Abstract
The localization in space and in time of proteins within the cytoplasm of eukaryotic cells is a central question of the cellular compartmentalization of metabolic pathways. The assembly of proteins within stable or transient complexes plays an essential role in this process. Here, we examined the subcellular localization of the multi-aminoacyl-tRNA synthetase complex in human cells. The sequestration of its components within the cytoplasm rests on the presence of the eukaryotic-specific polypeptide extensions that characterize the human enzymes, as compared with their prokaryotic counterparts. The cellular mobility of several synthetases, assessed by measuring fluorescence recovery after photobleaching, suggested that they are not freely diffusible within the cytoplasm. Several of these enzymes, isolated by tandem affinity purification, were copurified with ribosomal proteins and actin. The capacity of aminoacyl-tRNA synthetases to interact with polyribosomes and with the actin cytoskeleton impacts their subcellular localization and mobility. Our observations have conceptual implications for understanding how translation machinery is organized in vivo.
A wealth of data have suggested that the cytoplasm of human cells is highly organized. This is especially the case as far as translation machinery is concerned. Translation of genetic information involves several supramolecular assemblies, including the ribosome and multiprotein complexes involved in the initiation and elongation steps of the protein biosynthesis process (1). As opposed to their bacterial counterparts, aminoacyl-tRNA synthetases and elongation factors have been described as large molecular machines in human cells. Among the 20 aminoacyl-tRNA synthetases, nine of them (ArgRS, AspRS, GlnRS, GluRS, IleRS, LeuRS, LysRS, MetRS, and ProRS) are associated with the three structurally important proteins p18, p38, and p43 to form a multi-aminoacyl-tRNA synthetase (MARS)4 complex (2-4), and one of them (ValRS) forms the ValRS-EF1A-GEF assembly (VEGA) complex with the four subunits of elongation factor 1 (EF1A, and the guanine nucleotide exchange factors EF1Bα, EF1Bβ, and EF1Bγ) (5-7).
Global analysis of mRNA localization suggests that subcellular localization of proteins occurs at the level of translation, through mRNA localization (8). Colocalization of several components of the translation machinery, including ribosomes, mRNAs, initiation and elongation factors, and aminoacyl-tRNA synthetases can be easily observed in dendrites of neural cells (9, 10). Association of aminoacyl-tRNA synthetases and elongation factors with the cytoskeletal framework (11-13), and localization of aminoacyl-tRNA synthetases in the vicinity of ribosomes (14) have been described. Several pieces of evidence led to the suggestion that the cytoskeleton may serve as a scaffold to organize components of translation (15, 16). Direct evidence of in vivo compartmentalization of components of translation machinery remains elusive.
Recent studies have also indicated that some components of the translation machinery may lead a double life. Concerning components of the MARS complex, the involvement of GluProRS in the interferon-γ response (17), of LysRS in the regulation of transcription factors in the nucleus (18), of p43 in apoptosis (19-21), or of p38 in Parkinson disease (22) have been described. Thus, the knowledge of the cellular organization of the components of MARS in space and in time is a key factor for understanding the regulation of the activity of its components.
Using GFP fusion proteins, we analyzed the cellular localization of aminoacyl-tRNA synthetases and their cellular mobility by measuring fluorescence recovery after photobleaching (FRAP). We observed that these enzymes, associated or not within multienzyme complexes, are not freely diffusible in the cytoplasm of living cells. To evaluate the extent of macromolecular organization of the cell machinery, proteomic approaches have been conducted to identify protein interaction networks. High throughput two-hybrid analyses led to the description of the yeast (23) or human (24) interactome. However, assembly of multiprotein complexes often involves weak protein-protein interactions between two isolated partners of the complex and is difficult to depict by the two-hybrid approach. Affinity purification is a method of choice to isolate macromolecular complexes directly from cell extracts. The tandem affinity purification (TAP) strategy has been successfully employed to screen the yeast proteome organization (25) or to draw a connectivity map of factors involved in a defined transduction pathway in human (26). We isolated by TAP the MARS or VEGA complexes, and SerRS, a “free” synthetase. Apart from the major partners of the complexes, additional proteins were also found to copurify in substoichiometric amounts. Our data show that transient association of the components of translation with ribosomes and actin in the eukaryote cell is a key element of their subcellular organization.
EXPERIMENTAL PROCEDURES
Confocal Imaging—The complete cDNAs encoding human p18, p38, p43, ArgRS, LysRS, and AsnRS were introduced between the EcoRI and BamHI sites of pEGFP-N1 (BD Biosciences). Constructs of ΔNLysRS, ΔNAsnRS, and p43(ARF) fused to GFP in pEGFP-N1 correspond to LysRS, AsnRS, and p43 with N-terminal deletions of 56, 83, and 106 amino acid residues, respectively. The cDNAs encoding human MetRS and its derivatives were introduced either between the XhoI and EcoRI sites of pEGFP-C1 or between the EcoRI and AgeI sites of pEGFP-N1 (BD Biosciences). The different fusion proteins contained human MetRS sequences from amino acid residues 1-900 (MetRS), 215-900 (ΔNMetRS), 1-896 (MetRSΔK), 1-823 (MetRSΔC), 215-896 (ΔNMetRSΔK), or 215-823 (ΔNMetRSΔC). HeLa-ST cells were grown in F12 medium supplemented with 10% fetal calf serum. The cells were transfected with Effectene (Qiagen). The stability of the fusions expressed in HeLa cells was checked by Western blot analysis with anti-GFP antibodies. The cells were grown in 8-well Lab-Tek II chambers (Nalge Nunc International) and observed by confocal laser scanning microscopy using a Leica TCS SP2 confocal microscope equipped with an RSP 500 mirror. GFP was excited using laser line 488 nm of an argon laser (17 milliwatts) and detected at 500-535 nm. The oil objective used was 100× (NA 1.40), giving a resolution of 140 nm in the x,y plane and 240 nm along the z axis.
For FRAP experiments, three single scans were acquired at 1.7-s intervals to determine the initial fluorescence intensity, followed by a bleaching period of 16.86 s resulting from 10 scans at 1.7-s intervals (pulse of ∼10 ms/pixel). To monitor fluorescence recovery, 15 images were collected at 1.7-s intervals, followed by 12 images at 3.7-s intervals, and 12 images at 9.7-s intervals. The laser power was 17 milliwatts during the bleaching period and 1.7 milliwatts for imaging.
