκB-mediated pathways occurs largely by proteosomal degradation of IκBα (15) and potentially a regulatory site for innate anti-inflammatory responses mediated by Ado. The SUMO-1 modification of IκBα renders NFκB resistant to signal-induced degradation by blockade of its ubiquitination site Lys21/Lys22 and inhibition of NFκB dependent transcription (13). We addressed whether enhanced Ado receptor signaling resulting from H/R would directly increase the cytoplasmic pool of SUMO-1·IκBα complex, potentially influencing NFκB activation. We initially determined whether IκBα was SUMO-1 conjugated to a higher degree in H/R. To do this, we immunoprecipitated lysates from HeLa cells exposed to protocols of hypoxia and H/R using a SUMO-1 antibody and immunoblotted against IκBα. It has been established by Desterro et al. (13), specifically in HeLa cells coimmunoprecipitation of SUMO-1/IκBα results in a polypeptide of 55 kDa, which is consistent with the addition of a single molecule of SUMO-1 to IκBα. We confirmed the presence of a single 55-kDa band and observed a significantly increased association of SUMO-1 with IκBα in cells pre-exposed to H/R (Fig. 2A). This finding suggested that H/R promoted the SUMO-1 modification of IκBα, a mechanism potentially enhanced by Ado signaling. In addition, the degree of SUMO-1 modified IκBα was consistently decreased in hypoxia when compared with normoxic levels (Fig. 2A). This effect was attributed to the physiologic increases in IκBα degradation normally occurring during hypoxia, confirming the protective role of SUMO-1 modification of IκBα to ubiquitination and degradation by E3 SCF ubiquitin ligase (Fig. 2A). To further understand whether exogenous Ado stimulation would significantly increase the cytoplasmic SUMO-1·IκBα complex concentrations, we stimulated cells with NECA and probed the SUMOylated IκBα protein (55 kDa) levels by Western blotting. Stimulation with NECA produced a concentration-dependent increase in SUMO-1/IκBα levels with an EC50 of ∼100 nm (Fig. 2B). Moreover, the nonselective Ado receptor antagonist 8-phenyltheophylline completely inhibited the basal and NECA induced SUMOylation of IκBα (Fig. 2C). Furthermore, the specific Ado A2B receptor antagonist PSB 1115 nearly completely inhibited the basal SUMO-1/IκBα levels, yet partially inhibited the NECA-induced SUMOylation of IκBα, verifying that cooperative signaling is required among Ado receptor populations for most physiologic effects mediated by Ado (Fig. 2D).
FIGURE 2.
Adenosine signaling promotes SUMOylation of IκBα. A, HeLa cells were exposed to experimental conditions and lysates immunoprecipitated with anti-SUMO-1 antibody and Western blotted against IκBα. A single band of 55 kDa was observed, corresponding to SUMO-1/IκBα isoform. A significant (80%, p < 0.001) increase SUMOylation of IκBα was observed during H/R. Equal loading of samples is documented by actin levels. B, log dose response of NECA (10 nm to 10 μm: 30 min) in HeLa cells with an EC50 estimated to be 100 nm. C, incubation with 8-phenyltheophylline (100 nm; 30 min) resulted in complete inhibition of base-line and NECA-induced protein levels of SUMO-1/IκBα (55 kDa) studied by Western blotting. D, incubation with the Ado A2B specific receptor inhibitor PSB1115 showed ∼89% inhibition of base-line levels of SUMO-1/IκBα and 70% inhibition of the NECA-induced isoform. IP, immunoprecipitation; IB, immunoblot.
