Abstract
Angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) are secreted proteins that regulate triglyceride (TG) metabolism in part by inhibiting lipoprotein lipase (LPL). Recently, we showed that treatment of wild-type mice with monoclonal antibody (mAb) 14D12, specific for ANGPTL4, recapitulated the Angptl4 knock-out (-/-) mouse phenotype of reduced serum TG levels. In the present study, we mapped the region of mouse ANGPTL4 recognized by mAb 14D12 to amino acids Gln29–His53, which we designate as specific epitope 1 (SE1). The 14D12 mAb prevented binding of ANGPTL4 with LPL, consistent with its ability to neutralize the LPL-inhibitory activity of ANGPTL4. Alignment of all angiopoietin family members revealed that a sequence similar to ANGPTL4 SE1 was present only in ANGPTL3, corresponding to amino acids Glu32–His55. We produced a mouse mAb against this SE1-like region in ANGPTL3. This mAb, designated 5.50.3, inhibited the binding of ANGPTL3 to LPL and neutralized ANGPTL3-mediated inhibition of LPL activity in vitro. Treatment of wild-type as well as hyperlipidemic mice with mAb 5.50.3 resulted in reduced serum TG levels, recapitulating the lipid phenotype found in Angptl3-/- mice. These results show that the SE1 region of ANGPTL3 and ANGPTL4 functions as a domain important for binding LPL and inhibiting its activity in vitro and in vivo. Moreover, these results demonstrate that therapeutic antibodies that neutralize ANGPTL4 and ANGPTL3 may be useful for treatment of some forms of hyperlipidemia.
Lipoprotein lipase (LPL)5 plays a pivotal role in lipid metabolism by catalyzing the hydrolysis of plasma triglycerides (TGs). LPL is likely to be regulated by mechanisms that depend on nutritional status and on the tissue in which it is expressed (1–3). Two secreted proteins, angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4), play important roles in the regulation of LPL activity (4, 5). ANGPTL3 and ANGPTL4 consist of a signal peptide, an N-terminal segment containing coiled-coil domains, and a C-terminal fibrinogen-like domain. The N-terminal segment as well as full-length ANGPTL3 and ANGPTL4 have been shown to inhibit LPL activity, and deletion of the N-terminal segment of ANGPTL3 and ANGPTL4 resulted in total loss of LPL-inhibiting activity (6, 7). These observations clearly indicate that the N-terminal region of ANGPTL4 contains the functional domain that inhibits LPL and affects plasma lipid levels. The coiled-coil domains have been proposed to be responsible for oligomerization (8); however, it is not known whether the coiled-coil domains directly mediate the inhibition of LPL activity.
To define the physiological role of ANGPTL4 more clearly, we characterized the pharmacological consequences of ANGPTL4 inhibition in mice treated with the ANGPTL4-neutralizing monoclonal antibody (mAb) 14D12 (9). Injection of mAb 14D12 significantly lowered fasting TG levels in C57BL/6J mice relative to levels in C57BL/6J mice treated with an isotype-matched anti-KLH control (KLH) mAb (9). These reduced TG values were similar to decreases in fasting plasma TG levels measured in Angptl4 knock-out (-/-) mice. This study demonstrated that mAb 14D12 is a potent ANGPTL4-neutralizing antibody that is able to inhibit systemic ANGPTL4 activity and thereby recapitulate the reduced lipid phenotype found in Angptl4-/- mice. The readily apparent pharmacological effect of mAb 14D12 prompted new questions about the epitope recognized by mAb 14D12 and how this antibody-antigen binding event affected ANGPTL4 function as an LPL inhibitor.
Although ANGPTL4 is able to interact directly with LPL (10), it is not clear which amino acids within ANGPTL4 mediate this interaction. Here we show that amino acids Gln29–His53 of mANGPTL4 contain the epitope for mAb 14D12. This region, hereby designated specific epitope 1 (SE1), also defines a domain that mediates the interaction between ANGPTL4 and LPL and the subsequent inactivation of LPL. With this information we present evidence that ANGPTL3 also contains an SE1 region, and with antibodies specifically reactive with ANGPTL3 SE1 we examine whether the ANGPTL3 SE1 region is involved in LPL binding and inhibition. We also determined whether treatment of C57BL/6 mice with an anti-ANGPTL3 SE1 mAb can recapitulate the phenotype of lower serum TG and cholesterol levels found in Angptl3-/- mice. Finally we tested the therapeutic potential of an anti-ANGPTL3 SE1 mAb for treatment of hyperlipidemia in apolipoprotein E-/- (ApoE-/-) or low density lipoprotein receptor-/- (LDLr-/-) mice.
EXPERIMENTAL PROCEDURES
Reagents—Fatty acid-free bovine serum albumin (BSA), puromycin dihydrochloride, heparin sodium salt, and protease inhibitor mixtures were purchased from Sigma. FreeStyle 293 Expression Medium and FreeStyle 293 Expression Medium plus Glutamax-1 were from Invitrogen Corporation. Lipase substrate 1,2-O-dilauryl-rac-glycero-3-glutaric acid-(6′-methylresorufin) ester (DGGR) was purchased from the Fluka division of Sigma. Heparin-Sepharose 6 FF was purchased from GE Healthcare. All PCR primers were synthesized by the Sigma Genosys division of Sigma. All restriction endonucleases were from New England Biolab, Inc. (Beverly, MA). Taq DNA polymerase was from Roche Diagnostics Corporation (Indianapolis, IN). Ni-NTA affinity resin was from Qiagen Inc. (Valencia, CA).
Generation of Angptl3-/- Mice, Angptl4-/- Mice, and Mice Homozygous for the Angptl4 Variant E40K (Angptl4K/K) Allele—The Angptl3-/- mouse line was generated by homologous recombination.6 Exons 1–4 were disrupted with a targeting vector derived with the Lambda KOS system (11), as shown in supplemental Fig. S1 and described under supplemental Methods. LDLr-/- mice (catalog number 002207, The Jackson Laboratory, Bar Harbor, ME) were bred with Angptl3-/- mice to generate Angptl3-/-/LDLr-/- double knock-out mice. ApoE-/- mice (catalog number APOE-M, Taconic, Hudson, NY) were bred with Angptl3-/- mice to generate Angptl3-/-/ApoE-/- mice. The generation of Angptl4-/- mice has been described previously (9). The strategy for generating Angptl4K/K mice is shown in supplemental Fig. S2 and is described under supplemental Methods. The approach for characterizing Angptl4 mRNA in Angptl4K/K mice is also described under supplemental Methods.
Mouse Care and Study—All procedures involving animals were conducted in conformity with Institutional Animal Care and Use Committee guidelines in compliance with state and federal laws and the standards outlined (26). Mice were housed at 24 °C on a fixed 12-h light/12-h dark cycle and had free access to water and diet. LDLr-/- mice and Angptl3-/-/LDLr-/- mice were maintained on Clinton Diet (D12107, Research Diets, New Brunswick, NJ). Angptl4-/- mice and Angptl4K/K mice were studied while receiving a high fat diet (D12451, Research Diets). All other mice were maintained on regular chow (product 5021, Purina, St. Louis, MO).
Analysis of Serum Lipid Levels—Serum samples for lipid analysis were prepared from blood obtained from the retro-orbital plexus. Total TG levels were measured with a COBAS Integra 400 serum chemistry analyzer (Roche Diagnostics) or with a kit (Serum TG determination kit, catalog number TR0100, Sigma). Total cholesterol levels were measured with a COBAS Integra 400 serum chemistry analyzer (Roche Diagnostics) or with a kit (Wako Diagnostics, Richmond, VA; catalog number 439-17501).