Construction of Stable Cell Lines Expressing TAP-tagged Proteins—The TAP tag sequences were introduced into the mammalian expression vector pSG5 (2). The cDNAs encoding the proteins of interest were introduced into the EcoRI or BamHI sites of pSG5-CTAP to create fusion proteins carrying a TAP tag appended to the C-terminal extremity of the protein. The cDNAs encoding human p38 (GGGGGATCCATAATGCCGATGTACCAGGTA and GGGGGATCCCTTAAGGAGCTTGAGGG), SerRS (GGGGGATCCATAATGGTGCTGGATCTGGATT and GGGGGATCCAGCATCGGTGACCTCCAT), and EF1Bβ (GGGGGATCCATAATGGCTACAAACTTCCTA and GGGGGATCCGATCTTGTTGAAAGCTGCGA) were amplified by PCR with the two oligonucleotides indicated and cloned into pSG5-CTAP. The constructs were verified by DNA sequencing. HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mm glutamine, 100 μg/ml of penicillin and streptomycin. The cells were transfected with Effectene (Qiagen). Stable transformants were isolated by selection with G418.
Isolation of Complexes—Cells (3 billions) grown in 100 dishes of 140-mm diameter were washed with ice-cold phosphate-buffered saline, harvested at 4 °C by mechanical detachment with a cell lifter, washed with ice-cold phosphate-buffered saline, and resuspended in 10 ml of buffer A (10 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5 mm MgCl2, 10% glycerol, 0.1% Nonidet P-40, 1 mm dithiothreitol) containing 0.5% Triton X-100 and protease inhibitors (1 mm phenylmethylsulfonyl fluoride and 2.5 mm benzamidine). The lysate was clarified by centrifugation at 40,000 × g for 30 min at 4 °C, and the TAP-tagged proteins were purified by chromatography on IgG-Sepharose 6 Fast Flow and Calmoduline-Sepharose 4B (GE Healthcare), as described previously (2). The purified proteins were separated by SDS-PAGE and visualized by staining with Coomassie Blue R250 or silver nitrate. Polypeptides were identified by mass spectrometry after in-gel digestion and matrix-assisted laser desorption ionization time-of-flight mass spectrometry performed essentially as described (2, 27). The proteins were identified using the search program Mascot reducing the Swiss-Prot/TrEMBL and NCBI data bases to the Homo sapiens species.
Construction of Knock-down Strains for the p38 and p43 Components of MARS—Retroviral vectors expressing small interfering RNA targeting p38 or p43 were constructed in pLTRH1 (28), as described previously (2). HeLa cells were infected by viruses as described (28), and clones that stably expressed the corresponding small interfering RNAs (p38si, and p43si) were selected. The knock-down of the corresponding component of MARS was checked by Western blot analysis with anti-p38 or anti-p43 antibodies.
Sucrose Gradient Analysis of Polysomes—HeLa cells grown to 60% confluency in a dish (diameter of 8.5 cm) were incubated 5 min at 37 °C after addition of cycloheximide at 50 μg/ml, washed twice with 5 ml of ice-cold phosphate-buffered saline containing cycloheximide at 50 μg/ml, and lysed by addition of 150 μl of 10 mm Tris-HCl, pH 7.5, 35 mm KCl, 2 mm MgCl2, 0.5 mm dithiothreitol, 0.5% Triton X-100, and protease inhibitors (1 mm phenylmethylsulfonyl fluoride and 2.5 mm benzamidine). The extracts were subjected to centrifugation on 15-45% sucrose gradients containing 10 mm Tris-HCl, pH 7.5, 35 mm KCl, 2 mm MgCl2, 0.5 mm dithiothreitol. The gradients were centrifuged in a SW 41 Ti rotor (Beckman) at 27,000 rpm for 3 h and collected into 10 fractions, starting from the bottom of the tube. A254 was recorded. The fractions were subjected to Western blotting after precipitation by trichloroacetic acid/Triton, as described (2).
Antibodies and Western Blot Analysis—Monoclonal antibodies directed to GFP were from BD Biosciences. Polyclonal anti-p38, anti-p43, anti-GluProRS, anti-GlnRS, anti-MetRS, and anti-AspRS antibodies have been described previously (29). Western blot analyses were conducted with goat anti-rabbit or goat anti-mouse secondary antibodies conjugated with peroxidase (Chemicon) and the ECL detection reagents (GE Healthcare).
RESULTS
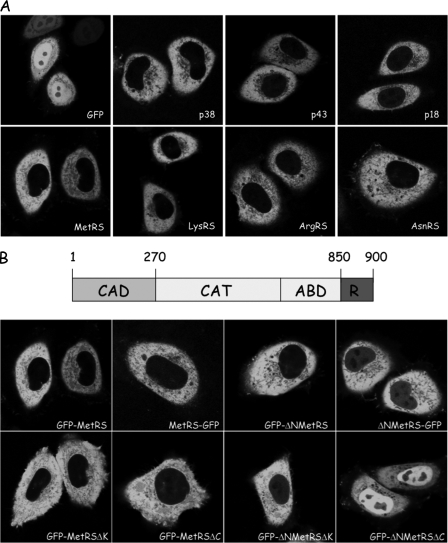
Subcellular Localization of Aminoacyl-tRNA Synthetases—The cellular localization of several components of MARS (MetRS, LysRS, ArgRS, p18, p38, and p43) and of AsnRS, a representative of free synthetases, was investigated. Transient transfection of HeLa cells with constructs that expressed those cDNA fused to the GFP clearly provided evidence that MARS and free aminoacyl-tRNA synthetase prominently localize in the cytoplasm (Fig. 1A).
FIGURE 1.
Subcellular localization of aminoacyl-tRNA synthetases in human cells. A, localization of GFP, of components of MARS (p38, p43, p18, MetRS, LysRS, and ArgRS), and of a representative of free enzymes (AsnRS) expressed in HeLa cells after transient transfection with pEGFP derivatives. B, subcellular localization of MetRS and of truncated derivatives. The structural organization of human MetRS is indicated above. CAD, the N-terminal, eukaryotic-specific complex association domain; CAT, the conserved catalytic domain; ABD, the conserved anticodon-binding domain; R, the eukaryotic-specific, repeated domain appended to the C terminus and containing an additional four-lysine cluster at the very C terminus (30). HeLa cells were transiently transfected with pEGFP-N1 or pEGFP-C1 plasmids expressing fusion proteins with MetRS (GFP-MetRS and MetRS-GFP, according to the polarity of the fusion), MetRS with a deletion of its N-terminal, eukaryotic-specific domain (GFP-ΔNMetRS and ΔNMetRS-GFP), with a deletion of its C-terminal, lysine-rich cluster (GFP-MetRSΔK), with a complete deletion of its C-terminal, helix-turn-helix domain (GFP-MetRSΔC), or with a combination of N- and C-terminal deletions (GFP-ΔNMetRSΔK and GFP-ΔNMetRSΔC). The cellular localization of GFP alone or of the fusion proteins was analyzed by confocal laser scanning microscopy. All of the images represent equatorial sections of the nuclei.