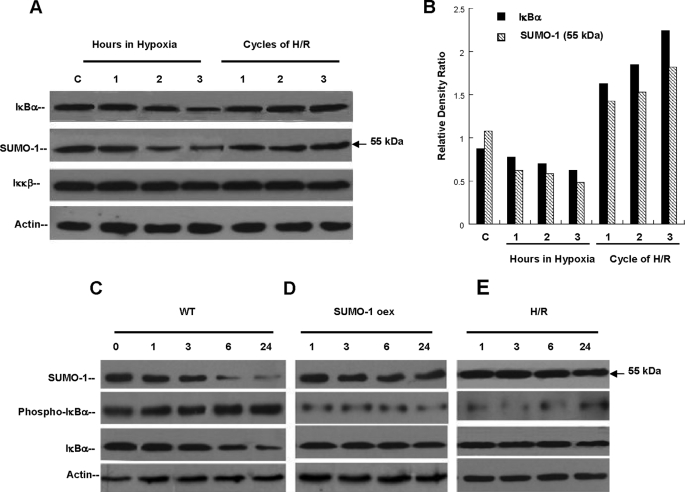
Hypoxia and Reoxygenation Prevents IκBα Ubiquitination via SUMO-1 Modification—To better define the functional attributes conferred differentially by H/R to IκBα degradation, we determined individual levels of SUMO-1/IκBα, IκBα, and the IκB kinase (Iκκβ) proteins by Western blotting. Here, cell lysates from sets of experiments comparing either incremental exposure to hypoxia or cycles of H/R were performed. As described under “Experimental Procedures,” one cell cycle of H/R consisted of 40 min of hypoxia (normobaric 1% O2), followed by 20 min of reoxygenation (21% O2) comparatively, hypoxia exposure was performed by increasing uninterrupted (60-min periods) exposure to 1% O2. This analysis revealed that a pool of SUMO-1/IκBα species exists in normoxic cells, because this isoform is part of the normal re-generative mechanism of IκBα, and also as negative feed back to limit NFκB activation (16). These studies established a time-dependent degradation of SUMO-1/IκBα and IκBα species in wild type cells exposed to continuous hypoxia (Fig. 3A). In contrast, cells exposed to sequential cycles of H/R demonstrated stable concentrations of SUMO-1/IκBα and of IκBα protein, corroborating the differential effect of H/R on IκBα degradation. Relative density ratios of IκBα and SUMO-1/IκBα demonstrate graphically the distinct effect of increasing cycles of H/R, clearly augmenting both isoforms in a cycle-dependent manner (Fig. 3B). We then explored the possibility that H/R could lead also to inhibition of Iκκβ by protein degradation and therefore result in the loss of phosphorylation as mechanism of inhibited degradation of SUMO-1/IκBα and IκBα during H/R. No significant differences in the protein levels of Iκκβ between continuous hypoxia and cycles of H/R were detected, suggesting that cells exposed to H/R conserved Iκκβ activity, providing a normal phosphorylation pattern. Unlike ubiquitination, SUMO-1 modification of IκBα does not require initial phosphorylation of Ser32 and Ser36 (13, 17); however, SUMO-1 modification prevents phosphorylation of Ser32/Ser36 by a mechanism not fully understood (13, 18). It is clear that SUMOylation of IκBα prevents proteosomal degradation by blockade of its ubiquitination (Lys21/Lys22) and its phosphorylation sites (Ser32/Ser36) (6, 19). To further examine whether enhanced SUMOylation of IκBα provided by Ado signaling would modify the phosphorylation pattern of IκBα Ser32/Ser36 sites as a mechanism of decreased degradation by hypoxia, we designed cell models of SUMO-1 overexpression. In wild type cells exposed to continuous hypoxia the native pools of SUMO-1/IκBα (55 kDa) and IκBα proteins demonstrated a time-dependent degradation over a period of 24 h with early phosphorylation of IκBα (Fig. 3C). Both SUMO-1/IκBα and IκBα protein levels show similar time-dependent degradation starting at 3 h of hypoxia with near complete degradation at 24 h, allowing NFκB activation. We then studied the degradation and phosphorylation characteristics of SUMO-1/IκBα and IκBα proteins in HeLa cells overexpressing the SUMO-1 full-length cDNA under continuous hypoxia. These experiments revealed a significant stabilization of SUMO-1/IκBα and IκBα proteins without appreciable evidence of degradation despite 24 h of hypoxia. In addition, the basal degree of phosphorylation of IκBα observed in cells overexpressing SUMO-1 was substantially lower than naive cells without significant change despite continuous hypoxia (Fig. 3D).
FIGURE 3.
Hypoxia and Reoxygenation prevents IκBα ubiquitination via SUMO-1 modification. A, HeLa cells exposed to 3 h of uninterrupted hypoxia (1% O2) or cycles of H/R. Hypoxia degraded SUMO-1/IκBα (p < 0.01) and unmodified IκBα (p < 0.05) in a similar pattern. No change in protein levels of Iκκβ was observed. In contrast, cycles of H/R totaling 3 h stabilized protein concentrations of SUMO-1/IκBα and prevented the degradation of IκBα with no change in Iκκβ. B, relative density ratios of unmodified and sumoylated IκBα were calculated during hypoxia and H/R. These ratios demonstrate the general degradation effect on both isoforms during continuous hypoxia and demonstrate the cycle-dependent increase provided by increasing cycles of H/R. C, wild type HeLa cells (WT) exposed to 24 h of hypoxia demonstrated significant degradation of SUMO-1/IκBα and unmodified IκBα with early (1 h) phosphorylation of IκBα. D, cells overexpressing the full-length SUMO-1 cDNA exposed to 24 h of hypoxia (1% O2) did not demonstrate evidence of SUMO-1/IκBα or unmodified IκBα degradation despite hypoxia. Relative levels of IκBα phosphorylation were significantly lower than in WT cells exposed to same conditions. E, cells treated with protocols of H/R followed by 24 h of hypoxia. Similarly to SUMO-1-overexpressing cells, H/R prevented degradation of SUMO-1/IκBα or unmodified IκBα and phosphorylation of IκBα.
