Abstract
Protrudin is a protein that contains a Rab11-binding domain and a FYVE (lipid-binding) domain and that functions to promote neurite formation through interaction with the GDP-bound form of Rab11. Protrudin also contains a short sequence motif designated FFAT (two phenylalanines in an acidic tract), which in other proteins has been shown to mediate binding to vesicle-associated membrane protein-associated protein (VAP). We now show that protrudin associates and colocalizes with VAP-A, an isoform of VAP expressed in the endoplasmic reticulum. Both the interaction between protrudin and VAP-A as well as the induction of process formation by protrudin were markedly inhibited by mutation of the FFAT motif. Furthermore, depletion of VAP-A by RNA interference resulted in mislocalization of protrudin as well as in inhibition of neurite outgrowth induced by nerve growth factor in rat pheochromocytoma PC12 cells. These defects resulting from depletion of endogenous rat VAP-A in PC12 cells were corrected by forced expression of (RNA interference-resistant) human VAP-A but not by VAP-A mutants that have lost the ability to interact with protrudin. These results suggest that VAP-A is an important regulator both of the subcellular localization of protrudin and of its ability to stimulate neurite outgrowth.
The molecular mechanisms that underlie neurite formation include both cytoskeletal remodeling and membrane trafficking. Membrane components are transported in a directional manner within the cell by a membrane recycling system, resulting in expansion of the surface area of the neurite. The small GTPase Rab11 regulates membrane recycling and constitutive exocytosis (1), and it is thought to contribute to neurite formation through regulation of directional membrane transport.
We have recently identified protrudin as a key regulator of Rab11-dependent membrane trafficking during neurite extension. Protrudin interacts with FKBP38 (also known as FKBP8) (2), which is a member of the immunophilin family of proteins that bind the immunosuppressant drug FK506 (3). FKBPs are multifunctional proteins that regulate the folding or export of other proteins as a result of their peptidyl-prolyl cis-trans-isomerase activity (4). Protrudin was found to interact with FKBP38, but not with other FKBP proteins such as FKBP12 or FKBP52 (5). Protrudin is hyperphosphorylated in Fkbp38-/- mice, which manifest abnormal extension of nerve fibers (5).
Protrudin contains a Rab11-binding domain (RBD11), two transmembrane domains (TM1 and TM2),2 an FFAT (two phenylalanines in an acidic tract) motif (6), a coiled-coil domain, and a FYVE domain (7). These structural characteristics suggested that protrudin might function in membrane trafficking, particularly in membrane recycling. The gene encoding ZFYVE27 (a synonym of human protrudin) was recently found to be mutated in a German family with an autosomal dominant form of hereditary spastic paraplegia (AD-HSP), which is characterized by selective degeneration of axons (8). The phenotype of the affected individuals is similar to that of patients with AD-HSP caused by mutation of spastin, a protein implicated in neuronal vesicular trafficking (9), and protrudin was shown to interact with spastin (8). These findings support the notion that protrudin plays a key role in Rab11-mediated directional membrane transport during neurite formation.
The subcellular localization of protrudin is dynamic. Whereas it is localized to the endoplasmic reticulum (ER) under basal conditions, nerve growth factor (NGF) triggers the translocation of protrudin from the ER, via recycling endosomes, to the tip of membrane protrusions in neuronal cells. Given that the FFAT motif is thought to serve as an ER targeting signal (6), this motif might be expected to contribute both to the localization of protrudin to the ER and to the regulation of neurite formation by this protein. The FFAT motif (consensus amino acid sequence of EFFDAXE, where X is any amino acid) is present in several lipid-binding proteins that are implicated in the transfer of lipids between the ER and other organelles such as the Golgi apparatus (10, 11). Vesicle-associated membrane protein-associated protein (VAP) interacts with these lipid-binding proteins through their FFAT motifs (6, 11, 12). The VAP-A and VAP-B isoforms of mammalian VAP are ER-resident type II membrane proteins (13) that are encoded by different genes (14); VAP-C is a splicing variant of VAP-B that lacks the membrane-spanning domain. VAP-A and VAP-B share ∼60% amino acid sequence identity, form homo- or heterodimers, and are expressed in many tissues (14-16). In addition to their localization to the ER (16), VAP-A and VAP-B are present in a wide range of intracellular membranes or membrane structures, including the Golgi, the ER-Golgi intermediate compartment (17), tight junctions (18), neuromuscular junctions (19), recycling endosomes, and the plasma membrane (20).
We have now identified VAP-A and VAP-B as proteins that interact with protrudin. Protrudin preferentially interacts with VAP-A via its FFAT motif, and this motif was found to be required for the protrudin-dependent formation of membrane protrusions in HeLa cells. In addition, depletion of VAP-A by RNA interference resulted in inhibition of NGF-induced neurite outgrowth in the PC12 rat pheochromocytoma cell line. This inhibition of neurite outgrowth was reversed by expression of human VAP-A but not by that of VAP-A mutants that have lost the ability to bind to protrudin. These results suggest that interaction of protrudin with VAP-A is important both for its ER retention and for its ability to stimulate neurite formation.
EXPERIMENTAL PROCEDURES
Construction of Plasmids—Construction of vectors encoding human protrudin and FKBP52 was described previously (2, 21). Complementary DNAs encoding mutants of protrudin were generated by the PCR with Prime Star polymerase (Takara, Ohtsu, Japan); those encoding VAP-A or VAP-B were generated by PCR from a human kidney cDNA library; and that encoding RAMP4 was generated by PCR from a mouse thyroid cDNA library. The VAP or RAMP4 cDNAs were subcloned into the pEF-BOS-2× HA or pEF-BOS-2× Myc vectors (kindly provided by Shigekazu Nagata, Kyoto University, Japan) or into pGEX-6P (Amersham Biosciences). DNA fragments encoding stem-loop-type short hairpin RNAs (shRNAs) specific for rat VAP mRNAs (VAP-A-shRNA 1, 5′-GGTAGCACATTCGGATAAACC-3′; VAP-A-shRNA 2, 5′-GCCGTGTCCTTCAGAGATAAT-3′; VAP-A-shRNA 3, 5′-GGATTCTTTCTAGGGAAATTC-3′; VAP-B-shRNA 1, 5′-GGGTCATTATAGGGAAGATCG-3′; VAP-B-shRNA 2, 5′-GGTGGTGCTGTTCTTTATTGT-3′; VAP-B-shRNA 3, 5′-GCCCGACACTTCCGATATGGA-3′); or human VAP-B mRNA (5′-GGGAGGAGAACAAGCAGTTCA-3′) were synthesized, attached to the U6 promoter, and subcloned into the vector pIRES-Venus-B (Venus cDNA was kindly provided by Atsushi Miyawaki, RIKEN, Japan), which contains an internal ribosome entry site and encodes the reporter protein Venus.