Immunization and Generation of Hybridomas—Angptl3-/- mice were immunized by first priming with hANGPTL3 protein and then boosting every 2–3 weeks with either hANGPTL3 protein or with a synthetic peptide containing SE1 of ANGPTL3 (amino acids E32-L57) conjugated to KLH. Immunogens for priming were emulsified in Complete Freund's adjuvant, whereas immunogens for boosting were emulsified in Incomplete Freund's adjuvant. After two to three boosts, the serum titers were monitored by ELISA. Once high titers were achieved, splenocytes were harvested from the immunized mice and fused with myeloma cells (P3/NSI/1-Ag4-1) using PEG 1500 as a fusion agent. The resulting cell fusion products were diluted into the hybridoma medium and seeded into 96-well tissue culture plates. After 1 day, hypoxanthine/aminopterin/thymidine medium was added and hybridoma cultures were maintained under selection for 10–14 days. The culture medium was changed every 3–4 days as necessary. After 10–14 days of selection and culture, hybridoma screening was performed by ELISA. Depending on the immunogen used, ELISA utilized either hANGPTL3 protein or both hANGPTL3 and ANGPTL3 peptide antigen. Confirmed ELISA-positive hybridomas were subcloned by limiting dilution, and the resulting subcloned lines for each hybridoma line were stored in liquid nitrogen using standard methods.
Hybridoma Scale-up and mAb Purification—Methods for hybridoma scale-up and mAb purification were performed as previously described (9). The anti-ANGPTL4 14D12 and anti-KLH mAbs have been previously described (9).
Epitope Mapping—For epitope mapping, linear expression cassettes for in vitro transcription and translation of His-tagged SUMO-mouse ANGPTL4 fusion proteins were synthesized by PCR amplification. First, a cDNA corresponding to a fragment of pET SUMO vector (Invitrogen) was PCR amplified with sense primer 5′-GAAATTAATACGACTCACTATAGGG-3′ and antisense primer 5′-ACCACCAATCTGTTCTCTGTGAGC-3′. This cDNA (T7-SUMO) spans the T7 promoter through the C-terminal end of the His-tagged SUMO open reading frame (ORF). For epitope mapping of mouse ANGPTL4 amino acids Gln24–Pro180 (accession number Q9Z1P8), the initial set of expression cassettes consisted of overlapping mouse ANGPTL4 cDNAs that encoded five 50-amino acid ORFs and four 25-amino acid ORFs (Fig. S3A). The cDNA fragments were PCR amplified from mouse ANGPTL4 cDNA with primers containing sequences overlapping either the 3′ end of the T7-SUMO cDNA or the 3′ end of the antisense T7 terminator primer (Table S1). The linear expression cassettes for in vitro transcription and translation were generated by bridge PCR amplification using the T7-SUMO cDNA, an Angptl4 cDNA, sense primer 5′-GAAATTAATACGACTCACTATAGGG-3′, and antisense T7 terminator primer 5′-AAAACCCCTCAAGACCCGTTTAGAGGCCCCAAGGGGTTGGGAGTAGAATGTTAAGGATTAGTTTATTA-3′.
To fine map the epitope of mAb 14D12, a series of 10 expression cassettes encoding overlapping His-tagged SUMO-mouse ANGPTL4 fusion proteins spanning amino acids Gln24–Ser93 were generated. The mouse ANGPTL4 cDNAs encode 25 amino acids that overlap by 20 amino acids (Fig. S3B). Each cDNA was PCR amplified with the primers containing the sequence overlaps described above (Table S2). The linear expression cassettes for in vitro transcription and translation were generated by bridge PCR amplification as described above.
SUMO fusion proteins containing the mouse ANGPTL4 peptides were synthesized by in vitro transcription and translation (RTS100 Escherichia coli HY Kit, Roche Applied Science) according to the manufacturer's instructions. The amount of in vitro synthesized proteins was normalized by Western blot analysis with anti-His tag antibody (Bethyl, Montgomery, TX). Epitope mapping was performed by Western blot analysis of the fusion proteins with mAb 14D12. A nearly identical strategy was used to map the epitope of mAbs reactive with ANGPTL3 (details not shown).
Expression and Purification of Full-length Recombinant ANGPTL3 and LPL—A cDNA encoding the entire ORF of human ANGPTL3 (hANGPTL3) (accession NP_055310.1) with an appended C-terminal His6 epitope tag was cloned into pIRESpuro2 vector (Clontech Laboratories, Inc., Mountain View, CA). Freestyle HEK293-F cells stably transfected with the expression vector encoding hANGPTL3 were cultured in Free-Style 293 Expression Medium according to the manufacturer's recommendations (Invitrogen). For production of recombinant protein, cells were typically seeded into medium at a density of 1 × 106/ml and cultured for 48–96 h at 37 °C in shaker flasks. The cells were then collected by centrifugation at 2000 × g for 20 min, and the conditioned medium containing secreted hANGPTL3 was filtered through 0.22-μm filter units and stored at -20 °C. Purification of hANGPTL3 from conditioned medium was performed as described previously (9).
A cDNA encoding human LPL (accession NP_000228.1) with an appended C-terminal FLAG epitope tag (GDYKD-DDDK) was cloned into pIRESpuro2 vector, and stably transfected cells and conditioned medium were generated as described above. Catalytically active LPL was prepared by heparin-Sepharose chromatography (12). All procedures were performed at 4 °C. The medium was thawed and supplemented with Tris-HCl (pH 7.4) to a final concentration of 10 mm and with Triton X-100 to a final concentration of 0.1%. The supplemented medium was applied to a 1.0-ml heparin-Sepharose column equilibrated in 10 ml of Buffer A (10 mm Tris-HCl, 0.15 m NaCl, 0.1% Triton X-100, pH 7.4) at a flow rate of 1 ml/min. The column was washed with 10 ml of Buffer A and then 10 ml of Buffer B (10 mm Tris-HCl, 0.6 m NaCl, 0.1% Triton X-100, pH 7.4). The column was then washed with Buffer C (10 mm Tris-HCl, 1 m NaCl, 0.1% Triton X-100, pH 7.4), collecting the eluate in 0.5-ml fractions. The fractions were assayed for LPL activity with the DGGR substrate as described below, and the peak fractions were pooled and stored at -80 °C.
Expression and Purification of N-terminal Fragments of Recombinant ANGPTL3 and ANGPTL4—Complementary DNAs encoding human ANGPTL3 amino acids Asp20–Pro243 (N′-hANGPTL3) and human ANGPTL4 amino acids Gly26–His176 (N′-hANGPTL4; accession number NP_647475.1) were cloned into pET24a(+) vector (EMD Chemicals Inc., Gibbstown, NJ). A cDNA encoding mouse ANGPTL4 amino acids Gly25–Leu185 (N′-mANGPTL4) was cloned into pET22b(+) vector (EMD Chemicals). Expression vectors were transformed into E. coli BL21(DE3). Protein expression was induced by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 1 mm once the cultures reached an A600 reading of 0.5. Induced cultures were incubated for 18 h at room temperature. Cells were collected by centrifugation at 2000 × g for 20 min at 4 °C, and the cell pellets were stored at -80 °C.
Recombinant proteins were purified with Ni-NTA resin according to the manufacturer's recommendations. All procedures were performed on ice or at 4 °C. Frozen bacterial cell pellets (2–4 g) were thawed and resuspended in 20 ml of Lysis Buffer (50 mm sodium phosphate, 500 mm sodium chloride, 10% glycerol, 20 mm imidazole, 1 mm benzamidine, 5 mm β-mercaptoethanol, 1% protease inhibitor mixture, 0.05% benzonase, pH 7.8). Lysozyme was added to the cell suspension at a final concentration of 1 mg/ml, and the mixture was incubated on ice for 30 min. The suspension was sonicated and then centrifuged at 10,000 × g for 30 min. The resulting supernatant was recovered and passed through a 0.22-μm filter unit. The filtered supernatant was added to 1 ml of washed Ni-NTA resin and incubated with agitation for 1 h at 4 °C. The resin was collected by centrifugation and the supernatant was removed. The packed resin was resuspended in 2 ml of Lysis Buffer and transferred to a column. The flow-through was discarded, and the column was washed with 2 ml of Lysis Buffer. The column was sequentially washed with 20 mm imidazole (50 ml), 50 mm imidazole (10 ml), and 100 mm imidazole (10 ml) in Wash Buffer (50 mm sodium phosphate, 500 mm sodium chloride, 10% glycerol, 1 mm benzamidine, 5 mm β-mercaptoethanol, pH 7.8). The protein was eluted from the column with 250 mm imidazole in Wash Buffer, collecting 1-ml fractions. Fractions were analyzed by SDS-PAGE, and the fractions containing the recombinant protein peak were pooled, dialyzed against Wash Buffer without benzamidine, and stored at -80 °C. Protein was quantified using Microplate BCA™ Protein Assay Kit-Reducing Agent Compatible (Pierce Biotechnology).