Because proof of principle that H/R through Ado signaling and increased SUMOylation modulates IκBα degradation, we exposed wild type HeLa cells to protocols of H/R followed by 24 h of continuous hypoxia. Similarly as found in cells overexpressing SUMO-1, SUMO-1/IκBα, and IκBα species were distinctively stabilized without evidence of proteosomal degradation despite hypoxia. Interestingly and as noted in overexpressing cells, the pattern of IκBα phosphorylation was similar to the one observed by SUMO-1 overexpression (Fig. 3E). No significant phosphorylation was detected in cells pre-exposed to H/R, despite 24 h of hypoxia. These results support the concept that H/R significantly halts IκBα degradation by SUMO-1 modification, preventing its phosphorylation by Iκκβ, potentially modulating NFκB-mediated transcription.
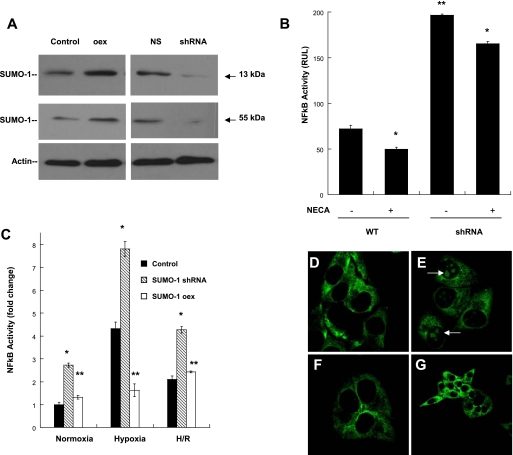
SUMO-1 Modification of IκBα Controls NFκB Activation— Guided by initial results suggesting that H/R trough Ado receptor signaling stabilized specifically the levels of SUMO-1/IκBα isoform, we extended the experiments to understand whether this effect will have functional repercussion on NFκB activation and mediated transcription. Initially SUMO-1 overexpression and SUMO-1 knockdown cell assays were designed to assess their transcriptional contribution to NFκB-mediated pathways (Fig. 4A). To this end we profiled the pattern of NFκB activation using the NFκB luciferase-reporter plasmid in SUMO-1 knockdown cells during hypoxia in the presence of NECA. As demonstrated in preliminary experiments using hypoxia, wild type cells demonstrate significant attenuation of NFκB activation in the presence of enhanced Ado signaling despite 24 h of hypoxia (Figs. 1B and 4B). In contrast SUMO-1 knockdown cells demonstrated a significantly increased NFκB activation (∼3-fold) during hypoxia when compared with wild type cells. NECA did in fact attenuate the degree of NFκB activation in hypoxic SUMO-1 knockdown cells; however, this level was 2.5-fold higher than native levels treated with NECA.
FIGURE 4.
SUMO-1 modification of IκBα controls NFκB activation. A, cell models of SUMO-1 overexpression (105%, increase in total SUMO-1 (13 kDa) and 70% increase in SUMO-1/IκBα (55 kDa) (p < 0.001) and knockdown (small hairpin RNA) results in appreciable (90% decrease in total SUMO-1 and SUMO-1/IκBα, p < 0.01) modulation of SUMO-1/IκBα and SUMO-1 protein levels. Nonspecific small hairpin RNA (NS) is shown as control. B, cells transfected with pNRE-Luc were examined for NFκB activity during hypoxia (24 h) in the presence of NECA (10 μm). WT cells demonstrated attenuation of NFκB activity during Ado receptor stimulation (*, p < 0.05). SUMO-1 knockdown cells exposed to same conditions evidence significant increased NFκB activity compared with WT cells (**, p < 0.01). C, expression levels of SUMO-1 regulate NFκB activity at base-line normoxia, during hypoxia and H/R. SUMO-1 knockdown cells demonstrated increase NFκB activity in all conditions (*, p < 0.001); however, the increase was more impressive during hypoxia (∼8-fold). Overexpressing cells showed attenuated NFκB activity in all conditions (**, p < 0.01). D and E, as confirmatory evidence of NFκB attenuation by H/R and adenosine signaling the profile of p65 translocation in normoxic conditions (D) was compared with 24 h of hypoxia exposure (E). Here, a significant nuclear localization was observed as shown by the arrows. In contrast, cells pre-exposed to H/R followed by hypoxia did not evidenced significant p65 translocation (F). G, cells coincubated with NECA demonstrated a predominant cytoplasmic localization of p65, despite hypoxia exposure. WT, wild type.