Protein Identification by Liquid Chromatography-MS/MS Analysis—Human protrudin tagged with the FLAG epitope at its NH2 terminus was transiently expressed in HEK293T cells and then purified together with associated proteins from cell lysates by immunoaffinity chromatography with anti-FLAG. Protrudin-associated proteins were digested with Lys-C endoproteinase, and the resulting peptides were analyzed with a nanoscale liquid chromatography-MS/MS system as described previously (22, 23). The peptide mixture was applied to a Mightysil-PR-18 (particle size, 1 μm; Kanto Chemical, Tokyo, Japan) fritless column (45 × 0.150-mm inner diameter) and fractionated over 30 min at a flow rate of 50 nl/min with a 0 to 40% gradient of acetonitrile in 0.1% formic acid. Eluted peptides were sprayed directly into a quadropole time-of-flight hybrid mass spectrometer (Q-Tof Ultima; Micromass, Manchester, UK). Mass spectrometry and MS/MS spectra were obtained in a data-dependent mode. Up to four precursor ions with an intensity above a threshold of 10 counts/s were selected for MS/MS analysis from each survey scan. All of the MS/MS spectra were compared with protein sequences in Swiss Prot and RefSeq (NCBI) with the use of batch processes of the Mascot software package (Matrix Science, London, UK). The criteria for match acceptance included the following: (i) if the match score exceeded the threshold by 10, identification was accepted without further consideration; (ii) if the difference between the score and the threshold was <10, or if a protein was identified on the basis of a single matched MS/MS spectrum, we manually confirmed the raw data before acceptance; and (iii) peptides assigned by fewer than three y series ions and those with a charge of +4 were eliminated regardless of their scores.
Cell Culture and Transfection—HEK293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). PC12 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum; they were treated with recombinant human NGF (Millipore, Billerica, MA) at 100 ng/ml in RPMI 1640 supplemented with 1% fetal bovine serum. PC12 cells were transfected with the use of a Nucleofector system (Amaxa Biosystems, Cologne, Germany), whereas other cell types were transfected with the use of the FuGENE 6 reagent (Roche Applied Science).
Antibodies—Rabbit polyclonal antibodies (for immunoblot analysis and immunoprecipitation) and a mouse monoclonal antibody (for immunofluorescence staining) specific for protrudin were generated in response to a His6-tagged recombinant protein corresponding to amino acids 206-335 of human protrudin. A mouse monoclonal antibody to VAP-A was obtained from BD Biosciences (San Jose, CA), and rabbit polyclonal antibodies to VAP-B were kindly provided by Masaaki Matsuoka (Keio University, Japan). A mouse monoclonal antibody to glutathione S-transferase (GST) was obtained from MBL (Nagoya, Japan); rabbit polyclonal antibodies (H-15) to His6 were from Santa Cruz Biotechnology (Santa Cruz, CA); antibodies to FLAG (mouse monoclonal M2 and rabbit polyclonal) and a mouse monoclonal antibody (9E10) to Myc were from Sigma; a mouse monoclonal antibody (HA11) to the HA epitope was from Research Diagnostics (Flanders, NJ); and rabbit polyclonal antibodies (HA-Y11) to HA were from Funakoshi (Tokyo, Japan). Rabbit polyclonal antibodies to calnexin were obtained from Stressgen (Victoria, Canada); a mouse monoclonal antibody to HSP90 (heat shock protein of 90 kDa) was from BD Biosciences; and a mouse monoclonal antibody to protein-disulfide isomerase (PDI) was from Affinity Bioreagents (Golden, CO). Alexa 488- or Alexa 546-conjugated goat antibodies to mouse or rabbit IgG were from Molecular Probes (Invitrogen).
Immunoblot Analysis and Immunoprecipitation—The cells were lysed in a solution containing 40 mm HEPES-NaOH (pH 7.6), 150 mm NaCl, 10% glycerol, 0.5% Triton X-100, 10 mm MgCl2, 1 mm Na3VO4, 25 mm NaF, 1 mm phenylmethylsulfonyl fluoride, aprotinin (10 μg/ml), leupeptin (10 μg/ml), and 10 μm MG132. After incubation for 10 min at 4 °C, the lysate was centrifuged at 20,400 × g for 10 min at 4 °C, and the protein concentration of the resulting supernatant was determined with the Bradford assay (Bio-Rad). Whole mouse brain was homogenized by 10 strokes (900 rpm) of a Potter homogenizer in a solution containing 20 mm HEPES-NaOH (pH 7.6), 0.32 m sucrose, 1 mm Na3VO4, 25 mm NaF, aprotinin (10 μg/ml), leupeptin (10 μg/ml), 10 μm MG132, 1 mm phenylmethylsulfonyl fluoride, and 1 mm EDTA. The homogenate was centrifuged at 1000 × g for 5 min at 4 °C, and the resulting supernatant was centrifuged at 60,000 × g for 2 h at 4 °C. The crude microsomal pellet was resuspended in a solution containing 40 mm HEPES-NaOH (pH 7.6), 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 10 mm MgCl2, 1 mm Na3VO4, 25 mm NaF, 1 mm phenylmethylsulfonyl fluoride, aprotinin (10 μg/ml), leupeptin (10 μg/ml), and 10 μm MG132, incubated for 10 min at 4 °C, and then centrifuged at 20,400 × g for 1 min at 4 °C. The protein concentration of the resulting supernatant was then determined with the Bradford assay.
The cell and mouse brain extracts were subjected to SDS-polyacrylamide gel electrophoresis, and the separated proteins were transferred to an Immobilon-P membrane (Millipore) and probed with primary antibodies. Immune complexes were detected with Super Signal reagents (Thermo Scientific, Rockford, IL). For immunoprecipitation, cell or mouse brain extracts were incubated with anti-FLAG or anti-protrudin and with protein G-Sepharose 4 Fast Flow (Amersham Biosciences).