Affinity Determination by Surface Plasmon Resonance— Affinity constants of mAbs for ANGPTL3 SE1 peptide and N′-ANGPTL4 were determined using a BIACORE® 3000 system (GE Healthcare) as described under supplemental Methods.
Measurement of LPL Activity with DGGR Substrate—LPL activity was determined in vitro with the fluorogenic substrate DGGR according to a modification of the method of Panteghini and co-workers (13). A 0.24 mm DGGR substrate solution was prepared by adding 10 mm DGGR in ethanol to 1.6 mm sodium tartrate (pH 4.0) and 0.5% Triton X-100. The solution was vortexed vigorously and then filtered through a 0.22-μm filter unit. This solution is stable for at least 2 weeks at room temperature.
Samples containing LPL were prepared in DGGR LPL Assay Buffer, with the following composition: 50 mm Tris-HCl, 0.12 m NaCl, 0.5% Triton X-100, 10 mg/ml BSA (pH 7.4). Reactions were performed at room temperature and were initiated by adding 90 μl of sample to 10 μl of 0.24 mm DGGR substrate. Fluorescence was monitored at 30-s intervals for 10 min with a Cytofluor Series 4000 Fluorescence Multi-well Plate Reader (Applied Biosystems, Foster City, CA) fitted with a 530/25 nm excitation filter and a 620/40 nm emission filter. The rate of product formation was typically determined after a 2-min lag and is expressed as the change in relative fluorescence units/min per 90 μl of sample.
Binding of ANGPTL4 with LPL—All incubations were performed at 4 °C unless otherwise stated. Purified recombinant human LPL was biotinylated by sulfo-NHS-LC-biotin (Pierce) according to the manufacturer's instructions. Wells of a Poly-Sorp 96-well plate (Nunc, Rochester, NY) were incubated with 50 μl of either Coating Buffer (0.1 m sodium carbonate, pH 9.2) or 50 nm N′-hANGPTL4 in Coating Buffer. After incubating overnight, the solution was removed, and the wells were incubated for 1 h with 200 μl of 1% BSA. The wells were then washed three times with 200 μl of washing buffer (phosphate-buffered saline, 0.05% Tween 20). The wells were incubated with 50 μl of 1% BSA containing either no LPL or 4 nm biotinylated LPL supplemented with no mAb, 66 nm anti-ANGPTL4 mAb, or 66 nm anti-KLH mAb. After 1 h, the wells were washed three times with washing buffer. HRP-conjugated streptavidin (Pierce) in phosphate-buffered saline was then added to each well and incubated for 30 min. The solution was then removed, and each well was washed three times as described above. TMB substrate (100 μl) was then added to each well and incubated at room temperature. After 10–30 min, 2 n sulfuric acid (100 μl) was added to each well. Bound LPL was quantitated by measuring the absorbance at 450 nm with a Bio-Tek Synergy HT plate reader (Winooski, VT). Specific N′-hANGPTL4-bound LPL was calculated by subtracting the LPL bound to uncoated wells from LPL bound to N′-hANGPTL4-coated wells.
Binding of ANGPTL3 with LPL—All incubations were performed at 4 °C unless otherwise stated. Wells of a MaxiSorp 96-well plate (Nunc, Rochester, NY) were incubated with 50 μl of either Coating Buffer or 100 nm N′-hANGPTL3 in Coating Buffer. After incubating overnight, the solution was removed, and the wells were incubated for 1 h with 200 μl of 1% BSA. The wells were then washed three times with 200 μl of washing buffer (phosphate-buffered saline, 0.05% Tween 20). The wells were incubated with 50 μl of 1% BSA containing either no LPL or 20 nm FLAG-tagged LPL supplemented with no mAb, 200 nm anti-ANGPTL3 mAb, or 200 nm anti-KLH mAb. After 1 h, 10 μl of 100 nm HRP-conjugated anti-FLAG antibodies (Sigma) were added to each well and incubated for 30 min. Unbound LPL and antibodies were removed from the plate by washing three times with ice-cold washing buffer. TMB substrate (50 μl) was then added to each well and incubated at room temperature. After 10–30 min, 2 n sulfuric acid (50 μl) was added to each well. Bound LPL was quantitated by measuring the absorbance at 450 nm with a Bio-Tek Synergy HT plate reader (Winooski, VT). Specific N′-hANGPTL3-bound LPL was calculated by subtracting the LPL bound to uncoated wells from LPL bound to N′-hANGPTL3-coated wells.
Calculations and Bioinformatic and Statistical Analyses— EC50 values were determined by nonlinear regression analysis with a sigmoidal dose-response (variable slope) equation (GraphPad Prism version 4.03 for Windows, GraphPad Software, San Diego, CA). Data are presented as the mean (±S.E.) unless otherwise stated. Amino acid sequence similarity between the N-terminal domains of ANGPTL4 and ANGPTL3 were assessed by BLAST (14) and Align X analysis (Vector NTI Suite 9 software package, Invitrogen). Comparisons between two groups were analyzed by unpaired Student's t test. Comparisons among multiple groups were analyzed by one-way analysis of variance followed by a post hoc test if statistical significance was less than 0.05.
RESULTS
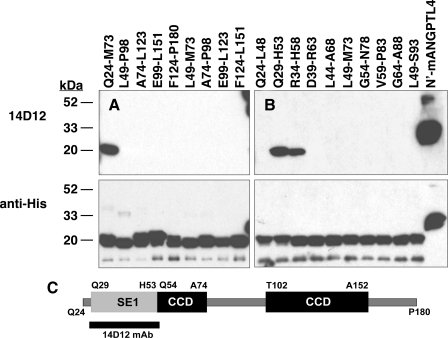
Identification of a Domain in ANGPTL4 Involved in LPL Inhibition—We previously identified a monoclonal antibody, 14D12, that reacts specifically with ANGPTL4 and neutralizes its LPL-inhibiting activity in vitro and in vivo (9). We reasoned that by mapping the epitope in ANGPTL4 that reacts with this antibody, we could identify a similar region within ANGPTL3 and explore the mechanism by which these two proteins inhibit LPL. Toward this goal, we expressed cDNAs encoding a series of overlapping peptides spanning mANGPTL4 amino acids Gln24–Pro180 (Fig. S3A) as His6-tagged SUMO fusion proteins and probed Western blots of these proteins with anti-mANG-PTL4 mAb 14D12. Our results showed that mAb 14D12 only bound to the fusion protein containing mANGPTL4 amino acids Gln24–Met73 (Fig. 1A).
FIGURE 1.
Epitope mapping of mAb 14D12. Western blots of His6-tagged SUMO fusion proteins containing overlapping peptides spanning mouse ANGPTL4 amino acids Gln24–Leu151 (A) or Gln24–Ser93 (B) were immunostained with either mAb 14D12 (top panel) or anti-His6 polyclonal IgGs (bottom panel) as described under “Experimental Procedures.” His6-tagged mouse ANGPTL4 amino acids Gly25–Leu185 (N′-mANGPTL4) was also blotted to serve as a positive control for immunostaining. The epitope for mAb 14D12 (SE1) is contained mainly within mouse ANGPTL4 amino acids Gln29–His53. A schematic representation (C) of N-terminal ANGPTL4 amino acids Gln24–Pro180 shows the location of SE1 relative to the coiled-coil domains (CCDs) of mouse ANGPTL4.