To investigate the role of complex assembly on the subcellular localization of an integral component of MARS, we analyzed the cellular distribution of different domains of MetRS fused to GFP. When GFP was added either to its N terminus (GFP-MetRS) or to its C terminus (MetRS-GFP), full-length MetRS was always excluded from the nucleus (Fig. 1B). As compared with its prokaryotic homologue, human MetRS possesses a large (214 amino acid residues) polypeptide extension appended to its N terminus (30). Deletion of this extension precludes association of MetRS to the complex (31). The GFP-ΔNMetRS fusion protein was essentially localized in the cytoplasm, but a faint nuclear fluorescence was clearly apparent (Fig. 1B). A ΔNMetRS-GFP fusion displayed a similar localization pattern (Fig. 1B). Thus, the cytoplasmic retention of MetRS does not rest only on its ability to associate with MARS.
A short (77 amino acid residues), eukaryotic-specific polypeptide extension is also appended to the C terminus of human MetRS. Neither of the N- and C-terminal extensions are necessary for the activity of MetRS, but the C-terminal domain improves the tRNA binding properties of MetRS (30). The RNA-binding domain of MetRS can be divided into two regions: a helix-turn-helix motif found in other aminoacyl-tRNA synthetases (32) and a lysine-rich cluster at the very C terminus. Deletion of this cluster (GFP-MetRSΔK) or of the complete C-terminal domain (GFP-MetRSΔC) did not alter cellular localization of MetRS (Fig. 1B). In these two constructs, the N-domain of MetRS is present, and the enzyme retains its ability to associate with MARS. The N- and C-terminal deletions were combined to give GFP-ΔNMetRSΔK and GFP-ΔNMetRSΔC. The GFP fusion with the minimal human MetRS (GFP-ΔNMetRSΔC), recapitulating the MetRS core enzyme found in eubacteria such as Aquifex aeolicus, was no more excluded from the nuclear compartment (Fig. 1B). We therefore conclude that protein-protein interactions, involving the N-domain of MetRS that mediates its association with MARS, and protein-RNA interactions, involving the C-domain of MetRS, are both required for cytoplasmic confinement of this enzyme.
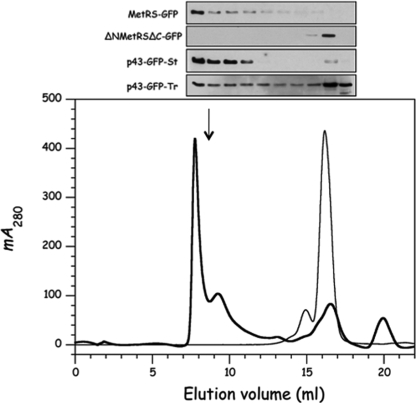
Subcellular Mobility of Human, Cytoplasmic Aminoacyl-tRNA Synthetases—The cytoplasmic mobilities of several aminoacyl-tRNA synthetases from MARS and of truncated derivatives that had lost their ability to associate within MARS were analyzed. These proteins were fused to GFP, and stable HeLa cell lines were isolated. We verified that MetRS-GFP from a crude extract of stable transformant of HeLa cells behaved as a high molecular weight entity, whereas the ΔNMetRSΔC derivative did not (Fig. 2). To probe the degree of mobility of these proteins in the cytoplasm of a living cell, we carried out FRAP. In our FRAP experiments, a defined region of the living cell was bleached (bleached region, filled circle in Fig. 3A), and the time course of fluorescence recovery was recorded in the bleached area. The kinetics of recovery is a measure of the mobility of the GFP-labeled protein in the living cell. In addition, we also recorded the variation of fluorescence in another region of the cell, located at the opposite of the bleached area (distal region, open circles in Fig. 3A). Initially, the two regions of the cell had a similar fluorescence intensity. After photobleaching, the time required to reach a new fluorescence equilibrium between the two distant regions of the cell is also a signal of the mobility of the protein throughout the cell.
FIGURE 2.
Structural behavior of GFP fusion proteins. Extracts of HeLa cells expressing GFP fusion proteins were analyzed on a Superose 6 HR 10/30 column equilibrated in 20 mm HEPES, pH 7.5, 200 mm NaCl, 2 mm dithiothreitol, and 0.001% Tween 20 and developed at a flow rate of 0.2 ml/min. Fractions of 0.5 ml were collected and analyzed by Western blotting with antibodies directed to GFP. A typical elution profile is shown (absorbance at 280 nm, thick line). The column was previously calibrated with bovine serum albumin (thin line); the arrow shows the elution volume of purified MARS. Extracts of stable HeLa cells expressing MetRS-GFP, ΔNMetRSΔC-GFP, or p43-GFP-St or from HeLa cells that transiently expressed p43-GFP-Tr were analyzed independently, and the fractions were probed with anti-GFP antibodies.
FIGURE 3.
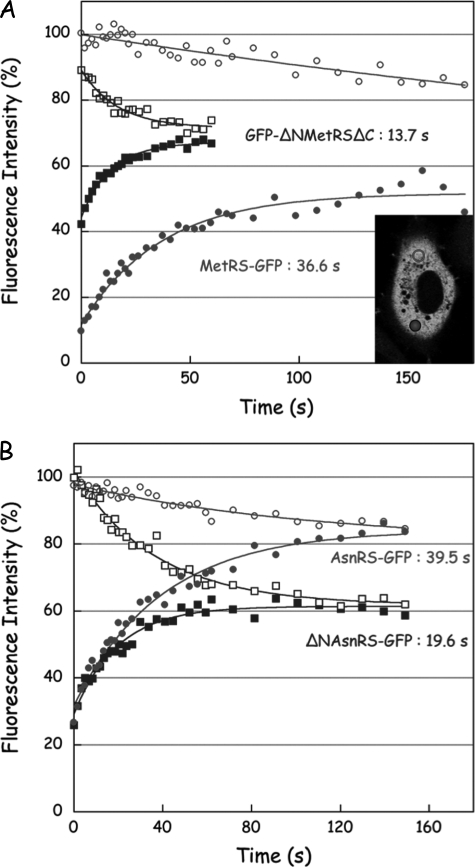
FRAP of aminoacyl-tRNA synthetases in the cytoplasm of living cell. FRAP was conducted on stably transformed cell lines. The areas of the bleach region where fluorescence recovery is monitored and of the distal region where fluorescence intensity is recorded in parallel are indicated with a closed and open symbols, respectively. The inset in A illustrates an experiment conducted with MetRS-GFP. Kinetics of fluorescence intensity variations in the bleach and distal regions are indicated with closed and open symbols, respectively. The experimental data were fitted to monoexponential curves to determine the half-times of fluorescence recovery. A, FRAP of MetRS-GFP and of GFP-ΔNMetRSΔC. B, FRAP of AsnRS-GFP and of ΔNAsnRS-GFP.