Immunofluorescence Staining—HeLa cells grown on glass coverslips were transfected with the use of the FuGENE 6 reagent and subsequently prepared for immunostaining. In brief, the cells were fixed for 15 min at room temperature with 4% formaldehyde in phosphate-buffered saline (PBS) and were then incubated for 1 h at room temperature first with primary antibodies in PBS containing 0.1% bovine serum albumin and 0.1% saponin and then with Alexa 488- or Alexa 546-labeled goat secondary antibodies at a dilution of 1:2000. The cells were finally stained with Hoechst 33258 (Wako, Osaka, Japan), covered with a drop of GEL/MOUNT (Biomeda, Foster City, CA) and examined with a confocal fluorescence microscope (Radiance 2000, Bio-Rad).
Transfected PC12 cells were grown on glass coverslips coated with poly-l-lysine and subsequently prepared for immunostaining. In brief, the cells were fixed for 10 min at room temperature with 4% formaldehyde in PBS and were then incubated for 1 h at room temperature first with primary antibodies in PBS containing 0.1% bovine serum albumin and 0.1% saponin and then with Alexa 546-labeled goat secondary antibodies at a dilution of 1:2000. The cells were then covered with a drop of GEL/MOUNT and examined with a confocal fluorescence microscope (Radiance 2000).
In Vitro Binding Assay—Recombinant GST- or His6-tagged proteins were expressed in and purified from Escherichia coli. Recombinant His6-protrudin(207-409) (1 μg in 12 μl of a solution containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) and GST-VAP-A or mutants thereof (0.5 μg in 600 μl of a solution containing 20 mm Tris-HCl, pH 7.5, and 150 mm NaCl) were mixed and then incubated at 4 °C for 1 h with rotation. After the addition of glutathione-Sepharose 4B beads (Amersham Biosciences), the mixture was incubated for an additional 1 h at 4 °C with rotation. The beads were then washed twice with 50 volumes of Tris-buffered saline, and the bound proteins were subjected to immunoblot analysis.
Quantitation of Process Formation—Transfected cells with processes whose length was greater than the diameter of the nucleus were counted. For evaluation of the effects of FKBP52, protrudin, or protrudin mutants, the cells were immunostained with the M2 antibody to FLAG, whereas the effects of VAP-A or VAP-B were evaluated by immunostaining with the HA-Y11 antibody to HA. Ten nonoverlapping photomicrographs of each sample were examined for cell protrusion with a confocal fluorescence microscope (Radiance 2000).
RESULTS
Protrudin Interacts with VAP-A via Its FFAT Motif—To investigate the physiological role of protrudin, we adopted a proteomics approach to identify proteins with which it is physically associated in cells. Human protrudin tagged with the FLAG epitope at its NH2 terminus was expressed in HEK293T cells and then purified together with associated proteins from cell lysates by immunoaffinity chromatography with anti-FLAG. Proteins in the column eluate were digested with Lys-C endoproteinase, and the resulting peptide fragments were analyzed directly with a highly sensitive liquid chromatography-MS/MS system. The protrudin-associated proteins identified by this approach included VAP-A and VAP-B/C (Table 1).
TABLE 1.
Identification of protrudin-associated proteins by proteomics analysis
HEK293T cells transiently expressing 3× FLAG-tagged human protrudin were subjected to immunoaffinity chromatography with anti-FLAG. Proteins in the column eluate were digested with Lys-C endoproteinase, and the resulting peptide fragments were analyzed directly with a highly sensitive liquid chromatography-MS/MS system. The proteins reproducibly detected in two independent experiments are listed. The percentage of sequence coverage for each protein is also shown.
| Protein | Sequence coverage |
|---|---|
| % | |
| Vesicle associated membrane protein-associated protein A (VAP-A) | 14.5 |
| Vesicle associated membrane protein-associated protein B and C (VAP-B/C) | 14.4 |
| T-complex protein 1, δ subunit (TCPD) | 9.2 |
| T-complex protein 1, γ subunit (TCPG) | 7.7 |
| T-complex protein 1, ε subunit (TCPE) | 6.3 |
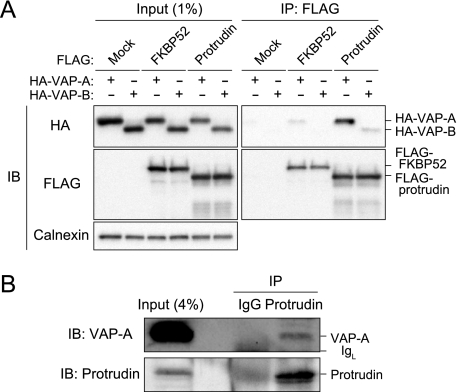
To confirm the interaction between protrudin and VAP, we performed a coimmunoprecipitation assay. Immunoprecipitates were prepared with anti-FLAG from lysates of HEK293T cells transfected with expression vectors both for FLAG-tagged protrudin (or FLAG-tagged FKBP52 (24) as a control) and for HA-tagged VAP-A or VAP-B. We previously showed that protrudin interacts with FKBP38, but not with FKBP12 or FKBP52 (5). The resulting precipitates were then subjected to immunoblot analysis with anti-HA and anti-FLAG (Fig. 1A). Both VAP-A and VAP-B were detected in the immunoprecipitates prepared from the cells expressing FLAG-protrudin, but the efficiency of immunoprecipitation for VAP-A was much greater than that for VAP-B.
FIGURE 1.
Interaction between protrudin and VAP. A, extracts of HEK293T cells transiently transfected with expression vectors for 3× FLAG-tagged protrudin or FKBP52 (negative control) and for 2× HA-tagged VAP-A or VAP-B were subjected to immunoprecipitation (IP) with anti-FLAG. The resulting precipitates, as well as a portion (1% of the input for immunoprecipitation) of the cell extracts, were subjected to immunoblot analysis (IB) with anti-HA, anti-FLAG, or anti-calnexin (loading control). B, mouse brain extract was subjected to immunoprecipitation with anti-protrudin or control rabbit IgG, and the resulting precipitates, as well as a portion (4% of the input for immunoprecipitation) of the tissue extract, were subjected to immunoblot analysis with anti-VAP-A and anti-protrudin. IgL, Ig light chain.
Similar analysis was performed to detect the potential interaction between endogenous proteins. Immunoprecipitates prepared from mouse brain extracts with anti-protrudin were subjected to immunoblot analysis with anti-VAP-A and anti-protrudin. Endogenous VAP-A was coprecipitated with endogenous protrudin (Fig. 1B). Together, these various data indicated that protrudin interacts with VAP-A in the physiological setting.