We further refined the epitope map for mAb 14D12 by expressing another series of 10 cDNAs encoding overlapping mANGPTL4 peptides fused with His-tagged SUMO protein and probed a Western blot of these proteins with mAb 14D12 (Fig. S3B). This series spanned mANGPTL4 amino acids Gln24–Ser93, enabling us to map the epitope within mANG-PTL4 amino acids Gln24–Met73 (Fig. 1A) with a resolution of about 5 amino acids. We found that mAb 14D12 bound strongly only with mANGPTL4 amino acids Gln29–His53 and somewhat less strongly with mANGPTL4 amino acids Arg34–His58 (Fig. 1B). Moreover, we found that mAb 14D12 did not bind with the flanking overlapping peptides (Fig. S3B), indicating that mANGPTL4 amino acids Leu49 through His53 and Arg34 through Asp39 are involved in binding with mAb 14D12. In addition, maximal antibody binding appears to require the 5 mANGPTL4 amino acids from Gln29 to Pro33, because the apparent binding affinity of mAb 14D12 was slightly lower for mANGPTL4 amino acids Arg34–His58 than for mANGPTL4 amino acids Gln29–His53. These results indicate that the epitope for mAb 14D12 resides within mANGPTL4 amino acids Gln29 through His53 (Fig. 1C). We define this region as ANGPTL4 SE1.
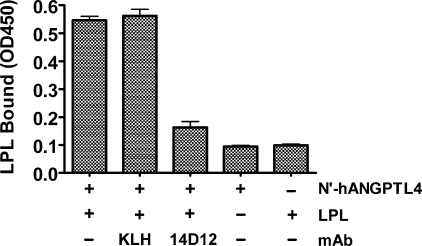
ANGPTL4 has been shown to interact directly with LPL using surface plasmon resonance (10); however, the domains involved in this interaction have not been identified. To test whether mAb 14D12 is able to inhibit binding of ANGPTL4 with LPL, we developed an ELISA-based ANGPTL4-LPL binding assay. For this experiment, biotinylated LPL with either mAbs KLH or 14D12 were added to N′-hANGPTL4-coated ELISA plates. After a 1-h incubation at 4 °C, the ELISA plates were washed and then probed with HRP-conjugated streptavidin to quantify the presence of biotinylated LPL bound to N′-hANGPTL4. Binding of LPL to N-hANGPTL4 was ∼4-fold lower in the presence of mAb 14D12 than in the presence of control mAb KLH (Fig. 2).
FIGURE 2.
The SE1 domain mediates binding of ANGPTL4 with LPL. Biotinylated recombinant LPL (4 nm) with or without mAb 14D12 or mAb KLH (66 nm) was added to N′-hANGPTL4 protein-coated ELISA plates. After a 1-h incubation at 4 °C, ELISA plates were washed and then probed with HRP-conjugated streptavidin to quantify the presence of biotinylated LPL bound to N′-hANGPTL4. Additional negative control wells included those not coated with N′-hANGPTL4 as well as N′-hANGPTL4-coated wells from which biotinylated recombinant LPL was omitted. Values for LPL bound are the mean ± S.E. of three determinations. The experiment shown in this figure is representative of at least three experiments.
Several other mAbs were identified that bind to the N terminus of ANGPTL4 in regions C-terminal to the SE1 domain (Table 1). With one exception, none of these mAbs reduced serum TGs in vivo more than 30% relative to the control group (data not shown). The lone exception, mAb 90B4, (a) showed a binding pattern to N′-hANGPTL4 peptides that was different from all other mAbs (Table 1); (b) was less effective than mAb 14D12 at lowering serum TG levels in vivo despite comparable serum mAb levels in vivo and higher affinity for N′-hANGPTL4 in vitro (Fig. S4); and (c) was unable to inhibit the binding of LPL to N-hANGPTL4 in vitro (Fig. S4). These results suggest that the SE1 region of ANGPTL4 is uniquely involved with binding to, and maximal inhibition of, LPL.
TABLE 1.
Epitope mapping of mAbs reactive with mouse ANGPTL4 A series of mAbs were tested for their reactivity with a series of overlapping peptides corresponding to mouse ANGPTL4 amino acids Gln24–Pro180. A “–” sign indicates no immunoreactivity and “+” sign indicates positive immunoreactivity. The column heading “ANGPTL4” corresponds to N′-mANGPTL4 (Gln25–Leu185; accession number Q9Z1P8).
|
mAb
|
ANGPTL4 peptide
|
|||||
|---|---|---|---|---|---|---|
| Gln24–Met73 | Leu49–Pro98 | Ala74–Leu123 | Glu99–Leu151 | Phe124–Pro180 | ANGPTL4 | |
| 7H8 | – | – | – | + | – | + |
| 8D8 | – | + | – | – | – | + |
| 9C10 | – | – | – | + | – | + |
| 11C11 | – | – | – | – | – | + |
| 14D12 | + | – | – | – | – | + |
| 18G3 | – | – | – | + | – | + |
| 20C9 | – | – | – | – | – | + |
| 90B4 | – | + | – | + | – | + |
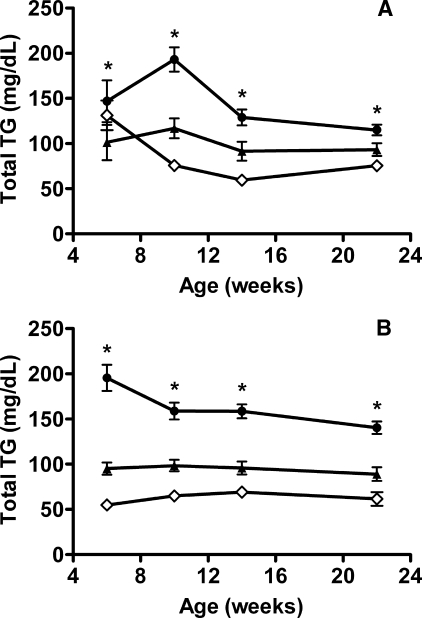
The ANGPTL4 E40K variant is associated with lower TG levels in humans (15), and Glu40 resides near the center of the SE1 domain in humans and mice. To examine whether the E40K variant is associated with lower TG levels in mice as well as humans, we generated mice expressing only this variant. Intercrosses of mice heterozygous for the Angptl4 E40K allele (Angptl4E/K) produced healthy Angptl4K/K progeny. Comparable levels of the ANGPTL4 transcript were present in the liver and kidney of wild-type (Angptl4E/E) and Angptl4K/K littermates, and sequence analysis revealed that the E40K variant was the only ANGPTL4 transcript present in Angptl4K/K mice (data not shown). When serum TG levels were examined in Angptl4K/K, Angptl4E/K, and Angptl4E/E mice maintained on a high fat diet, we found these levels to be notably lower in Angptl4E/K and Angptl4K/K mice than in Angptl4E/E littermates. The effect of one or two copies of the Angplt4 E40K allele on serum TG levels was similar to the effect observed in Angptl4 heterozygous null (+/-) and Angptl4-/- mice (Fig. 3).
FIGURE 3.
Activity of ANGPTL4 E40K variant in vivo. A, TG levels in Angptl4E/E (closed circles; n = 16), Angptl4E/K (closed triangles; n = 8), and Angptl4K/K (open diamonds; n = 12) mice on chow diet at 6 weeks of age and after 4, 8, and 16 weeks on a high fat diet. *, Angptl4E/E mice versus Angptl4E/K or Angptl4K/K mice (p < 0.02). B, TG levels in Angptl4+/+ (closed circles; n = 17), Angptl4+/- (closed triangles; n = 8), and Angptl4-/- (open diamonds; n = 9) mice on chow diet at 6 weeks of age and after 4, 8, and 16 weeks on a high fat diet. *, Angptl4+/+ mice versus Angptl4+/- or Angptl4-/- mice (p < 0.01). Total TG values are the mean ± S.E.