Next, we compared the profile of NFκB activation in cells exposed to hypoxia and protocols of H/R of both SUMO-1 overexpressing and knockdown cells. In hypoxia, SUMO-1 knockdown cells demonstrated significantly increased NFκB activation (∼4-fold) when compared with native cells, confirming the regulatory effect of SUMO-1 to IκBα degradation. In contrast SUMO-1-overexpressing cells showed substantial attenuation of NFκB activation during hypoxia and close to normoxic levels. As documented in previous experiments, wild type cells exposed to protocols of H/R followed by 24 h of hypoxia demonstrated significant attenuation of NFκB activation (p < 0.05); however, this attenuation was less impressive in SUMO-1 knockdown cells, returning the NFκB activity to similar levels expressed by wild type hypoxic cells (Fig. 4C). SUMO-1-overexpressing cells pre-exposed to H/R followed by 24 h of hypoxia returned the NFκB activation levels to identical levels observed in wild type cells. These results demonstrate that transcriptional levels of SUMO-1 directly control NFκB activation by increasing specifically the intracellular pool of SUMO-1 modified IκBα, an isoform resistant to proteosomal degradation attenuating NFκB activity. To obtain additional evidence of the effect of adenosine on NFκB activation, we examined the cytoplasmic-to-nuclear translocation of the p65 subunit NFκB in cells exposed to hypoxia, H/R, and NECA. This experiment demonstrates that in contrast to normoxia (Fig. 4D); 24 h of hypoxia induced a significant nuclear translocation of p65 (Fig. 4E). In cells pre-exposed to H/R, hypoxia did not significantly increased p65 nuclear translocation (Fig. 4F). Further, cells coincubated with NECA demonstrated a predominant cytoplasmic localization of p65 similar to the one observed during H/R (Fig. 4G).
SUMO-1 Modification of IκBα Controls NFκB-mediated Transcription—To further confirm that modulation of SUMO-1 cellular levels will significantly influence downstream NFκB-mediated pathways and to document a physiologically relevant role, we profiled IL-6 mRNA transcript levels by real time PCR. IL-6 is transcriptionally regulated at the promoter level by NFκB through a unique binding sequence (20, 21) and is up-regulated by known pro-inflammatory stimuli using NFκB activation (tumor necrosis factor-α, lipopolysaccharide) (22, 23). We used wild type cell levels of IL-6 mRNA expression as a control during normal oxygen conditions. We then used our models of SUMO-1 knockdown and overexpression to determine whether SUMO-1 would directly control IL-6 expression. Interestingly, under hypoxic conditions, SUMO-1 knockdown cells demonstrated a vigorous expression of IL-6 transcripts (∼6-fold, p < 0.005) compared with the native response of wild type cells (Fig. 5A). In contrast, SUMO-1-overexpressing cells demonstrated a significant reduction in IL-6 expression, to levels below the ones observed for wild type hypoxic cells. To verify that the effect of SUMO-1 on NFκB-mediated pathways was not limited to cultured cells, we extended these findings to models of hypoxia and H/R in vivo (for details see “Experimental Procedures”). Here we used lung tissue from CD 73 null mice and C57BL6 adult mice. CD 73 null mice lack the ecto-5′-nucleotidase (CD 73), a critical enzyme for extracellular Ado generation (2). In previous studies we have demonstrated the pulmonary-specific pro-inflammatory effects of CD 73 gene deletion (24) and selected the lung tissue as suitable model to also understand SUMO-1 regulation in vivo. Initially we profiled IL-6 mRNA transcripts from lung tissue from wild type and CD 73 null mice exposed to environmental conditions of hypoxia and cycles of H/R. Wild type animals demonstrated significantly increased IL-6 expression (p < 0.05) during hypoxia and attenuation (p < 0.05) when exposed to cycles of H/R, consistent with results in vitro documenting similar patterns of NFκB activity (Fig. 5B). We then compared IL-6 mRNA levels in CD73 null mice exposed to hypoxia and H/R. These results demonstrated that in CD 73 (-/-) lung tissue, the level of expression of IL-6 was significantly (∼11-fold, p < 0.01) increased at base-line normoxia with a substantial (∼12-fold, p < 0.01) increased expression exerted by hypoxic conditions. No significant attenuation of IL-6 expression was observed by exposure to H/R (p = 0.2). Further, to demonstrate that Ado signaling is required in vivo to maintain regulatory levels of SUMO-1, we compared protein levels of SUMO-1/IκBα of wild type lung tissue lysates. These experiments demonstrated that levels SUMO-1/IκBα species are significantly enhanced by H/R compared with normoxia and significantly degraded by exposure to hypoxia. Remarkably, tissue levels of SUMO-1/IκBα of CD 73 null mice were considerably lower in all experimental conditions without appreciable increase as expected by H/R (Fig. 5C). These results demonstrate that the inability to generate extracellular Ado significantly affects levels of SUMO-1/IκBα and downstream regulation of NFκB-mediated transcription.
FIGURE 5.