After photobleaching of MetRS-GFP (Fig. 3A), the recovery in the bleached region reached a new equilibrium after 180 s, with a half-time of 36.6 s. This half-time of recovery is very long, as compared with the value of 4.9 s obtained for GFP alone (Table 1), a protein known to be freely diffusible in the cytoplasm of human cells. Moreover, a new equilibrium was not reached between the distal areas of the cell, even after 180 s of recovery, suggesting that MetRS-GFP exchanged very slowly between the different regions of the cell. The fluorescence intensity in the distal region remained constant during photobleaching and slowly decreased during the recovery period, with a half-time of 249 ± 60 s. The low mobility of MetRS-GFP could be the consequence of its association with MARS, and/or of the interaction of MARS with some subcellular components, that would preclude its free diffusion. When GFP-ΔNMetRSΔC was photobleached (Fig. 3A), fluorescence recovery in the bleached region was observed after 60 s, with a half-time of 13.7 s, approximately three times faster that with MetRS-GFP. In the mean time, the fluorescence of GFP-ΔNMetRSΔC also rapidly decreased in the distal region (half-time of 16.9 ± 2.7 s), and a new fluorescence equilibrium was reached after ∼50 s of recovery. These results are consistent with the facts that ΔNMetRSΔC has lost its ability to associate with MARS (deletion of its N terminus) and to associate with nucleic acids (deletion of its C terminus). Similar measurements of fluorescence recovery were conducted after photobleaching of other components of MARS. Stable transformants of HeLa cells expressing LysRS, p43, or p38 fused to GFP were obtained and displayed half-times of recovery similar to those observed with MetRS-GFP (Table 1). These data are consistent with the finding that all of these proteins are members of the same multienzyme complex. As expected, deletion of the N terminus of LysRS had no effect on its half-time of recovery (Table 1), in good agreement with the observation that association of LysRS to MARS does not involve its N-terminal polypeptide extension (33).
TABLE 1.
Kinetics of fluorescence recovery after photobleaching
The standard errors were determined from at least three independent data sets.
| Stablea | Transientb | |
|---|---|---|
| s | s | |
| GFP | 4.9 ± 1.1 | |
| MetRS | 38.5 ± 7.2 | 14.4 ± 1.6 |
| LysRS | 32.5 ± 8.7 | 22.4 ± 5.6 |
| p43 | 33.1 ± 6.1 | 17.9 ± 5.2 |
| p38 | 32.3 ± 3.7 | 16.7 ± 4.9 |
| ΔNMetRSΔC | 12.1 ± 3.9 | 7.4 ± 0.8 |
| ΔNLysRS | 32.7 ± 9.9 | 16.5 ± 4.8 |
| p43(ARF) | 8.1 ± 2.1 | 11.0 ± 2.5 |
| AsnRS | 39.5 ± 8.3 | 33.1 ± 12.5 |
| ΔNAsnRS | 19.6 ± 5.2 | 19.8 ± 3.3 |
The values were determined from stable cell lines.
The values were determined 24 h after transfection.
We also analyzed the cellular mobility of derivatives of MARS that have lost their ability to associate within the particle. The p43 subunit of MARS is a procytokine that is matured by proteolytic processing during apoptosis (21). After cleavage, its C-terminal domain, p43(ARF) is released from the cell and is converted into p43(EMAPII), a putative cytokine (19, 20). When FRAP analysis was conducted with p43(ARF)-GFP instead of p43-GFP, the half-time of recovery was much lower (8.1 s for p43(ARF), as compared with 33.1 s for p43; Table 1), consistent with the fact that p43(ARF) is released from MARS.
In all cases, when transient transformants of HeLa cells were analyzed by FRAP, the half-times of fluorescence recovery were significantly lower as compared with those observed with stable transformants, especially in the cases of proteins that associate with MARS (Table 1). This reflects the heterogeneous behavior of the proteins overexpressed after transient transformation of HeLa cells that were in part associated with MARS, and predominantly present as free enzymes because the endogenous partner proteins were present in substoichiometric amounts (see the analysis of stable or transient transformants of HeLa cells with p43-GFP shown in Fig. 2).
The mobility of AsnRS, a free enzyme not associated within stable multiprotein complexes, in the cytoplasm of a living cell was also assessed by FRAP. The half-life of recovery of AsnRS-GFP (39.5 s) was much higher than that observed for GFP (4.9 s) or for ΔNAsnRS-GFP (19.6 s), a derivative of AsnRS with a deletion of its N-terminal, tRNA-binding domain (34) (Fig. 3B and Table 1). Thus, the limited mobility of aminoacyl-tRNA synthetases in the cytoplasm of human cells is not restricted to those that form stable multiprotein complexes. The kinetics of recovery in the distal region of the cell was much faster for ΔNAsnRS-GFP (26.6 ± 3.2s) than for AsnRS-GFP (113 ± 44 s). However, in contrast with the components of MARS, for AsnRS-GFP and ΔNAsnRS-GFP a new fluorescence equilibrium was reached between the bleached and the distal regions of the cell after 150 s of recovery. This suggests that the mechanisms of subcellular confinement differ to some extent between MARS and the free synthetases. The multiple potential binding sites displayed by MARS as compared with a single synthetase may favor a tighter binding.
Copurification of Aminoacyl-tRNA Synthetases and Elongation Factor 1 with Ribosomes and Actin—TAP was used to isolate putative cellular partners of components of the translation machinery. A tag made of protein A and calmodulin-binding peptide and containing a TEV cleavage site just in between was appended to p38, the scaffold protein of the MARS complex, to SerRS, a free synthetase, or to EF1Bβ, a component of the VEGA complex. Each individually tagged protein was expressed in HeLa cells after stable integration of the DNA constructs. Tandem affinity purification was conducted at low salt concentration (50 mm NaCl) starting from 2-3 billion cells.