ANGPTL3 Contains an SE1 Domain—Our epitope mapping study indicates that the SE1 region of ANGPTL4 likely defines a region necessary for inhibiting LPL activity. To determine whether any other angiopoietin or angiopoietin-like family members contain this region, we searched the NCBI non-redundant protein data base with mANGPTL4 amino acids Met1–Leu185, lacking the fibrinogen-like conserved domain, using the BLASTP program (14). The only angiopoietin or angiopoietin-like protein other than ANGPTL4 with a significant alignment score (E value <10) was ANGPTL3. Similar results were observed when the NCBI non-redundant data base was searched with hANGPTL4 amino acids Met1–Leu181. Moreover, reciprocal results were obtained with mouse or human ANGPTL3 as the query sequence. Alignment of the N-terminal portions of mouse and human ANGPTL4 and ANGPTL3 protein sequences are shown in Fig. 4. Whereas the N-terminal regions of ANGPTL3 and ANGPTL4 are less than 20% similar overall, the SE1 peptide of ANGPTL4 and its corresponding sequence in ANGPTL3 (Glu32–His55) are 56% identical and 68% similar.
FIGURE 4.
Alignment of N-terminal regions of mouse and human ANGPTL3 and ANGPTL4. The predicted sequences of N-terminal mouse (Mm) or human (Hs) ANGPTL3 and ANGPTL4 were aligned using the Align X program of the Vector NTI Suite 9 package (Invitrogen). Identical residues are shown in red. The boxed amino acids contain the SE1 region of ANGPTL4 and the putative SE1 region of ANGPTL3.
Because ANGPTL3 appears to contain an SE1 segment and because the overall domain organization of ANGPTL4 and ANGPTL3 are similar, we hypothesized that the SE1 region in ANGPTL3 likely binds with LPL and inhibits its activity by a mechanism similar to that of ANGPTL4 (10). Toward testing this hypothesis, we produced a monoclonal antibody, 5.50.3, which was reactive with the SE1 region within mouse and human ANGPTL3 and displayed high affinity for the SE1 peptide antigen (Fig. S5).
Inhibition of LPL by ANGPTL3 Requires the SE1 Domain— To determine whether the SE1 region of ANGPTL3 is involved in inhibition of LPL, we preincubated 5 nm recombinant human LPL with 100 nm hANGPTL3 in the presence of varying concentrations of either mAb 5.50.3 or control mAb KLH and then assayed the mixture for LPL activity. In the absence of any mAb, hANGPTL3 inhibited LPL by ∼50%. Control mAb KLH at concentrations as high as 400 nm had no effect on the LPL-inhibiting activity of ANGPTL3. However, mAb 5.50.3 prevented inhibition of LPL by hANGPTL3 with an EC50 of 22 nm and an EC90 of 120 nm (Fig. 5A).
FIGURE 5.
Neutralization of ANGPTL3 activity by mAb 5.50.3 in vitro. A, mAb KLH (open circles) or mAb 5.50.3 (closed circles) at concentrations ranging between 0 and 1000 nm were incubated with 100 nm hANGPTL3 and ∼5 nm LPL for 60 min at room temperature and then assayed for LPL activity with lipase substrate DGGR. B, mAbs at 400 nm were incubated with 200 nm hANGPTL3 and LPL for 60 min at room temperature and then assayed for LPL activity with lipase substrate DGGR. The concentration of hANGPTL3 (52.7 kDa) and LPL (51.4 kDa) is that of the monomeric recombinant protein. The concentration of mAb (150 kDa) is that of the bivalent mAb. LPL activity values are the mean ± S.E. of three determinations. The experiments shown in panels A and B are representative of at least three experiments.
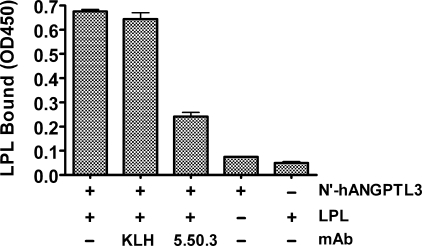
We then compared mAb 5.50.3 with mAbs 6C6 and 8E2, two mAbs that bind N′-hANGPTL3 epitopes C-terminal to and independent of the SE1 region (Table 2), for their ability to neutralize ANGPTL3-mediated inhibition of LPL. In this experiment, 5 nm LPL was preincubated with 200 nm hANG-PTL3 in the presence or absence of 400 nm mAb 5.50.3, mAb 6C6, mAb 8E2, or control mAb KLH and then assayed for LPL activity. Whereas 400 nm mAb 5.50.3 completely neutralized inhibition of LPL by 200 nm hANGPTL3, neither mAb 6C6, mAb 8E2, nor control mAb KLH at 400 nm had any effect on neutralizing the LPL-inhibiting activity of hANGPTL3 (Fig. 5B). These results show that mAb 5.50.3 is capable of neutralizing the LPL-inhibiting activity of ANGPTL3 in vitro and suggest that the SE1 region is necessary for inhibition of LPL by ANGPTL3.
TABLE 2.
Epitope mapping of mAbs reactive with mouse ANGPTL3 A series of mAbs were tested for their reactivity with overlapping peptides corresponding to mouse ANGPTL3 amino acids Ser17–Met116. A “–” sign indicates no immunoreactivity and “+” sign indicates positive immunoreactivity. The column heading “ANGPTL3” corresponds to N′-mANGPTL3 (Ser17–Arg221; accession NP_038941.1).
|
mAb
|
ANGPTL3 peptide
|
|||
|---|---|---|---|---|
| Ser17–Gly66 | Asp42–Glu91 | Gln67–Met116 | ANGPTL3 | |
| 3E11 | – | + | + | + |
| 21H9 | – | + | + | + |
| 6C6 | – | + | – | + |
| 34B2 | – | + | + | + |
| 8E2 | – | + | + | + |
| 30H2 | – | – | + | + |
| 1.251.1 | – | + | – | + |
| 1.173.1 | – | + | – | + |
| 1.135.1 | – | + | – | + |
| 1.424.1 | – | + | – | + |
| 1.431.1 | – | + | – | + |
| 5.50.3 | + | – | – | + |
| 1.431.1 | – | + | – | + |
The SE1 Domain Is Involved in Binding of LPL with ANGPTL3—To investigate whether ANGPTL3, like ANGPTL4, directly binds with LPL, we developed an ELISA-based ANGPTL3-LPL binding assay. In this experiment, FLAG-tagged LPL with no mAb, mAb KLH, or mAb 5.50.3 were added to N′-hANGPTL3-coated ELISA plates. After a 1-h incubation at 4 °C, the ELISA plates were washed and then probed with HRP-conjugated anti-FLAG antibody to quantify the presence of FLAG-tagged LPL binding to N′-hANG-PTL3. LPL binding was readily detected in N′-hANGPTL3-coated wells but not in uncoated wells (Fig. 6). Addition of mAb 5.50.3 decreased binding of LPL to N′-hANGPTL3 by more than 60%, whereas addition of mAb KLH had no effect on binding. These results indicate that the SE1 region of ANGPTL3, like that of ANGPTL4, is involved with the binding and inhibition of LPL.
FIGURE 6.
The SE1 domain mediates binding of ANGPTL3 with LPL. Recombinant FLAG-tagged LPL with or without mAb 5.50.3 or mAb KLH was added to N′-hANGPTL3 protein-coated ELISA plates. After a 1-h incubation at 4 °C, ELISA plates were washed and then probed with HRP-conjugated anti-FLAG antibody to quantify binding of FLAG-tagged LPL with N′-hANGPTL3. Additional negative control wells included those not coated with N′-hANGPTL3 as well as N′-hANGPTL3 coated wells from which recombinant FLAG-tagged LPL were omitted. LPL bound values are the mean ± S.E. of three determinations. The experiment shown in this figure is representative of at least three experiments.