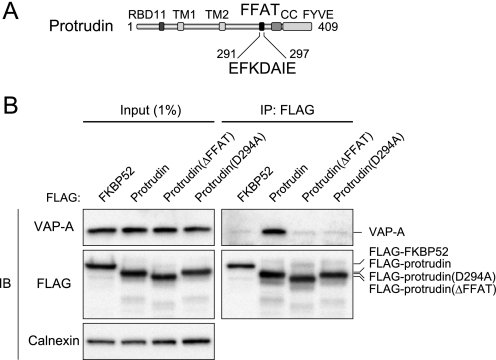
Given that VAP interacts with the FFAT motif of certain proteins (11, 25), we examined whether the binding of VAP-A to protrudin might be dependent on the FFAT motif of protrudin. We constructed two mutants of human protrudin, ΔFFAT and D294A (Fig. 2A); the former lacks the entire FFAT motif (EFKDAIE), and the latter contains Ala instead of Asp294, which is required for binding of proteins to the FFAT motif (6). The amount of endogenous VAP-A associated with either mutant in HEK293T cells was greatly reduced compared with that associated with the wild-type protein, even though the expression levels of the protrudin mutants were similar to that of the wild-type protein (Fig. 2B). The residual interaction observed between VAP-A and the protrudin mutants was likely attributable to the fact that VAP and FFAT motif-containing proteins form a 2:2 tetramer (26), with the result that the protrudin mutants may be present in complexes of the endogenous wild-type protrudin with VAP-A. These data thus suggested that the interaction of protrudin with VAP-A is dependent on its FFAT motif.
FIGURE 2.
Role of the FFAT motif in the interaction of protrudin with VAP-A. A, domain organization of human protrudin. B, extracts of HEK293T cells transiently transfected with an expression vector for 3× FLAG-tagged protrudin, protrudin mutants (ΔFFAT or D294A), or FKBP52 were subjected to immunoprecipitation with anti-FLAG. The resulting precipitates, as well as a portion (1% of the input for immunoprecipitation) of the cell extracts, were subjected to immunoblot analysis with anti-VAP-A, anti-FLAG, or anti-calnexin.
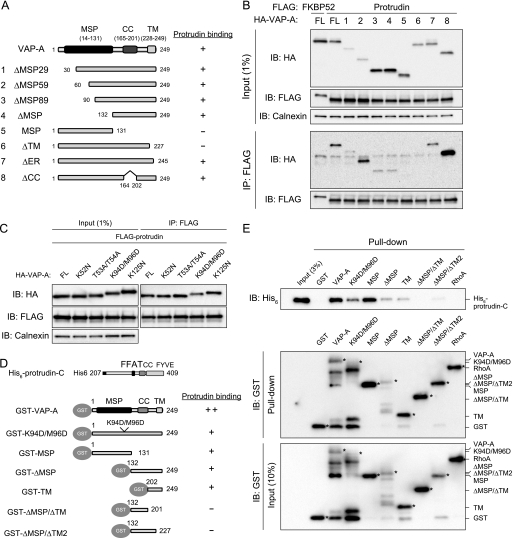
VAP-A Interacts with Protrudin in the ER via Its MSP and TMs—VAP-A contains an major sperm protein (MSP) domain, a coiled-coil (CC) domain, and a single TM. We next investigated which of these domains are required for interaction with protrudin by generating a series of deletion mutants of VAP-A (ΔMSP29, ΔMSP59, ΔMSP89, ΔMSP, MSP, ΔTM, ΔER, and ΔCC) and examining their ability to associate with FLAG-protrudin in a coimmunoprecipitation assay (Fig. 3, A and B). Deletion of 59 amino acids from the NH2 terminus of VAP-A (ΔMSP59) did not appear to affect its interaction with protrudin, whereas deletion of 89 amino acids from the NH2 terminus (ΔMSP89) or of the entire MSP domain (ΔMSP) greatly reduced the efficiency of such binding. The COOH-terminal transmembrane domain of VAP-A appeared essential for the interaction with protrudin, given that the deletion mutants MSP and ΔTM did not show binding. The COOH terminus of VAP-A contains a KDEL-like sequence (KFIL), which functions as an ER retention signal (27). A mutant (ΔER) that lacks this sequence associated with protrudin, albeit with a reduced efficiency. The coiled-coil domain, which is thought to contribute to protein-protein interaction, was not necessary for binding of VAP-A to protrudin. We also prepared a series of point mutants of VAP-A. In particular, mutation of Lys94 and Met96 (equivalent to Lys87 and Met89 of the rat protein), residues that have been shown to be critical for binding to other FFAT motif-containing proteins, attenuated (but did not abolish) the interaction between VAP-A and protrudin (Fig. 3C), whereas other mutations (K52N, T53A/T54A, or K125N) corresponding to residues that are important for binding of the yeast VAP homolog Scs2p to other FFAT motif-containing proteins (28) did not affect the interaction of VAP-A with protrudin.
FIGURE 3.
Delineation of the regions of VAP-A responsible for interaction with protrudin. A, domain organization of human VAP-A and structure of deletion mutants thereof. A summary of the ability of the mutants to bind protrudin as determined in B is shown on the right. B, full-length (FL) VAP-A or the mutants thereof shown in A fused at their NH2 termini to the 2× HA tag were expressed in HEK293T cells together with 3× FLAG-tagged protrudin or FKBP52. The cell extracts were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as a portion (1% of the input for immunoprecipitation) of the cell extracts, were subjected to immunoblot (IB) analysis with anti-HA, anti-FLAG, or anti-calnexin. C, full-length human VAP-A and the indicated mutants thereof fused at their NH2 termini to the 2× HA tag were expressed together with 3× FLAG-tagged protrudin in HEK293T cells. The cell extracts were subjected to immunoprecipitation with anti-FLAG, and the resulting precipitates, as well as a portion (1% of the input for immunoprecipitation) of the cell extracts, were subjected to immunoblot analysis with anti-HA, anti-FLAG, or anti-calnexin. D, structure of VAP-A mutants fused to GST at their NH2 termini and a summary of their ability to bind to a His6-tagged COOH-terminal fragment of protrudin comprising residues 207-409 (protrudin-C) as determined in E. E, His6-tagged protrudin-C was incubated with the GST-tagged VAP-A mutants shown in D or with GST-RhoA or GST as negative controls, and the binding mixtures were then subjected to precipitation with glutathione-conjugated beads. The bead-bound proteins (Pull-down), as well as a portion (3 or 10% of the input for precipitation) of the binding mixtures, were subjected to immunoblot analysis with anti-His6 and anti-GST. The asterisks indicate bands corresponding to the GST-tagged proteins.