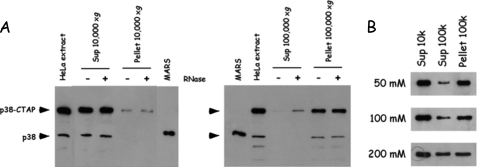
During isolation of MARS by the TAP tag procedure, we observed that centrifugation of the cell lysate at high speed (100,000 × g) resulted in the recovery of MARS essentially in the pellet fraction (Fig. 4A). Treatment of the 10,000 × g supernatant with RNaseA only slightly improved the recovery of MARS in the supernatant of the high speed centrifugation (Fig. 4A). The fraction of MARS associated with the 100,000 × g pellet decreased when increasing NaCl concentration (Fig. 4B). Thus, TAP was performed after centrifugation of the cell extract at low speed, and at low ionic strength, conditions intended to recover transient protein partners weakly associated with the stable MARS complex.
FIGURE 4.
Association of aminoacyl-tRNA synthetases with high molecular weight fractions in cellular extracts of HeLa cells. A, extracts from HeLa cells expressing p38-CTAP were subjected to low speed (10,000 × g) or high speed (100,000 × g) centrifugation without (-) or with (+) prior incubation with RNase A (50 μg/ml, for 30 min, at 4 °C). The presence of p38 and p38-CTAP in the different fractions (supernatant and pellet) was analyzed by Western blotting with anti-p38 antibodies. The intensity of the polypeptide p38-CTAP is enhanced ∼7-fold as compared with p38, because of the presence of the tag containing protein A. B, effect of the NaCl concentration in the extraction buffer on the recovery of p38 in the supernatant of high speed centrifugation.
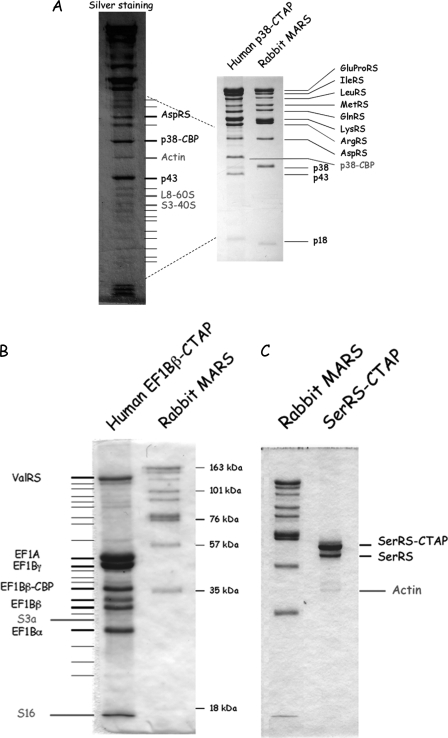
The MARS complex recovered with p38-CTAP essentially contained the major polypeptides identified as bona fide MARS components, but other minor polypeptides were also recovered (Fig. 5A). Among the polypeptides copurified with p38-CTAP, observed by SDS-PAGE and silver nitrate staining, the identity of some of them could be unambiguously determined by mass spectrometry. Actin and ribosomal proteins were present in the purified fraction (Fig. 5A).
FIGURE 5.
Copurification of aminoacyl-tRNA synthetases with actin and with ribosomes. A, the polypeptides copurified with p38-CTAP were analyzed by SDS-PAGE and stained with Coomassie Blue (right) or silver nitrate (left). The identity of some of the low abundance polypeptides was determined by mass spectrometry. Polypeptides that could not be identified are marked with thin lines. B and C, protein complexes isolated using EF1Bβ-CTAP (B) or SerRS-CTAP (C) were analyzed as described above. The MARS complex purified from rabbit liver by conventional procedure (29) is indicated as a standard.
To address the possibility that aminoacyl-tRNA synthetases and other components of the translation machinery could be transiently associated with ribosomes and/or with actin within the cell, we also constructed stable HeLa cell lines that expressed seryl-tRNA synthetase, a dimeric synthetase that had always been described as a free enzyme. When seryl-tRNA synthetase was isolated from HeLa cells by the TAP tag procedure, the tagged subunit was preferentially recovered (homodimers) along with the untagged subunit that could form heterodimers with the tagged subunit (Fig. 5C). A minor polypeptide was identified to actin, but no trace of ribosomal subunits was observed.
A stable HeLa cell line that expressed the β subunit of the elongation factor EF1B complex, which had been described to form a complex with valyl-tRNA synthetase, was also constructed. When the β subunit of EF1B was purified by the TAP tag procedure, five major polypeptides were recovered, corresponding to the three EF1Bα, EF1Bβ, and EF1Bγ subunits of the EF1B complex, to elongation factor EF1A, and to valyl-tRNA synthetase (Fig. 5B). This complex corresponds to the stable VEGA complex, already described (5, 7). Minor polypeptides were also recovered, and some of them could be identified to ribosomal proteins. Because actin comigrates with EF1Bβ-CBP, its presence or absence in the purified fraction could not be assessed.
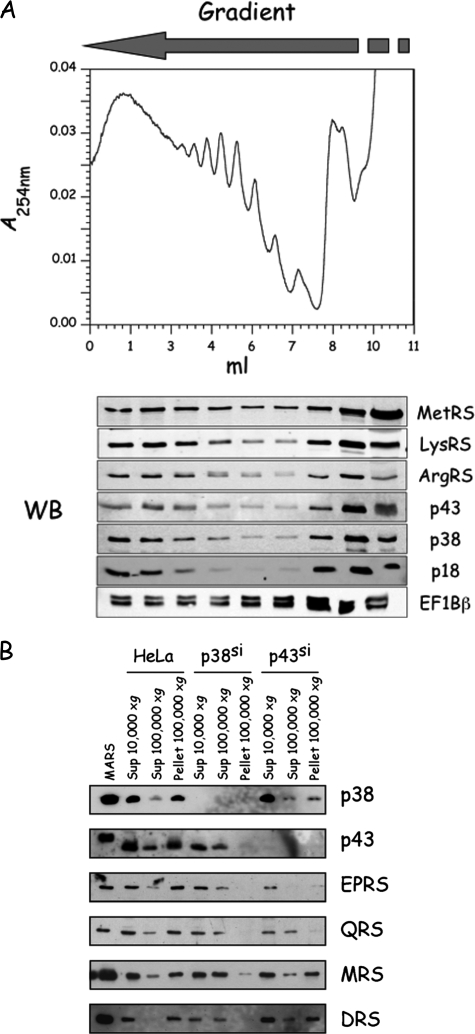
Association of Aminoacyl-tRNA Synthetases and Elongation Factor 1 with Polyribosomes—To get more clues on the association of aminoacyl-tRNA synthetases and of elongation factor 1 with ribosomes, polysomes from exponentially growing, wild-type HeLa cells were fractionated by sucrose gradient centrifugation, and cosedimentation of these components of the translation machinery was examined by Western blot analysis (Fig. 6A). Antibodies directed to MetRS, LysRS, ArgRS, p43, p38, and p18 unambiguously showed that a fraction of MARS (∼30%) cosediments with polysomes. Antibodies to EF1Bβ also revealed cosedimentation of VEGA with polysomes (Fig. 6A).