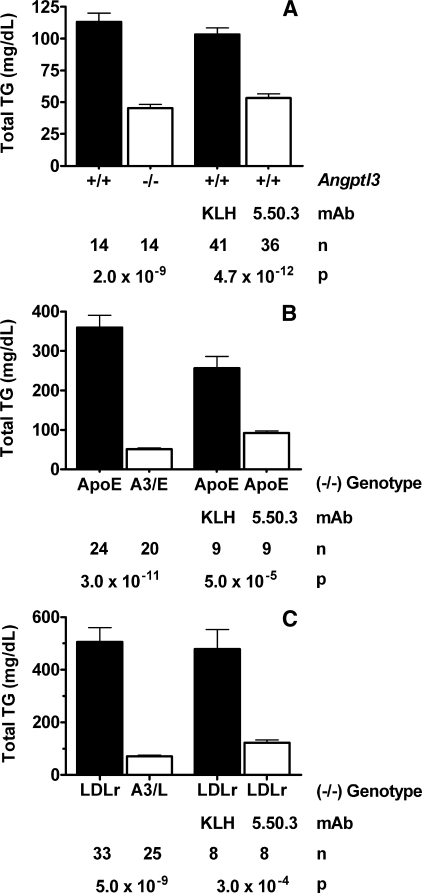
Preventing Inhibition of LPL by ANGPTL3 in Vivo Lowers Serum Lipids in Mouse Models of Hyperlipidemia—The results of our studies thus far show that the SE1 region of ANGPTL3 is involved in inhibiting LPL activity. We reasoned that antibodies that react with the SE1 region and thereby block inhibition of LPL activity by ANGPTL3 might also increase LPL activity and decrease circulating TG levels in vivo. To test this hypothesis, we first generated Angptl3-/- mice to serve as controls that would demonstrate, in vivo, the effect of complete ANGPTL3 absence on serum lipid levels. Angptl3+/- intercrosses produced healthy progeny, and Mendelian ratios at weaning indicated normal viability of Angptl3-/- mice. We next treated C57BL/6J mice with either control mAb KLH or anti-ANGPTL3 SE1 mAb 5.50.3 and then compared the TG levels of the mAb-treated mice with those of Angptl3-/- mice and their wild-type (+/+) littermates. Serum TG levels were 60% lower in Angptl3-/- mice than in Angptl3+/+ littermates and 48% lower in mice treated with mAb 5.50.3 than in mice treated with control mAb KLH (Fig. 7A). These results suggest that mAb 5.50.3 efficaciously prevents inhibition of LPL by ANGPTL3 in vivo.
FIGURE 7.
Neutralization of ANGPTL3 activity by mAb 5.50.3 in vivo. A, comparison of serum TG levels between Angptl3-/- mice and Angptl3+/+ mice or between C57BL/6J mice treated with mAb 5.50.3 and C57BL/6J mice treated with control mAb KLH. Mice were injected intraperitoneally with 30 mg/kg mAb 5.50.3 or mAb KLH on day 0. Blood was then drawn from fed mice for serum TG level measurements on day 4. B, comparison of serum TG levels between ApoE-/- mice and Angptl3-/-/ApoE-/- (A3/E) mice or between ApoE-/- mice treated with mAb 5.50.3 and ApoE-/- mice treated with control mAb KLH. Mice were injected intraperitoneally with 30 mg/kg mAb 5.50.3 or mAb KLH on days 0 and 4. Blood was then drawn from fed mice for serum TG measurements on day 8. C, comparison of serum TG levels between LDLr-/- mice and Angptl3-/-/LDLr-/- (A3/L) mice or between LDLr-/- mice treated with mAb 5.50.3 and LDLr-/- mice treated with control mAb KLH. Mice were injected intraperitoneally with 30 mg/kg mAb 5.50.3 or mAb KLH on days 0 and 4. Blood was then drawn from fed mice for serum TG measurements on day 8. Total TG values are the mean ± S.E. The sample number (n) and p values are shown in the graphs below each comparison.
We then asked whether preventing inhibition of LPL activity by ANGPTL3 is a viable mechanism for lowering serum lipid levels in mouse models of hyperlipidemia. To address this question, we treated ApoE-/- or LDLr-/- mice with either control mAb KLH or mAb 5.50.3 and determined the effect of treatment on serum TG and cholesterol levels. The TG and cholesterol levels of these mAb-treated mice were also compared, respectively, to those of Angptl3-/-/ApoE-/- mice or Angptl3-/-/LDLr-/- mice. Serum TG levels were 86% lower in Angptl3-/-/ApoE-/- mice than in ApoE-/- littermates and 64% lower in ApoE-/- mice treated with mAb 5.50.3 than in ApoE-/- mice treated with mAb KLH (Fig. 7B). Similarly, serum TG levels were 86% lower in Angptl3-/-/LDLr-/- mice than in LDLr-/- littermates and 75% lower in LDLr-/- mice treated with mAb 5.50.3 than in LDLr-/- mice treated with mAb KLH (Fig. 7C). Serum cholesterol levels were 49% lower in Angptl3-/-/ApoE-/- mice than in ApoE-/- littermates and 27% lower in ApoE-/- mice treated with mAb 5.50.3 than in ApoE-/- mice treated with mAb KLH (data not shown). Similarly, serum cholesterol levels were 60% lower in Angptl3-/-/LDLr-/- mice than in LDLr-/- littermates and 41% lower in LDLr-/- mice treated with mAb 5.50.3 than in LDLr-/- mice treated with mAb KLH (data not shown). In Angptl3-/-/LDLr-/- mice and in LDLr-/- mice treated with mAb 5.50.3, the lower cholesterol levels were due to less circulating VLDL/IDL/LDL cholesterol (data not shown). Overall, these results suggest that monoclonal antibodies that prevent ANGPTL3-mediated inhibition of LPL activity can dramatically lower plasma TGs and also lower cholesterol under certain conditions that cause hyperlipidemia.
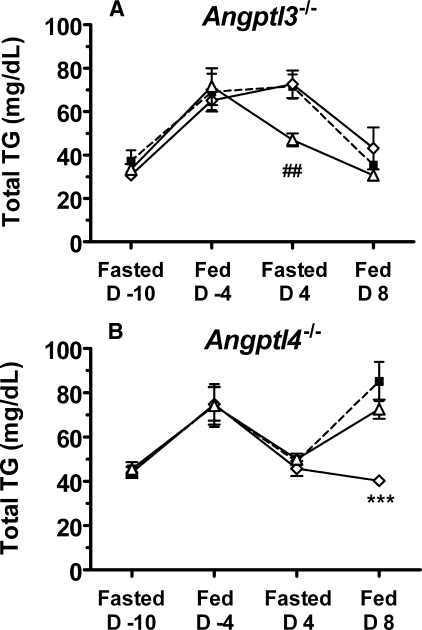
The SE1 regions of ANGPTL3 and ANGPTL4 are quite similar, raising the possibility that the SE1 mAbs might recognize both ANGPTLs in vivo. To determine the specificity of mAbs 14D12 and 5.50.3 in vivo, we treated Angptl3-/- mice or Angptl4-/- mice with either mAb 14D12 or 5.50.3 and measured serum TG levels under fed and fasting conditions (Fig. 8). Whereas treatment of Angptl3-/- mice with anti-ANGPTL4-SE1 mAb 14D12 lowered fasting serum TG levels, treatment of Angptl3-/- mice with mAb 5.50.3 had no effect on serum TG levels (Fig. 8A). Similarly, treatment of Angptl4-/- mice with anti-ANGPTL3-SE1 mAb 5.50.3 lowered fed serum TG levels, whereas treatment of Angptl4-/- mice with mAb 14D12 had no significant effect on serum TG levels (Fig. 8B). These results suggest that mAb 14D12 is specifically reactive with ANGPTL4 and mAb 5.50.3 is specifically reactive with ANGPTL3 in vivo.
FIGURE 8.