SUMO-1 modification of IκBα controls NFκB-mediated transcription. A, wild type, knockdown, and overexpressing SUMO-1 cells were assayed for IL-6 mRNA transcript levels using real time PCR at base-line normoxia or after 24 h of hypoxia. Control and SUMO-1 knockdown cells demonstrated remarkable increase in IL-6 expression in hypoxia (*, p < 0.005) compared with Normoxic cells. SUMO-1-overexpressing cells showed impressive attenuation of IL-6 expression despite hypoxia (**, p < 0.005). B, IL-6 mRNA levels of whole lung tissue lysates from C57BL/6 or CD 73 null mice exposed to hypoxia or in vivo protocols of H/R. In wild type mice (WT), H/R induced attenuation of IL-6 expression (*, p < 0.05, compared with hypoxia) similar to the effect observed in vitro. In CD 73 (-/-) tissue, the absence of Ado signaling produced a substantial increase (10-fold; **, p < 0.01) in base-line normoxic levels of IL-6 and 11-fold (***, p < 0.01, compared with normoxic WT) increase during hypoxia. C, protein levels of SUMO-1/IκBα. WT animals exposed to H/R showed restoration of SUMO-1/IκBα compared with degraded levels in animals exposed to hypoxia (p < 0.01). Basal levels of SUMO-1/IκBα in CD 73 null mice were substantially lower (p < 0.001) compared with WT animals and no increase was observed despite H/R.
In summary, we present a functional model by which sequential exposure to hypoxia followed by reoxygenation activates general Ado signaling increasing protein levels of SUMO-1. This general effect, in turn results in the specific SUMOylation of IκBα, inhibiting its phosphorylation and ubiquitination. The SUMOylated IκBα is resistant to hypoxia-induced proteosomal degradation, attenuating NFκB activation and mediated gene transcription.
DISCUSSION
Adenosine generation and Ado-mediated signaling has been described to regulate cellular mechanisms aimed at adaptation to a number of stress conditions (25). In addition, Ado attenuates NFκB activation by modulation of the catalytic activity of the E3 SCF ubiquitin ligase responsible for proteosomal degradation of IκBα and nuclear translocation of the p50/p65 subunits of NFκB. We have demonstrated that Ado receptor stimulation predominantly mediated by the Ado A2B subtype regulates anti-inflammatory signaling and modulates IκBα degradation by Cullin-1 deneddylation suppressing NFκB activation(4). In the current study we assessed the role of Ado signaling on recognized modulators of IκBα degradation. Our studies revealed initially that cycles of hypoxia and reoxygenation resulted in accumulation of Ado with distinct effect on NFκB activation. Recent studies have demonstrated that individual receptor signaling regulate cellular and tissue effects of pro-inflammatory stimulus and mediate normal physiological responses (26). The generation of extracellular Ado during hypoxic conditions has been studied in detail (27), and the phenotypical consequences of its absence is described in particular organs (24). Although much about the generation, degradation, and uptake of Ado has been reviewed, the specific mechanism of enhanced Ado production during hypoxia followed by reoxygenation is not entirely clear. It is possible that H/R would affect both the activity of de novo enzymatic cascade (CD39/CD73) and/or its degradation by Ado deaminase. Clearly, either mechanism would potentially result in increased selective Ado receptor stimulation and NFκB attenuation. We further studied the possibility that Ado signaling would also modulate additional mechanisms involved in IκBα degradation.
As a normal mechanism of regulation, NFκB is activated by proteosomal degradation of IκBα and subsequently induces the synthesis of new IκBα protein in the nucleus of the cell (15). This newly synthesized IκBα is modified by SUMO-1 and transported back to the cytoplasm (16). The SUMOylated isoform is resistant to proteosomal degradation by at least two mechanisms. SUMO-1 conjugation of IκBα by Ubc9 occurs to the unphosphorylated form IκBα using its ubiquitination site Lys21/Lys22. Ubc9 conjugates a single molecule of ubiquitinin, further blocking E1 and E2 ubiquitination activity. Whether the initial SUMO-1 modification per se prevents phosphorylation of IκBα by structural blockade of its neighboring phosphorylation site Ser32/Ser36 or recruits another molecule occluding this site remains to be determined. It is clear however that SUMOylation of IκBα results in occupancy of the effective ubiquitination site, preventing phosphorylation and further degradation by the E3 ligase. In untreated cells we identified a single band of ∼55 kDa by immunoprecipitation of SUMO-1 and Western blotting to IκBα. This band corresponded to the SUMO-1/IκBα isoform also observed by Desterro et al. (10) in HeLa cells and different from the unmodified IκBα (43 kDa). The SUMO-1/IκBα was significantly increased in cells exposed to H/R and appeared degraded after exposure to hypoxia. These findings were consistent with our hypothesis that Ado signaling provided by H/R could potentially modulate the SUMO-1 modification of IκBα. Next we determined that exogenous Ado analogs would preferentially increase the SUMO-1/IκBα isoform in a dose-dependent fashion. We confirmed the effect of Ado on the concentrations of SUMO-1/IκBα isoform by incubation with both general and specific Ado receptor antagonists consistent with reports of pro-inflammatory responses induced by Ado receptor inhibition (28).