FIGURE 6.
Aminoacyl-tRNA synthetases cosediment with ribosomes. A, polysome profile of HeLa cells. Polysomes from exponentially growing HeLa cells were subjected to sucrose gradient centrifugation. A254 was recorded, and the fractions were analyzed by Western blotting (WB) with antibodies directed to MetRS, LysRS, ArgRS, p43, p38, and p18 from MARS, or to EF1Bβ from the ValRS-EF1 complex. B, extracts of HeLa cells were subjected to low speed (10,000 × g) or high speed (100,000 × g) centrifugation, and the supernatants and pellets were analyzed by Western blotting, as indicated. HeLa cells growing in a p38si or p43si background were also analyzed in parallel.
Association of components of the translation machinery with actin and/or polysomes is consistent with their recovery in the post-ribosomal pellet (Fig. 4A). To further understand the mode of association of MARS with other subcellular components, we analyzed the content of the post-ribosomal supernatants and pellets obtained after centrifugation of crude lysates from HeLa cells that expressed small interfering RNA against p38 or p43 (Fig. 6B). We showed previously that MARS can be described as the assembly of two subcomplexes, subcomplex I containing MetRS, IleRS, LeuRS, GluProRS, and p18, and subcomplex II containing ArgRS, GlnRS, and p43, and of the dimeric proteins AspRS and LysRS, which are linked via the scaffold protein p38 (2). The various components of MARS, isolated from wild-type HeLa cells, were recovered in the pellet fraction but did not associate with the pellet fraction when MARS was dissociated into its various subcomplexes by silencing of its p38 component (Fig. 6B). Especially, subcomplex I and subcomplex II were recovered in the supernatant of high speed centrifugation (Fig. 6B). After silencing of the p43 component of MARS, subcomplex II is disrupted, and none of its components are recovered in the pellet fraction, but subcomplex I, that is now associated with p38, LysRS, and AspRS (2), is mainly recovered in the pellet fraction (Fig. 6B). Thus, association of MARS with subcellular components, such as actin and polysomes, does not absolutely require a native MARS particle but necessitates that a nearly complete particle is formed (subcomplex I, plus LysRS, AspRS, and p38).
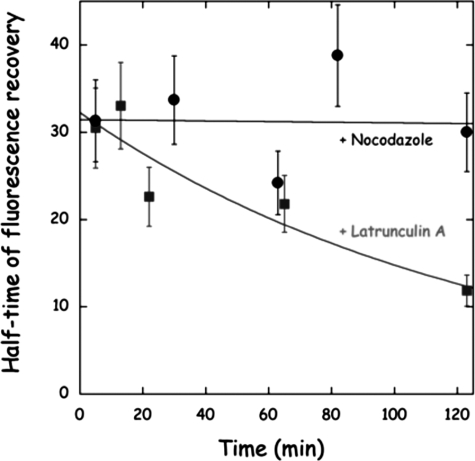
Cellular Mobility of Aminoacyl-tRNA Synthetases after Disruption of Actin Network—Because aminoacyl-tRNA synthetases were shown to copurify with actin after TAP tag isolation, we surmised that disruption of the actin-cytoskeleton might result in an increased cellular mobility of the synthetases. HeLa cells that stably expressed MetRS-GFP were subjected to treatment with nocodazole, a drug known to disrupt microtubules, or with latrunculin A, a drug that disrupts actin-filaments. As expected, addition of latrunculin A, but not of nocodazole, resulted in an increased mobility of MetRS-GFP (Fig. 7). This result is consistent with the finding that MARS is an actin-binding particle and suggests that actin filaments are involved in organizing translation machinery in human cells.
FIGURE 7.
Effect of latrunculin A on the mobility of MetRS in living cell. HeLa cells that stably expressed MetRS-GFP were subjected to treatment with nocodazole (1 μg/ml) or latrunculin A (1 μm), and the mobility of MetRS-GFP was assessed by FRAP. Following addition of the drugs, at the times indicated, half-times of fluorescence recovery were determined as specified in the legend to Fig. 3.
DISCUSSION
We have performed an in vivo analysis of the subcellular organization of some components of the translation machinery in human cells. The fusion of the native form of MetRS with GFP displays an exclusive cytoplasmic localization, as previously observed by immunofluorescence microscopy (11, 12). In these earlier experiments and in the present analysis, no evidence for nucleolar localization of MetRS was obtained, as claimed in another study (35). Nuclear exclusion of MetRS is observed even when the MetRS-GFP fusion is overexpressed, by transient transfection, as compared with the other components of MARS. Thus, the full-length form of MetRS is efficiently excluded from the nucleus, independently of its association within MARS. In human, two polypeptide extensions are appended to the core catalytic domain of MetRS: a N-terminal domain involved in association with MARS and a C-terminal domain with RNA binding properties (30). Deletion of only one of these domains did not impact the cellular localization of MetRS, but the core domain alone could distribute evenly throughout the cell, including the nucleus (but not the nucleolus). Thus, the cytoplasmic sequestration of human MetRS involves protein-protein interactions (assembly within MARS via its N-domain) and protein-nucleic acid interactions (with the C-domain). These data suggest that the eukaryotic-specific polypeptide extensions of aminoacyl-tRNA synthetases are involved in their association with other cellular components and in the subcellular organization of translation in human cells. In the case of p43, we observed previously that the native form of p43, a component of MARS, is exclusively localized in the cytoplasm of living cells, whereas its C-terminal derivatives, p43(EMAPII) and p43(ARF), released from MARS by proteolytic cleavage occurring after onset of apoptosis, are also recovered in the nucleus (21).