Specificity of 5.50.3 and 14D12 mAbs in vivo. A, TG levels in serum from 12-week-old Angptl3-/- mice were determined after fasting (16 h) on day -10 and in fed state on day -4. The mice were then treated with mAb 14D12 (open triangles), 5.50.3 (open diamonds), or KLH (closed squares) (30 mg/kg) on day 0 and again on day 4. Serum TG levels were determined in fasted mice on day 4 before the second antibody injection and again in fed mice on day 8. Values are the mean (±S.E.) of 10 mice per treatment group. ##, mAb KLH or mAb 5.50.3 versus mAb 14D12, p < 0.01. B, TG levels in serum from 24-week-old Angptl4-/- mice were determined after fasting on day -10 and in fed state on day -4. The mice were then treated with mAb 14D12 (open triangles), 5.50.3 (open diamonds), or KLH (closed squares) (30 mg/kg) on day 0 and again on day 4. Serum TG levels were determined in fasted mice on day 4 before the second antibody injection and again in fed mice on day 8. Total TG values are the mean ± S.E. of 8 mice per treatment group. ***, mAb KLH or mAb 14D12 versus mAb 5.50.3, p < 0.001.
Effect of Non-SE1 Binding ANGPTL3 mAbs on Serum TG Levels in Vivo—Several mAbs were identified, including mAb 6C6 and mAb 8E2, that bind to the N terminus of ANGPTL3 in regions C-terminal to and independent of the SE1 domain (Table 2). Despite their ability to react with ANGPTL3, the serum TG lowering efficacy of these mAbs was more than 2-fold less than that for mAb 5.50.3 (data not shown).
DISCUSSION
ANGPTL3 and ANGPTL4 regulate lipid metabolism by inhibiting the activity of LPL, which in turn elevates plasma lipid levels (5, 15–17). Previously, we showed that anti-ANGPTL4 mAb 14D12 neutralized the inhibition of LPL by ANGPTL4 and that mice treated with this mAb recapitulated the phenotype of Angptl4-/- mice (9). These in vitro and in vivo experiments suggested that mAb 14D12 recognizes a region in ANGPTL4 that is important for its function as an inhibitor of LPL activity. By identifying the ANGPTL4 epitopes recognized by mAb 14D12, we set out to gain insight into how ANGPTL4 inhibits LPL activity and to identify similar domains within other angiopoietin or angiopoietin-like family members that might inhibit LPL. We showed that the epitope recognized by mAb 14D12 is contained within mANGPTL4 amino acids Gln29–His53, which we designate as SE1. This region is located between the signal peptide and the first of two predicted coiled-coil domains (Gly54–Ala74 and Thr102–Ala152). The importance of the SE1 region in the function of ANGPTL4 is supported not only by the neutralizing activity of mAb 14D12 but also by the discovery of a loss-of-function mutation, E40K, which is located near the center of the SE1 region. The ANGPTL4 E40K variant is associated with lower serum TG levels in humans (15), and the LPL inhibitory potency of the human E40K variant is markedly decreased in vitro (18). We showed here that mice with the E40K variant also have lower serum TG levels in vivo, consistent with the human data. Thus, the SE1 region of ANGPTL4 likely defines a domain involved in inhibition of LPL.
Among all angiopoietin and angiopoietin-like family members other than ANGPTL4, we found that only ANGPTL3 contains an SE1-like region. To determine whether the SE1 region of ANGPTL3 also participated in LPL inhibition, we produced mAb 5.50.3, which reacts specifically with this SE1 region. This anti-ANGPTL3 mAb potently neutralized the LPL-inhibitory activity of ANGPTL3 in vitro and inhibited the binding of ANGPTL3 with LPL, just as anti-ANGPTL4 mAb 14D12 inhibited the binding of ANGPTL4 to LPL. mAb 5.50.3 is also an effective inhibitor of ANGPTL3 in vivo. Treatment of C57BL/6J mice with mAb 5.50.3 was capable of lowering serum TGs nearly to levels found in Angptl3-/- mice. Moreover, mAb 5.50.3 treatment also increases LPL activity in post-heparin plasma.7 Thus, these in vitro and in vivo results strongly support our conclusion that the SE1 region, present in both ANGPTL4 and ANGPTL3, defines a functional domain involved in inhibition of LPL activity.
Recently, Romeo and colleagues (19) reported on mutations in ANGPTL3 and ANGPTL4 outside of the SE1 domain that also appear to decrease the LPL-inhibitory activity of these proteins. We identified several mAbs that bind to the N terminus of ANGPTL3 or ANGPTL4 in regions that are C-terminal to the SE1 domain and that include some of these reported mutations, yet none of these mAbs were able to reduce serum TG levels comparably to SE1 binders. In addition, when tested, none of these mAbs were able to inhibit the binding of LPL to hANGPTL3 or hANGPTL4, in contrast to the SE1 binders. Together, these data suggest that the conserved SE1 domain in ANGPTL3 and ANGPTL4 plays a necessary and critical role in inhibiting LPL activity. However, our results do not preclude the participation of N-terminal amino acids other than those in SE1 in inhibition of LPL.
Serum TG levels of fed Angptl4-/- mice were significantly lowered by anti-ANGPTL3 mAb 5.50.3, and serum TG levels of fasted Angptl3-/- mice were significantly lowered by anti-ANGPTL4 mAb 14D12. Neither mAb had any effect in mice lacking the ANGPTL targeted by that mAb. These data indicate that mAb 5.50.3 specifically binds ANGPTL3 and mAb 14D12 specifically binds ANGPTL4. The data also underscore the increased activity of serum ANGPTL3 in the fed state and increased activity of ANGPTL4 in the fasted state, consistent with previous observations in rodents (5, 9). Finally, these data suggest that a significantly greater decrease in serum TG can be achieved with maximal inhibition of both ANGPTLs together compared with maximal inhibition of either ANGPTL individually.
Previously, we reported that both Angptl4-/- suckling and adult mice fed a high-fat diet displayed reduced viability associated with lipogranulomatous lesions of the intestines, draining lymphatics, and mesenteric lymph nodes (9). Treating high-fat diet-fed C57BL/6J, ApoE-/-, or LDLr-/- mice with anti-ANGPTL4 mAb 14D12 recapitulated the histopathologic phenotype found in Angptl4-/- mice. Although the underlying mechanism for this phenotype is not clearly understood, the association of high-fat diet intake with these lesions in mice suggests that ANGPTL4 may be required for down-regulating intestinal LPL activity to prevent premature release of fatty acids from intestinal lipids before or during lymphatic transport. In the present study we did not observe reduced viability or mesenteric histopathology in Angptl3-/- mice or in high-fat diet-fed C57BL/6J, ApoE-/-, or LDLr-/- mice treated with mAb 5.50.3, the neutralizing anti-ANGPTL3 mAb (data not shown). These results indicate that ANGPTL3, unlike ANGPTL4, is not involved in LPL-mediated lipid metabolism in mouse intestinal lymphoid tissues.
Recent studies have shown that genetic variants of ANGPTL3 correlate with plasma lipid concentrations and risk of coronary artery disease and atherosclerosis in humans (19–22). Moreover, two large-scale prospective studies have shown that higher non-fasting TG levels were associated with an increased incidence of cardiovascular events independent of established major risk factors (total or LDL cholesterol levels) (23, 24). In contrast, the association of fasting TG levels with increased cardiovascular risk was weakened after accounting for the effects of established major risk factors. The mechanisms for the association between non-fasting TG levels and cardiovascular disease remain unclear; however, it seems likely that increased postprandial TG levels, reflecting a higher peak or delay in clearance of TG-rich particles, can lead to an accumulation of atherogenic lipid particles (25). These observations suggest that new TG-lowering medications may have value in the treatment of cardiovascular diseases. In this study, we demonstrated that mAb 5.50.3 is capable of dramatically improving the lipid profile of ApoE-/- and LDLr-/- mice, two models of hyperlipidemia. These results suggest that ANGPTL3-neutralizing mAbs can treat hyperlipidemia by preventing inactivation of LPL, constituting a novel mechanism for treatment of human cardiovascular disease.
Supplementary Material
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Tables S1–S2, and additional data.