To examine the direct interaction between VAP-A and protrudin in vitro, we performed a pulldown assay. Recombinant GST-tagged VAP-A and six mutants thereof were produced in bacteria and tested for their ability to bind to recombinant His6-tagged protrudin (Fig. 3, D and E). The VAP-A mutants K94D/M96D, MSP, ΔMSP, and TM bound to protrudin, although the efficiency of binding appeared to be lower than that with full-length VAP-A. In contrast, the mutants ΔMSP/ΔTM and ΔMSP/ΔTM2 (as well as the negative controls GST and GST-RhoA) did not interact with protrudin. These results suggested that both the MSP and transmembrane domains of VAP-A are required for binding to protrudin.
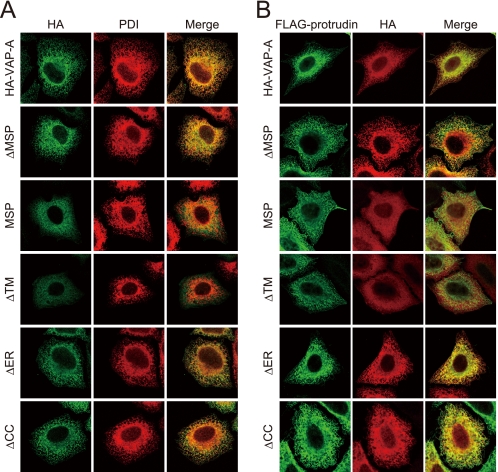
The VAP-A mutant MSP did not interact with protrudin in vivo, although it did so in vitro. One possible explanation of this discrepancy was that the subcellular distribution of the VAP-A mutant differs from that of protrudin. We therefore performed immunofluorescence analysis of HeLa cells expressing various HA-tagged VAP-A constructs and FLAG-tagged protrudin. The immunofluorescence signals of wild-type VAP-A as well as of VAP-A mutants that retain an intact transmembrane domain merged with those of the ER marker protein PDI (Fig. 4A) (29), suggesting that they reside in the ER. These VAP-A constructs also colocalized with protrudin (Fig. 4B). In contrast, VAP-A mutants that lack an intact transmembrane domain (MSP and ΔTM) were distributed throughout the cytoplasm and only partially colocalized with protrudin or PDI. The distribution of protrudin appeared to be in part independent of coexpressed VAP-A mutants, probably because a substantial amount of endogenous VAP-A is present in these cells; protrudin presumably associates with the endogenous VAP-A and thereby localizes to the ER. These results thus suggested that both the MSP domain and the transmembrane domain of VAP-A contribute to the interaction with the FFAT motif of protrudin and that the transmembrane domain is also necessary for ER retention, which is a prerequisite for the interaction with protrudin.
FIGURE 4.
Subcellular localization of VAP-A mutants. A, HeLa cells expressing 2× HA-tagged VAP-A and deletion mutants thereof were fixed and processed for immunofluorescence analysis with anti-HA (green) and anti-PDI (red). The cells were examined with a confocal microscope. The merged images are also shown. B, HeLa cells expressing 3× FLAG-protrudin and 2× HA-tagged VAP-A or the indicated mutants thereof were fixed and processed for immunofluorescence staining with anti-FLAG (green) and anti-HA (red). The merged images are also shown.
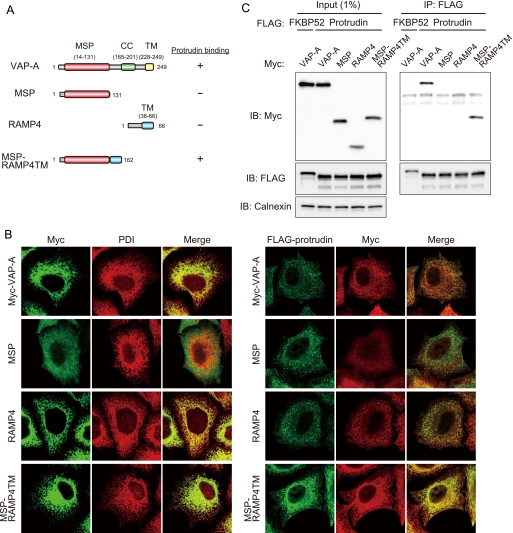
To examine whether the MSP domain of VAP-A is intrinsically capable of interacting with protrudin in vivo, we generated a chimeric protein, designated MSP-RAMP4TM (Fig. 5A). RAMP4 is a small tail-anchored protein that exposes the NH2 and COOH termini to the cytoplasmic and lumenal sides of the ER membrane, respectively (30). MSP-RAMP4TM was constructed by fusion of the MSP domain of VAP-A and the COOH-terminal transmembrane domain of RAMP4. Immunofluorescence analysis of HeLa cells expressing FLAG-tagged protrudin as well as Myc epitope-tagged forms of VAP-A, MSP, RAMP4, or MSP-RAMP4TM revealed that VAP-A, RAMP4, and MSP-RAMP4TM colocalized with the ER marker protein PDI (Fig. 5B), suggesting that these proteins reside in the ER. VAP-A and MSP-RAMP4TM also colocalized with protrudin (Fig. 5B). We next examined the ability of MSP-RAMP4TM to associate with protrudin in a coimmunoprecipitation assay. VAP-A and MSP-RAMP4TM interacted with protrudin, whereas MSP and RAMP4 did not (Fig. 5C). There results thus suggested that the MSP domain of VAP-A alone is able to interact with protrudin if it is localized to the ER. They are also consistent with the in vitro binding data showing that the MSP domain of VAP-A associates with protrudin in solution (Fig. 3E).
FIGURE 5.
Role of the MSP domain in the interaction of VAP-A with protrudin. A, structure of human VAP-A, the MSP mutant thereof, RAMP4, and the chimeric protein MSP-RAMP4TM as well as a summary of their abilities to bind protrudin as determined in C. B, left panel, HeLa cells expressing 2× Myc-tagged VAP-A, MSP, RAMP4, or MSP-RAMP4TM were fixed and processed for immunofluorescence analysis with anti-Myc (green) and anti-PDI (red). The merged images are also shown. B, right panel, HeLa cells expressing 3× FLAG-protrudin and 2× Myc-tagged VAP-A, MSP, RAMP4, or MSP-RAMP4TM were fixed and processed for immunofluorescence staining with anti-FLAG (green) and anti-Myc (red). The merged images are also shown. C, VAP-A, MSP, RAMP4, or MSP-RAMP4TM fused at their NH2 termini to the 2× Myc tag were expressed in HEK293T cells together with 3× FLAG-tagged protrudin or FKBP52. The cell extracts were subjected to immunoprecipitation (IP) with anti-FLAG, and the resulting precipitates, as well as a portion (1% of the input for immunoprecipitation) of the cell extracts, were subjected to immunoblot analysis with anti-Myc, anti-FLAG, or anti-calnexin. IB, immunoblot.