In addition to the bona fide components of MARS, the TAP-isolated complexes also contained ribosomal proteins and actin. They were present as minor components in the purified fractions, as expected for transiently interacting proteins. Among the many low molecular mass polypeptides recovered in the purified fraction, only few of them could be unambiguously identified by mass spectrometry. Identification of specific ribosomal proteins by mass spectrometry suggests that the whole ribosome was present in the fraction but does not imply a direct interaction between the particular component of the ribosome that has been identified and the TAP-tagged protein used during the purification. The ribosomal protein(s) interacting with aminoacyl-tRNA synthetases or with elongation factors remain(s) to be deciphered. Interaction of synthetases and elongation factors with ribosomes is exemplified by their cosedimentation with polyribosomes. Among the 20 aminoacyl-tRNA synthetases, potent association of PheRS with ribosomes was already reported (36, 37), whereas association of several of these enzymes with the detergent-insoluble fraction of the cell has been observed by immunofluorescence microscopy (11-13).
The observation that several components of the translation apparatus are ribosome- and/or actin-binding proteins, are confined to the cytoplasm when the eukaryote-specific polypeptide extensions are present, display a low cytoplasmic mobility in their native state, or are sensitive to agents that disrupt actin-filament integrity has conceptual implications for our understanding of subcellular organization of translation in the eukaryote cell. Two types of localized polyribosomes have been described in the cytoplasm of the eukaryote cell. Some mRNA are attached to membranes, especially to the ER membrane, through the newly synthesized proteins that recognize the signal recognition particle through their signal sequence, cross the membrane, and are secreted. A second class of mRNA are anchored to actin microfilaments through a zip code contained within the mRNA itself (38). Various studies have shown that integrity of actin microfilaments is required for an efficient protein synthesis (15, 39).
Macromolecular crowding of the cytoplasm is believed to promote intermolecular associations (40). Thus, the measure of the diffusion of macromolecules is the result of the rates of dissociation and reassociation to other molecules. Our FRAP experiments showed that the movement of native aminoacyl-tRNA synthetases in the cytoplasm of living human cells has kinetic properties not consistent with a free diffusion model. These data are in agreement with the observation that components of the translation machinery are not rapidly released from cells subjected to a gentle permeabilization treatment that does not disturb the integrity of the actin cytoskeleton (16). One surprising observation arising from FRAP on components of MARS was the different diffusion kinetics observed for short range and long range effects. Although fluorescence recovery was already quite slow in the bleach region, the time required to reach a new fluorescence equilibrium between distant regions of the cell was even much longer. This result suggests that the population of MARS could be heterogeneous in the cell cytoplasm, with one subpopulation diffusing much more slowly than the other. Although experimental data of fluorescence recovery could be satisfactory fitted to monoexponential kinetics, suggesting a single diffusion mechanism, it is not possible to exclude the possibility that different populations of MARS with very distinct diffusion kinetics would give the same global diffusion pattern. The existence of several populations of MARS is exactly what would be expected considering that translation is organized around the different types of mRNA that have been described: the free, membrane-bound, or actin-associated mRNA. More precise and quantitative examinations of MARS diffusion in living cells, with sophisticated motion analysis, could lead to a more precise view of the subcellular organization of the translation machinery in mammalian cells.
Acknowledgments
We thank Michael Härtlein for the gift of the plasmids carrying human cDNA for SerRS and AsnRS. We also thank Spencer Brown and Susanne Bolte for access to the confocal microscope facility (Institut des Sciences du Végétal, Gif-sur-Yvette, France).
This work was supported by grants from CNRS, the Association pour la Recherche sur le Cancer, and La Ligue.
Footnotes
The abbreviations used are: MARS, multi-aminoacyl-tRNA synthetase; FRAP, fluorescence recovery after photobleaching; TAP, tandem affinity purification; VEGA, ValRS-EF1A-GEF assembly; GFP, green fluorescent protein.
References
- 1.Kapp, L. D., and Lorsch, J. R. (2004) Annu. Rev. Biochem. 73 657-704 [DOI] [PubMed] [Google Scholar]
- 2.Kaminska, M., Havrylenko, S., Decottignies, P., Gillet, S., Le Maréchal, P., Negrutskii, B., and Mirande, M. (2009) J. Biol. Chem. 284 6053-6060 [DOI] [PubMed] [Google Scholar]
- 3.Lee, S. W., Cho, B. H., Park, S. G., and Kim, S. (2004) J. Cell Sci. 117 3725-3734 [DOI] [PubMed] [Google Scholar]
- 4.Mirande, M. (2005) in The Aminoacyl-tRNA Synthetases (Ibba, M., Francklyn, C., and Cusack, S., eds) pp. 298-308, Landes Bioscience, Georgetown, TX
- 5.Bec, G., Kerjan, P., Zha, X. D., and Waller, J. P. (1989) J. Biol. Chem. 264 21131-21137 [PubMed] [Google Scholar]
- 6.Motorin, Y. A., Wolfson, A. D., Orlovsky, A. F., and Gladilin, K. L. (1988) FEBS Lett. 238 262-264 [DOI] [PubMed] [Google Scholar]
- 7.Negrutskii, B. S., Shalak, V. F., Kerjan, P., El'skaya, A. V., and Mirande, M. (1999) J. Biol. Chem. 274 4545-4550 [DOI] [PubMed] [Google Scholar]