During hypoxia SUMO-1 is transcriptionally controlled by HIF-1 α and may play key role on its molecular stability and transcriptional activity (29–31). A recent study has documented the existence of a domain in RING finger and WD repeat-containing protein induced by hypoxia, enhancing SUMO-1 modification of IκBα and stabilizing HIF-1 α activity (32). Although SUMO-1 is activated early in hypoxic conditions (33), the SUMO-1/IκBα isoform is clearly degradated in a similar pattern to the unmodified IκBα isoform. Interestingly, in normal cells despite potential transcriptional induction of SUMO-1 and protein modification, the SUMO-1/IκBα was not exempt of proteosomal degradation by severe hypoxia. This effect may be cell type-specific and also dependent on different oxygen concentrations. In contrast cells exposed to H/R evidenced increasing concentrations of SUMO-1/IκBα protein with stabilization of IκBα isoform in a cycle-dependent fashion consistent with findings of Ado accumulation and NFκB attenuation also in cycle-dependent fashion. Next we determined the relative resistance to proteosomal degradation provided by SUMO-1 overexpression and H/R after prolonged exposure (24 h) of hypoxia, because the earlier results demonstrated significant attenuation in NFκB activation even after 24 h of hypoxia followed H/R. In both instances the SUMO-1/IκBα and unmodified IκBα proteins demonstrated resistance to proteosomal degradation and stabilization despite severe hypoxia. Although H/R involves periods of hypoxia and potentially induction of SUMO-1 protein mediated by HIF-1 α as observed in hypoxia alone, its effect on IκBα degradation and NFκB activation is quite distinct from uninterrupted hypoxia. Studies documenting the influence of SUMOylation of diverse pathways (peroxisome proliferator-activated receptor γ) (34) may explain additional potential anti-inflammatory effects also influencing the NFκB pathways; however, in our experiments exposure to uninterrupted hypoxia did not prevent IκBα proteosomal degradation nor attenuation of NFκB. In addition to hypoxia, H/R promotes Ado signaling, specifically increasing the SUMOylated IκBα isoform and modifying the pattern of IκBα phosphorylation and proteosomal degradation. This effect was similarly observed in cells overexpressing SUMO-1.
The findings were further tested in vivo to document a relevant and distinct effect of Ado signaling to SUMO-1 modification and NFκB-mediated transcription. Because of the known regulatory effect of NFκB activity to IL-6, we studied levels of expression of mRNA transcripts in wild type and CD 73 null mice paralleled with proteins levels of SUMO-1/IκBα. These results demonstrated that under the influence of normal Ado signaling in wild type mice, H/R significantly attenuates the expression of IL-6 because of suppression of NFκB activity. The absence of Ado in CD 73 (-/-) lung tissue resulted in increased expression of IL-6 and loss of the regulatory effect provided by H/R. Protein levels of SUMOylated IκBα isoform in lung tissue of CD 73 null mice and the lack of augmentation despite H/R provided confirmatory evidence of the relevance of Ado signaling to the normal pathways responsible for restoring base-line and Ado-stimulated levels of SUMO-1/IκBα. The pulmonary phenotype observed in the CD 73 null mice has been attributed to the absence of anti-inflammatory input provided by A2B and A1 receptor subtypes (24). Studies in A2B receptor null mice have shown pulmonary-specific defects on barrier function and cell migration necessary to build immune and inflammatory responses (35). Enhanced Ado receptor signaling provides inactivation of the E3 ligase by Cullin-1 deneddylation and decreased IκBα degradation by SUMO-1 modification, effectively inhibiting NFκB activation during severe hypoxia.