Mutation of the FFAT Motif Attenuates Protrudin Function—Expression of FLAG-protrudin in HeLa cells results in the generation of long processes (2) (Fig. 6A). To evaluate the effect of the interaction of protrudin with VAP-A on the process forming activity of protrudin, we first examined process formation in HeLa cells expressing FFAT mutants of protrudin (ΔFFAT and D294A) that have lost the ability to bind VAP-A. Protrusion was observed in <5% of cells expressing FKBP52 (negative control), whereas ∼20% cells expressing wild-type protrudin showed process formation. In contrast, the efficiency of process formation was significantly reduced in cells expressing either FFAT mutant of protrudin compared with that in cells expressing the wild-type protein (Fig. 6, A-C). The residual process forming activity of the FFAT mutants was likely attributable to the formation of VAP-protrudin complexes containing both wild-type and mutant protrudin, as mentioned above. These results suggested that the FFAT motif of protrudin is necessary for the ability of the protein to induce process formation.
FIGURE 6.
Mutation of the FFAT motif impairs the ability of protrudin to induce process formation in HeLa cells. A, HeLa cells transfected with an expression vector for 3× FLAG-tagged protrudin or the ΔFFAT mutant thereof were subjected to immunofluorescence staining with anti-FLAG (green). The nuclei were also stained with Hoechst 33258 (blue). B and C, HeLa cells transfected with expression vectors for 3× FLAG-tagged FKBP52, protrudin, protrudin(ΔFFAT), or protrudin(D294A) for 36 h (B) or 48 h (C) were stained with anti-FLAG (green), and the proportion of transfected cells with protrusions was determined. The data are the means ± S.E. of values from three independent experiments. p < 0.01 (one-way analysis of variance). *, p < 0.05 (Tukey-Kramer test). D, HeLa cells transfected with expression vectors for 3× FLAG-tagged protrudin and 2× HA-tagged VAP-A or VAP-B (or the corresponding empty vectors) were stained with anti-FLAG (green) or anti-HA (red), and the proportion of transfected cells with protrusions was determined. The data are the means ± S.E. of values from three independent experiments. *, p < 0.005 (Student's t test). E, HeLa cells were transfected for 48 h with an expression vector for VAP-A-shRNA 3 (or with the corresponding empty vector, pIRES-Venus-B) as well as with a vector for 3× FLAG-protrudin, after which cell extracts were subjected to immunoblot (IB) analysis with anti-VAP-A, anti-VAP-B, anti-FLAG, or anti-calnexin. F, quantitation of protrusion formation in cells treated as in E. The data are the means ± S.E. of values from three independent experiments. Independent experiments were performed on different days (B-D and F).
We next examined the effect of VAP overexpression on process formation. Neither VAP-A nor VAP-B alone induced process formation in HeLa cells (Fig. 6D). However, the expression of VAP-A slightly but significantly increased the stimulatory effect of protrudin on process formation. VAP-B did not exhibit such an effect, consistent with the observation that the extent of binding of VAP-B to protrudin was much reduced compared with that of VAP-A (Fig. 1A).
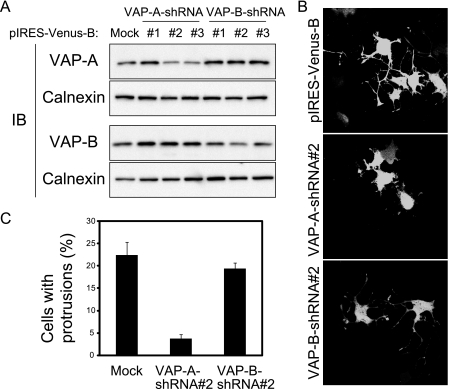
We also examined the effect of VAP-A depletion on the process forming activity of protrudin by RNA interference with a VAP-A shRNA (VAP-A-shRNA 3) that targets both rat and human VAP-A mRNAs (Fig. 6E). Depletion of VAP-A resulted in marked inhibition of the process forming activity of protrudin (Fig. 6F). These results thus suggested that the interaction of protrudin with VAP-A is important for the process forming activity of protrudin.
VAP-A Is Essential for Protrudin-dependent Neurite Outgrowth—We next investigated whether VAP might be necessary for neurite extension in neurons by examining NGF-induced neurite outgrowth in PC12 cells depleted of VAP-A or VAP-B by RNA interference. Three different shRNAs specific for VAP-A mRNA or for VAP-B mRNA were tested for their ability to deplete the corresponding protein (Fig. 7A), and the most effective shRNAs (VAP-A-shRNA 2 and VAP-B-shRNA 2) were examined for their effects on NGF-induced neurite formation in PC12 cells. Depletion of VAP-A, but not that of VAP-B, resulted in marked inhibition of NGF-induced neurite outgrowth (Fig. 7, B and C). Moreover, depletion of VAP-A in PC12 cells eventually led to cell death (data not shown), as previously observed in rat primary neurons (31) These results thus suggested that VAP-A is essential for neurite formation and cell survival in neurons.
FIGURE 7.
Depletion of VAP-A results in inhibition of NGF-induced neurite outgrowth in PC12 cells. A, PC12 cells were transfected with expression vectors encoding shRNAs specific for VAP-A or VAP-B mRNAs, or with the corresponding empty vector (pIRES-Venus-B), for 48 h, after which cell extracts were prepared and subjected to immunoblot (IB) analysis with anti-VAP-A, anti-VAP-B, or anti-calnexin. B, PC12 cells transfected with the indicated vectors as in A were stimulated with NGF for 24 h and then examined for Venus fluorescence. C, the proportion of transfected cells with protrusions in experiments similar to that shown in B was quantified. The data are the means ± S.E. of values from three independent experiments, which were performed on different days.