Footnotes
The abbreviations used are: LPL, lipoprotein lipase; ANGPTL3, angiopoietin-like 3; ANGPTL4, angiopoietin-like 4; hANGPTL3, recombinant human angiopoietin-like 3 protein (full-length); N′-hANGPTL3, recombinant human angiopoietin-like 3 amino acids Asp20–Pro243; N′-mANGPTL4, mouse ANGPTL4 amino acids Gly25–Leu185; KLH, keyhole limpet hemocyanin; mAb KLH, isotype-matched control mAb against KLH; ORF, open reading frame; LDLr, low density lipoprotein receptor; ApoE, apolipoprotein E; -/-, homozygous null; +/-, heterozygous null; +/+, wild-type littermate; Angptl4K/K, homozygous for the Angptl4 E40K variant allele; Angptl4E/K, heterozygous for the Angptl4 E40K variant allele; Angptl4E/E, Angptl4 wild-type littermate from parents heterozygous for Angptl4 E40K variant allele; TG, triglyceride; mAb, monoclonal antibody; SE1, specific epitope 1; BSA, bovine serum albumin; Ni-NTA, nickel-nitrilotriacetic acid; ELISA, enzyme-linked immunosorbent assay; HRP, horseradish peroxidase; DGGR, 1,2-O-dilauryl-rac-glycero-3-glutaric acid-(6′-methylresorufin) ester.
The Angptl3 and Angptl4 knock-out mice were produced in collaboration between Genentech, Inc., and Lexicon Pharmaceuticals, Inc., to analyze the function of about 500 secreted and transmembrane proteins.
E-C. Lee, U. Desai, J. Gay, N. Wilganowski, B. P. Zambrowicz, G. Landes, D. R. Powell, and W. K. Sonnenburg, unpublished observation.
References
- 1.Goldberg, I. J., and Merkel, M. (2001) Front. Biosci. 6 D388-D405 [DOI] [PubMed] [Google Scholar]
- 2.Merkel, M., Eckel, R. H., and Goldberg, I. J. (2002) J. Lipid Res. 43 1997-2006 [DOI] [PubMed] [Google Scholar]
- 3.Preiss-Landl, K., Zimmermann, R., Hammerle, G., and Zechner, R. (2002) Curr. Opin. Lipidol. 13 471-481 [DOI] [PubMed] [Google Scholar]
- 4.Koishi, R., Ando, Y., Ono, M., Shimamura, M., Yasumo, H., Fujiwara, T., Horikoshi, H., and Furukawa, H. (2002) Nat. Genet. 30 151-157 [DOI] [PubMed] [Google Scholar]
- 5.Koster, A., Chao, Y. B., Mosior, M., Ford, A., Gonzalez-DeWhitt, P. A., Hale, J. E., Li, D., Qiu, Y., Fraser, C. C., Yang, D. D., Heuer, J. G., Jaskunas, S. R., and Eacho, P. (2005) Endocrinology 146 4943-4950 [DOI] [PubMed] [Google Scholar]
- 6.Ono, M., Shimizugawa, T., Shimamura, M., Yoshida, K., Noji-Sakikawa, C., Ando, Y., Koishi, R., and Furukawa, H. (2003) J. Biol. Chem. 278 41804-41809 [DOI] [PubMed] [Google Scholar]
- 7.Ge, H., Cha, J. Y., Gopal, H., Harp, C., Yu, X., Repa, J. J., and Li, C. (2005) J. Lipid Res. 46 1484-1490 [DOI] [PubMed] [Google Scholar]
- 8.Ge, H., Yang, G., Yu, X., Pourbahrami, T., and Li, C. (2004) J. Lipid Res. 45 2071-2079 [DOI] [PubMed] [Google Scholar]
- 9.Desai, U., Lee, E. C., Chung, K., Gao, C., Gay, J., Key, B., Hansen, G., Machajewski, D., Platt, K. A., Sands, A. T., Schneider, M., Van Sligtenhorst, I., Suwanichkul, A., Vogel, P., Wilganowski, N., Wingert, J., Zambrowicz, B. P., Landes, G., and Powell, D. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11766-11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukonina, V., Lookene, A., Olivecrona, T., and Olivecrona, G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17450-17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wattler, S., Kelly, M., and Nehls, M. (1999) BioTechniques 26 1150-1156, 1158, 1160 [DOI] [PubMed] [Google Scholar]
- 12.Bergo, M., Olivecrona, G., and Olivecrona, T. (1996) Biochem. J. 313 893-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panteghini, M., Bonora, R., and Pagani, F. (2001) Ann. Clin. Biochem. 38 365-370 [DOI] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romeo, S., Pennacchio, L. A., Fu, Y., Boerwinkle, E., Tybjaerg-Hansen, A., Hobbs, H. H., and Cohen, J. C. (2007) Nat. Genet. 39 513-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizugawa, T., Ono, M., Shimamura, M., Yoshida, K., Ando, Y., Koishi, R., Ueda, K., Inaba, T., Minekura, H., Kohama, T., and Furukawa, H. (2002) J. Biol. Chem. 277 33742-33748 [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, K., Shimizugawa, T., Ono, M., and Furukawa, H. (2002) J. Lipid Res. 43 1770-1772 [DOI] [PubMed] [Google Scholar]
- 18.Shan, L., Yu, X. C., Liu, Z., Hu, Y., Sturgis, L. T., Miranda, M. L., and Liu, Q. (2008) J. Biol. Chem. 284 1419-1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romeo, S., Yin, W., Kozlitina, J., Pennacchio, L. A., Boerwinkle, E., Hobbs, H. H., and Cohen, J. C. (2008) J. Clin. Investig. 119 70-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer, C. J., Sanna, S., Jackson, A. U., Scuteri, A., Bonnycastle, L. L., Clarke, R., Heath, S. C., Timpson, N. J., Najjar, S. S., Stringham, H. M., Strait, J., Duren, W. L., Maschio, A., Busonero, F., Mulas, A., Albai, G., Swift, A. J., Morken, M. A., Narisu, N., Bennett, D., Parish, S., Shen, H., Galan, P., Meneton, P., Hercberg, S., Zelenika, D., Chen, W. M., Li, Y., Scott, L. J., Scheet, P. A., Sundvall, J., Watanabe, R. M., Nagaraja, R., Ebrahim, S., Lawlor, D. A., Ben-Shlomo, Y., Davey-Smith, G., Shuldiner, A. R., Collins, R., Bergman, R. N., Uda, M., Tuomilehto, J., Cao, A., Collins, F. S., Lakatta, E., Lathrop, G. M., Boehnke, M., Schlessinger, D., Mohlke, K. L., and Abecasis, G. R. (2008) Nat. Genet. 40 161-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathiresan, S., Melander, O., Guiducci, C., Surti, A., Burtt, N. P., Rieder, M. J., Cooper, G. M., Roos, C., Voight, B. F., Havulinna, A. S., Wahlstrand, B., Hedner, T., Corella, D., Tai, E. S., Ordovas, J. M., Berglund, G., Vartiainen, E., Jousilahti, P., Hedblad, B., Taskinen, M. R., Newton-Cheh, C., Salomaa, V., Peltonen, L., Groop, L., Altshuler, D. M., and Orho-Melander, M. (2008) Nat. Genet. 40 189-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatsuda, S., Shoji, T., Shinohara, K., Kimoto, E., Mori, K., Fukumoto, S., Koyama, H., Emoto, M., and Nishizawa, Y. (2007) J. Vasc. Res. 44 61-66 [DOI] [PubMed] [Google Scholar]
- 23.Nordestgaard, B. G., Benn, M., Schnohr, P., and Tybjaerg-Hansen, A. (2007) J. Am. Med. Assoc. 298 299-308 [DOI] [PubMed] [Google Scholar]
- 24.Bansal, S., Buring, J. E., Rifai, N., Mora, S., Sacks, F. M., and Ridker, P. M. (2007) J. Am. Med. Assoc. 298 309-316 [DOI] [PubMed] [Google Scholar]
- 25.Zilversmit, D. B. (1995) Clin. Chem. 41 153-158 [PubMed] [Google Scholar]
- 26.Institute of Laboratory Animal Research, Commission on Life Sciences, National Research Council (1996) Guide for the Care and Use of Laboratory Animals, The National Academic Press, Washington, DC
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.