Overall we present a mechanistic model by which episodes of brief hypoxia followed by reoxygenation result in accumulation of a nondegradable form of IκBα mediated by Ado signaling. The cytoplasmic SUMO-1/IκBα represent normally a pool of nondegradable IκBα present to restore the levels of unmodified IκBα continuously metabolized by pro-inflammatory stimulus. In addition, once NFκB has been activated and its transcriptional effect has been initiated, the SUMO-1/IκBα to IκBα ratio favors inhibition of proteosomal degradation, limiting further unnecessary NFκB activation. It appears that the cyclic accumulation of SUMO-1/IκBα species provides especial physiologic advantages if achieved before exposure to severe pro-inflammatory stimulus. Our model was initiated with exposure to hypoxia, inducing phosphorylation, polyubiquitination, and degradation of IκBα with subsequent nuclear translocation of NFκB. Acute termination of hypoxic conditions allowed accumulation of Ado, facilitating the SUMO-1 modification of IκBα. Subsequent cycles of H/R permitted further accumulation of SUMO-1/IκBα also possibly because the short periods of hypoxia used in the H/R cycle only partially degraded both SUMO-1/IκBα and unmodified IκBα. At the end of three cycles and during reoxygenation, enough SUMO-1/IκBα had been accumulated attenuating NFκB activation (Fig. 6). We acknowledge the possibility that both transcriptional induction of SUMO-1 protein by hypoxia and Ado signaling generated during reoxygenation promoted the magnitude of SUMOylation of IκBα observed in our model. It is clear that without a reoxygenation phase, the potential hypoxia-mediated increase in SUMO-1 did not prevent proteosomal degradation of IκBα. The particular properties to IκBα degradation provided by H/R proved of potent physiological relevance and provide novel regulatory pathways distinctive from purely hypoxic conditions and basis for further interpretation of post-hypoxic induced metabolism and cell signaling.
FIGURE 6.
Part 1, mechanistic model of enhanced SUMOylation of IκBα during H/R. Part 2, initial exposure to hypoxia results in phosphorylation of IκBα at Ser32/Ser36 by Iκκβ. Part 3, phosphorylated IκBα is targeted for polyubiquitination by the E3 SCF ubiquitin-ligase and proteosomal degradation. Part 4, p65/p50 subunits of NFκB readily translocate to the nucleus and initiate transcriptional induction of NFκB-dependent genes, simultaneously p65 inducing expression of the IκBα gene. Part 5, the new generated unphosphorylated form of IκBα is transported back to the cytoplasm and available for SUMO-1 modification under the influence of Ado signaling. Part 6, the pool of SUMOylated IκBα increase with subsequent cycles of H/R under the influence increasing concentrations of Ado. High cytoplasmic concentrations of SUMOylated IκBα present as a result of H/R prevent further NFκB activation, because the SUMOylated IκBα is resistant to proteosomal degradation.
Supplementary Material
This work was supported by Funds from the Robert M. Smith Award, the Department of Anesthesiology, Perioperative and Pain Medicine, Children's Hospital, and the Eleanor and Miles Shore 50th Anniversary Fellowship Program for Scholars in Medicine, Children's Hospital Boston, and Harvard Medical School (to J. C. I.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: Ado, adenosine; SUMO, small ubiquitin-like modifier; H/R, hypoxia followed by reoxygenation; IL, interleukin; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; NECA, N-ethylcarbamido-adenosine; PSB 1115, 4-(2,3,6,7-tetrahydro-2,6-dioxo-1-propyl-1H-purin-8-yl)benzenesulfonic acid.
References
- 1.Furuta, G. T., Turner, J. R., Taylor, C. T., Hershberg, R. M., Comerford, K., Narravula, S., Podolsky, D. K., and Colgan, S. P. (2001) J. Exp. Med. 193 1027-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Synnestvedt, K., Furuta, G. T., Comerford, K. M., Louis, N., Karhausen, J., Eltzschig, H. K., Hansen, K. R., Thompson, L. F., and Colgan, S. P. (2002) J. Clin. Investig. 110 993-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckle, T., Fullbier, L., Wehrmann, M., Khoury, J., Mittelbronn, M., Ibla, J., Rosenberger, P., and Eltzschig, H. K. (2007) J. Immunol. 178 8127-8137 [DOI] [PubMed] [Google Scholar]
- 4.Khoury, J., Ibla, J. C., Neish, A. S., and Colgan, S. P. (2007) J. Clin. Investig. 117 703-711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy, M. N., Howe, K., Etkin, L. D., Solomon, E., and Freemont, P. S. (1996) Oncogene 13 971-982 [PubMed] [Google Scholar]