Given that human and rat VAP-A proteins share ∼97% amino acid sequence identity, we investigated whether the defect in neurite outgrowth in PC12 cells depleted of VAP-A might be corrected by introduction of human VAP-A. Human VAP-A was resistant to shRNA-mediated interference because of a 4-nucleotide difference in the sequence corresponding to VAP-A-shRNA 2 between the human and rat mRNAs. Human VAP-A indeed restored the ability of NGF to induce neurite formation in PC12 cells that had been depleted of endogenous rat VAP-A (Fig. 8). Cell death induced by VAP-A depletion was also suppressed by expression of human VAP-A (data not shown). These results indicated that human VAP-A is functionally interchangeable with rat VAP-A.
FIGURE 8.
Interaction of protrudin with VAP-A is required for NGF-induced neurite outgrowth in PC12 cells. A, PC12 cells were transfected for 48 h with an expression vector encoding both VAP-A-shRNA 2 and Venus as well as with a vector for the indicated 2× HA-tagged mutants of human VAP-A, after which the cells were stimulated with NGF for 48 h and then subjected to immunofluorescence analysis with anti-HA (red). Venus fluorescence images (green) and merged images are also shown. B, quantitation of protrusion formation in cells treated as in A. The data are the means ± S.E. of values from three independent experiments, which were performed on different days.
We next tested the ability of VAP-A mutants to restore the induction of neurite outgrowth by NGF in PC12 cells depleted of endogenous VAP-A. Whereas the human VAP-A mutants K94D/M96D and ΔMSP, both of which interact with protrudin, restored NGF-induced neurite outgrowth to the same extent as did wild-type VAP-A, the mutant ΔTM, which does not reside in the ER and has lost the ability to bind to protrudin, failed to restore this effect of NGF (Fig. 8). These results suggested that the interaction of protrudin with VAP-A is likely indispensable for the neurite-extending function of protrudin. Alternatively, it remains possible that the failure of the ΔTM mutant to restore NGF-induced neurite formation in VAP-A-depleted PC12 cells might be explained by the transmembrane domain being essential for VAP-A localization and function rather than for interaction with protrudin.
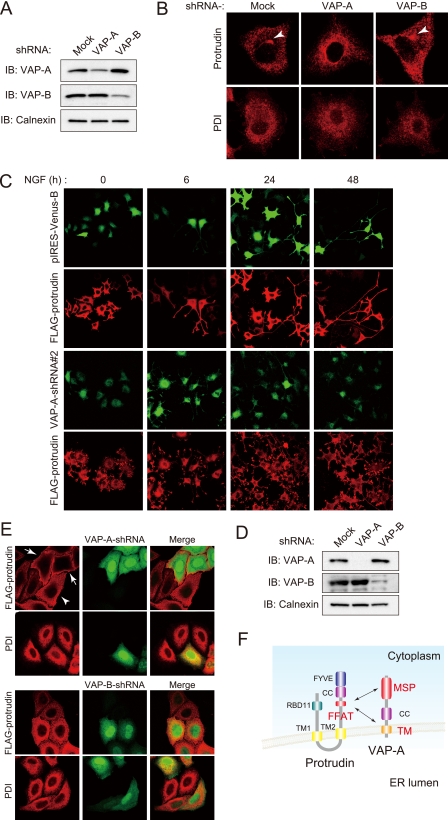
VAP-A Is Required for Protrudin Localization—Protrudin resides in the ER of PC12 cells in the absence of NGF, whereas it is translocated to recycling endosomes in response to NGF stimulation (2). To investigate whether VAP-A might affect the subcellular distribution of protrudin in PC12 cells, we examined cells depleted of VAP-A or VAP-B by RNA interference (Fig. 9A). Immunofluorescence staining of control cells stimulated with NGF for 6 h revealed that endogenous protrudin was concentrated in the region containing recycling endosomes, whereas such transport of protrudin from the ER to recycling endosomes was not evident in cells depleted of VAP-A (Fig. 9B). Examination of VAP-A-depleted cells expressing FLAG-protrudin at longer times of exposure to NGF revealed that FLAG-protrudin became diffusely distributed throughout the cell body and that neurite extension was inhibited (Fig. 9C). In contrast, depletion of VAP-B affected neither the localization of protrudin nor neurite extension (Fig. 9B and data not shown). We examined the effect of VAP-A or VAP-B depletion (Fig. 9D) on the distribution of FLAG-protrudin in more detail in HeLa cells. Immunofluorescence corresponding to FLAG-protrudin was apparent only in the periphery of cells depleted of VAP-A, whereas it appeared to be localized to the ER in control cells and VAP-B-depleted cells (Fig. 9E). The localization of PDI was unaffected by VAP-A or VAP-B depletion. These results suggested that VAP-A is indispensable for ER retention of protrudin in HeLa cells, which is important for its neurite extending function.
FIGURE 9.
Depletion of VAP-A results in an abnormal subcellular distribution of protrudin. A, PC12 cells were transfected for 48 h with an expression vector for VAP-A-shRNA 2 or VAP-B-shRNA 2 or with the corresponding empty vector (pIRES-Venus-B). The cell extracts were then prepared and subjected to immunoblot (IB) analysis with anti-VAP-A, anti-VAP-B, or anti-calnexin. B, PC12 cells transfected as in A were stimulated with NGF for 6 h and then subjected to immunofluorescence analysis with anti-protrudin (red) or anti-PDI (red). Arrowheads indicate the pericentrosomal region. C, PC12 cells transfected as in A but in the additional presence of an expression vector for 3× FLAG-protrudin were stimulated with NGF for the indicated times and then subjected to immunofluorescence staining with anti-FLAG (red). The cells were also monitored for Venus fluorescence (green). D, HeLa cells were transfected for 48 h with an expression vector for VAP-A-shRNA 3 or for an shRNA specific for human VAP-B (or with the corresponding empty vector, pIRES-Venus-B) as well as with a vector for 3× FLAG-protrudin, after which cell extracts were subjected to immunoblot analysis with anti-VAP-A, anti-VAP-B, or anti-calnexin. E, HeLa cells transfected as in D were stained with anti-FLAG (red) or anti-PDI (red). The cells were also monitored for Venus fluorescence (green). Arrows indicate cells depleted of VAP-A (Venus positive); the arrowhead indicates a cell not depleted of VAP-A (Venus negative). The merged images are also shown. F, a model of VAP-A function in the regulation of protrudin.