- 8.Lecuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., Hughes, T. R., Tomancak, P., and Krause, H. M. (2007) Cell 131 174-187 [DOI] [PubMed] [Google Scholar]
- 9.Barbarese, E., Koppel, D. E., Deutscher, M. P., Smith, C. L., Ainger, K., Morgan, F., and Carson, J. H. (1995) J. Cell Sci. 108 2781-2790 [DOI] [PubMed] [Google Scholar]
- 10.Tiedge, H., and Brosius, J. (1996) J. Neurosci. 16 7171-7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, C. V., Yang, D. C., and Pollard, T. D. (1983) J. Cell Biol. 96 1138-1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirande, M., Le Corre, D., Louvard, D., Reggio, H., Pailliez, J. P., and Waller, J. P. (1985) Exp. Cell Res. 156 91-102 [DOI] [PubMed] [Google Scholar]
- 13.Sanders, J., Brandsma, M., Janssen, G. M. C., Dijk, J., and Möller, W. (1996) J. Cell Sci. 109 1113-1117 [DOI] [PubMed] [Google Scholar]
- 14.Popenko, V. I., Ivanova, J. L., Cherny, N. E., Filonenko, V. V., Beresten, S. F., Wolfson, A. D., and Kisselev, L. L. (1994) Eur. J. Cell Biol. 65 60-69 [PubMed] [Google Scholar]
- 15.Gross, S. R., and Kinzy, T. G. (2007) Mol. Cell Biol. 27 1974-1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudder, A., Nathanson, L., and Deutscher, M. P. (2003) Mol. Cell Biol. 23 9318-9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampath, P., Mazumder, B., Seshadri, V., Gerber, C. A., Chavatte, L., Kinter, M., Ting, S. M., Dignam, J. D., Kim, S., Driscoll, D. M., and Fox, P. L. (2004) Cell 119 195-208 [DOI] [PubMed] [Google Scholar]
- 18.Lee, Y. N., and Razin, E. (2005) Mol. Cell Biol. 25 8904-8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, J., Fan, Y. G., Haehnel, I., Brett, J., Greenberg, S., Clauss, M., Kayton, M., Houck, K., Kisiel, W., Seljelid, R., Burnier, J., and Stern, D. (1994) J. Biol. Chem. 269 9774-9782 [PubMed] [Google Scholar]
- 20.Ko, Y. G., Park, H., Kim, T., Lee, J. W., Park, S. G., Seol, W., Kim, J. E., Lee, W. H., Kim, S. H., Park, J. E., and Kim, S. (2001) J. Biol. Chem. 276 23028-23033 [DOI] [PubMed] [Google Scholar]
- 21.Shalak, V., Guigou, L., Kaminska, M., Wautier, M. P., Wautier, J. L., and Mirande, M. (2007) J. Biol. Chem. 282 10935-10943 [DOI] [PubMed] [Google Scholar]
- 22.Corti, O., Hampe, C., Koutnikova, H., Darios, F., Jacquier, S., Prigent, A., Robinson, J. C., Pradier, L., Ruberg, M., Mirande, M., Hirsch, E., Rooney, T., Fournier, A., and Brice, A. (2003) Hum. Mol. Genet. 12 1427-1437 [DOI] [PubMed] [Google Scholar]
- 23.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S., and Rothberg, J. M. (2000) Nature 403 623-627 [DOI] [PubMed] [Google Scholar]
- 24.Rual, J. F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G. F., Gibbons, F. D., Dreze, M., Ayivi-Guedehoussou, N., Klitgord, N., Simon, C., Boxem, M., Milstein, S., Rosenberg, J., Goldberg, D. S., Zhang, L. V., Wong, S. L., Franklin, G., Li, S., Albala, J. S., Lim, J., Fraughton, C., Llamosas, E., Cevik, S., Bex, C., Lamesch, P., Sikorski, R. S., Vandenhaute, J., Zoghbi, H. Y., Smolyar, A., Bosak, S., Sequerra, R., Doucette-Stamm, L., Cusick, M. E., Hill, D. E., Roth, F. P., and Vidal, M. (2005) Nature 437 1173-1178 [DOI] [PubMed] [Google Scholar]
- 25.Gavin, A. C., Aloy, P., Grandi, P., Krause, R., Boesche, M., Marzioch, M., Rau, C., Jensen, L. J., Bastuck, S., Dumpelfeld, B., Edelmann, A., Heurtier, M. A., Hoffman, V., Hoefert, C., Klein, K., Hudak, M., Michon, A. M., Schelder, M., Schirle, M., Remor, M., Rudi, T., Hooper, S., Bauer, A., Bouwmeester, T., Casari, G., Drewes, G., Neubauer, G., Rick, J. M., Kuster, B., Bork, P., Russell, R. B., and Superti-Furga, G. (2006) Nature 440 631-636 [DOI] [PubMed] [Google Scholar]
- 26.Bouwmeester, T., Bauch, A., Ruffner, H., Angrand, P. O., Bergamini, G., Croughton, K., Cruciat, C., Eberhard, D., Gagneur, J., Ghidelli, S., Hopf, C., Huhse, B., Mangano, R., Michon, A. M., Schirle, M., Schlegl, J., Schwab, M., Stein, M. A., Bauer, A., Casari, G., Drewes, G., Gavin, A. C., Jackson, D. B., Joberty, G., Neubauer, G., Rick, J., Kuster, B., and Superti-Furga, G. (2004) Nat. Cell Biol. 6 97-105 [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850-858 [DOI] [PubMed] [Google Scholar]
- 28.Barton, G. M., and Medzhitov, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14943-14945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirande, M., Cirakoglu, B., and Waller, J. P. (1982) J. Biol. Chem. 257 11056-11063 [PubMed] [Google Scholar]
- 30.Kaminska, M., Shalak, V., and Mirande, M. (2001) Biochemistry 40 14309-14316 [DOI] [PubMed] [Google Scholar]
- 31.Mirande, M., Kellermann, O., and Waller, J. P. (1982) J. Biol. Chem. 257 11049-11055 [PubMed] [Google Scholar]
- 32.Cahuzac, B., Berthonneau, E., Birlirakis, N., Guittet, E., and Mirande, M. (2000) EMBO J. 19 445-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, J. C., Kerjan, P., and Mirande, M. (2000) J. Mol. Biol. 304 983-994 [DOI] [PubMed] [Google Scholar]
- 34.Beaulande, M., Kron, M., and Härtlein, M. (2001) FEBS Lett. 494 170-174 [DOI] [PubMed] [Google Scholar]
- 35.Ko, Y. G., Kang, Y. S., Kim, E. K., Park, S. G., and Kim, S. (2000) J. Cell Biol. 149 567-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pailliez, J. P., and Waller, J. P. (1984) J. Biol. Chem. 259 15491-15496 [PubMed] [Google Scholar]
- 37.Ussery, M. A., Tanaka, W. K., and Hardesty, B. (1977) Eur. J. Biochem. 72 491-500 [DOI] [PubMed] [Google Scholar]
- 38.Czaplinski, K., and Singer, R. H. (2006) Trends Biochem. Sci. 31 687-693 [DOI] [PubMed] [Google Scholar]
- 39.Stapulionis, R., Kolli, S., and Deutscher, M. P. (1997) J. Biol. Chem. 272 24980-24986 [DOI] [PubMed] [Google Scholar]
- 40.Luby-Phelps, K. (1994) Curr. Opin. Cell Biol. 6 3-9 [DOI] [PubMed] [Google Scholar]