- 6.Matunis, M. J., Coutavas, E., and Blobel, G. (1996) J. Cell Biol. 135 1457-1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muraoka, A., Maeda, A., Nakahara, N., Yokota, M., Nishida, T., Maruyama, T., and Ohshima, T. (2008) Biochem. Biophys. Res. Commun. 377 1031-1035 [DOI] [PubMed] [Google Scholar]
- 8.Kroetz, M. B. (2005) Yale J. Biol. Med. 78 197-201 [PMC free article] [PubMed] [Google Scholar]
- 9.Jennewein, C., Kuhn, A. M., Schmidt, M. V., Meilladec-Jullig, V., von Knethen, A., Gonzalez, F. J., and Brune, B. (2008) J. Immunol. 181 5646-5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desterro, J. M., Thomson, J., and Hay, R. T. (1997) FEBS Lett. 417 297-300 [DOI] [PubMed] [Google Scholar]
- 11.Johnson, E. S., and Blobel, G. (1997) J. Biol. Chem. 272 26799-26802 [DOI] [PubMed] [Google Scholar]
- 12.Saitoh, H., Sparrow, D. B., Shiomi, T., Pu, R. T., Nishimoto, T., Mohun, T. J., and Dasso, M. (1998) Curr. Biol. 8 121-124 [DOI] [PubMed] [Google Scholar]
- 13.Desterro, J. M., Rodriguez, M. S., and Hay, R. T. (1998) Mol. Cell 2 233-239 [DOI] [PubMed] [Google Scholar]
- 14.Mikolajczyk, J., Drag, M., Bekes, M., Cao, J. T., Ronai, Z., and Salvesen, G. S. (2007) J. Biol. Chem. 282 26217-26224 [DOI] [PubMed] [Google Scholar]
- 15.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14 649-683 [DOI] [PubMed] [Google Scholar]
- 16.Arenzana-Seisdedos, F., Turpin, P., Rodriguez, M., Thomas, D., Hay, R. T., Virelizier, J. L., and Dargemont, C. (1997) J. Cell Sci. 110 369-378 [DOI] [PubMed] [Google Scholar]
- 17.Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995) J. Biol. Chem. 270 17442-17456 [DOI] [PubMed] [Google Scholar]
- 18.Hay, R. T., Vuillard, L., Desterro, J. M., and Rodriguez, M. S. (1999) Philos. Trans. R. Soc. Lond. B Biol. Sci. 354 1601-1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matunis, M. J., Wu, J., and Blobel, G. (1998) J. Cell Biol. 140 499-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsusaka, T., Fujikawa, K., Nishio, Y., Mukaida, N., Matsushima, K., Kishimoto, T., and Akira, S. (1993) Proc. Nat;. Acad. Sci. U. S. A. 90 10193-10197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Libermann, T. A., and Baltimore, D. (1990) Mol. Cell. Biol. 10 2327-2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang, Y. H., Lin, J. X., and Vilcek, J. (1990) Mol. Cell. Biol. 10 3818-3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crisostomo, P. R., Wang, Y., Markel, T. A., Wang, M., Lahm, T., and Meldrum, D. R. (2008) Am. J. Physiol. 294 C675-682 [DOI] [PubMed] [Google Scholar]
- 24.Thompson, L. F., Eltzschig, H. K., Ibla, J. C., Van De Wiele, C. J., Resta, R., Morote-Garcia, J. C., and Colgan, S. P. (2004) J. Exp. Med. 200 1395-1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colgan, S. P., Eltzschig, H. K., Eckle, T., and Thompson, L. F. (2006) Purinergic Signalling 2 351-360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckle, T., Faigle, M., Grenz, A., Laucher, S., Thompson, L. F., and Eltzschig, H. K. (2008) Blood 111 2024-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eltzschig, H. K., Ibla, J. C., Furuta, G. T., Leonard, M. O., Jacobson, K. A., Enjyoji, K., Robson, S. C., and Colgan, S. P. (2003) J. Exp. Med. 198 783-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reutershan, J., Vollmer, I., Stark, S., Wagner, R., Ngamsri, K. C., and Eltzschig, H. K. (2008) Faseb J. 23 473-482 [DOI] [PubMed] [Google Scholar]
- 29.Berta, M. A., Mazure, N., Hattab, M., Pouyssegur, J., and Brahimi-Horn, M. C. (2007) Biochem. Biophys. Res. Commun. 360 646-652 [DOI] [PubMed] [Google Scholar]
- 30.Bae, S. H., Jeong, J. W., Park, J. A., Kim, S. H., Bae, M. K., Choi, S. J., and Kim, K. W. (2004) Biochem. Biophys. Res. Commun. 324 394-400 [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. W., Bae, S. H., Jeong, J. W., Kim, S. H., and Kim, K. W. (2004) Exp. Mol. Med. 36 1-12 [DOI] [PubMed] [Google Scholar]
- 32.Carbia-Nagashima, A., Gerez, J., Perez-Castro, C., Paez-Pereda, M., Silberstein, S., Stalla, G. K., Holsboer, F., and Arzt, E. (2007) Cell 131 309-323 [DOI] [PubMed] [Google Scholar]
- 33.Shao, R., Zhang, F. P., Tian, F., Anders Friberg, P., Wang, X., Sjoland, H., and Billig, H. (2004) FEBS Lett. 569 293-300 [DOI] [PubMed] [Google Scholar]
- 34.Pascual, G., Fong, A. L., Ogawa, S., Gamliel, A., Li, A. C., Perissi, V., Rose, D. W., Willson, T. M., Rosenfeld, M. G., and Glass, C. K. (2005) Nature 437 759-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckle, T., Grenz, A., Laucher, S., and Eltzschig, H. K. (2008) J. Clin. Investig. 118 3301-3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.