DISCUSSION
Protrudin was discovered on the basis of its ability to induce process formation in non-neuronal cells, and this activity was shown to be essential for neurite outgrowth in neuronal cells (2, 5). Protrudin resides in the ER of neuronal cells in the absence of NGF, but it translocates to recycling endosomes in response to NGF stimulation and is ultimately transported to the tip of newly formed neurites (2). We have now investigated the molecular mechanism that underlies this translocation of protrudin by attempting to isolate proteins that interact with it and regulate its subcellular localization. Proteomics analysis led to the identification of VAP-A and VAP-B as proteins that interact with protrudin. These proteins are thought to regulate the localization of proteins that contain an FFAT motif through interaction with this motif (11). VAP-A and VAP-B were thus candidates for regulators of protrudin localization. Indeed, we found that VAP-A interacts with protrudin through the FFAT motif of the latter and that it is required for the ER retention of protrudin in HeLa cells. In contrast, the NGF-induced transport of protrudin from the ER to recycling endosomes in PC12 cells was shown to be dependent on VAP-A, as was the process forming activity of protrudin. Although the reason for the difference in the effects of VAP-A depletion on protrudin localization between HeLa and PC12 cells (Fig. 9, B and E) remains unclear, it might be attributable to the difference in the extent of VAP-A depletion, which was partial in PC12 cells but almost complete in HeLa cells (Fig. 9, A and D) or to a difference in the inherent properties of the two cell types. Consistent with the latter notion, VAP-A depletion eventually results in cell death in PC12 cells but not in HeLa cells (data not shown). Depletion of VAP-A also triggers cell death in primary cultured rat hippocampal neurons (31).
The FFAT motif-containing proteins CERT (11), OSBP (25), and Nir-2 (12) interact with VAP and possess lipid binding activity. The interaction of VAP with the FFAT motifs of these proteins is required for their mediation of lipid transport between intracellular membrane systems. For example, in addition to an FFAT motif, CERT contains a Start domain, which interacts with ceramide and serves to mediate ER-Golgi trafficking of ceramide. Protrudin also contains a FYVE domain, which is thought to mediate the association of proteins with phosphatidylinositol 3-phosphate (32). These common characteristics of FFAT motif-containing proteins suggest that protrudin might contribute to vesicular transport through its association with lipids in the vesicular membrane and that this role might be regulated by VAP.
The MSP domain of VAP-A has been shown to be responsible for the binding of VAP-A to the FFAT motif of its target molecules. The crystal structure of a complex of VAP-A with the FFAT motif revealed that Lys87 and Met89 of rat VAP-A are important for the interaction (26). We have now shown that the corresponding residues of human VAP-A (Lys94 and Met96) contribute to its interaction with protrudin. In contrast, mutation of other residues of human VAP-A corresponding to those implicated in the interaction of VAP-A with the FFAT motif, including Lys52, Thr53, Thr54, and Lys125, was found not to affect the binding of human VAP-A to protrudin. However, the fact that VAP and FFAT motif-containing proteins form a 2:2 tetramer (26) makes it difficult to interpret the results of such binding analyses in cells because of the presence of the endogenous wild-type protein. In addition to the MSP domain, the transmembrane domain of VAP-A also appeared to be important for the interaction with protrudin (Fig. 9F). The manner of the interaction between VAP-A and protrudin may thus differ somewhat from that of the interaction between VAP-A and other FFAT motif-containing proteins.
Dysfunction of VAP or protrudin is implicated in human neurological disorders. A mutation in the MSP domain (P56S) of human VAP-B has been associated with familial amyotrophic lateral sclerosis (33, 34). This condition is characterized by the death of motor neurons in the cerebral cortex, brainstem, and spinal cord, eventually resulting in muscle weakness and atrophy followed by death caused by respiratory failure 2-5 years after disease onset. The P56S mutation affects the interaction of VAP-B with other cellular proteins (35). ZFYVE27 (protrudin) was also found to be mutated in a German family with AD-HSP, which is characterized by the selective degeneration of axons (8). The phenotype of the affected individuals is similar to that of patients with AD-HSP caused by mutation of spastin, a protein implicated in the trafficking of vesicular cargo in neurons (9). Protrudin is thought to interact with spastin via its COOH-terminal FYVE domain (8). Further molecular analysis of VAP and protrudin may thus provide insight into the pathogenesis of these neurological disorders as well as a basis for the development of new therapeutic agents.
Acknowledgments
We thank S. Nagata for vectors; A. Miyawaki for Venus cDNA; M. Matsuoka for anti-VAP-B; A. Hamasaki, N. Nishimura, and other laboratory members for technical assistance; and A. Ohta and M. Kimura for help in preparation of the manuscript.
Footnotes
The abbreviations used are: TM, transmembrane domain; AD-HSP, autosomal dominant hereditary spastic paraplegia; ER, endoplasmic reticulum; NGF, nerve growth factor; VAP, vesicle-associated membrane protein-associated protein; shRNA, short hairpin RNA; MS/MS, tandem mass spectrometry; GST, glutathione S-transferase; HA, hemagglutinin epitope; PDI, protein-disulfide isomerase; PBS, phosphate-buffered saline; MSP, major sperm protein.
References
- 1.Zerial, M., and McBride, H. (2001) Nat. Rev. Mol. Cell Biol. 2 107-117 [DOI] [PubMed] [Google Scholar]
- 2.Shirane, M., and Nakayama, K. I. (2006) Science 314 818-821 [DOI] [PubMed] [Google Scholar]
- 3.Barik, S. (2006) Cell Mol. Life Sci. 63 2889-2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton, G. S., and Steiner, J. P. (1998) J. Med. Chem. 41 5119-5143 [DOI] [PubMed] [Google Scholar]
- 5.Shirane, M., Ogawa, M., Motoyama, J., and Nakayama, K. I. (2008) Genes Cells 13 635-651 [DOI] [PubMed] [Google Scholar]
- 6.Loewen, C. J., Roy, A., and Levine, T. P. (2003) EMBO J. 22 2025-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillooly, D. J., Simonsen, A., and Stenmark, H. (2001) Biochem. J. 355 249-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mannan, A. U., Krawen, P., Sauter, S. M., Boehm, J., Chronowska, A., Paulus, W., Neesen, J., and Engel, W. (2006) Am. J. Hum. Genet. 79 351-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salinas, S., Carazo-Salas, R. E., Proukakis, C., Schiavo, G., and Warner, T. T. (2007) J. Neurosci. Res. 85 2778-2782 [DOI] [PubMed] [Google Scholar]
- 10.Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003) Nature 426 803-809 [DOI] [PubMed] [Google Scholar]
- 11.Kawano, M., Kumagai, K., Nishijima, M., and Hanada, K. (2006) J. Biol. Chem. 281 30279-30288 [DOI] [PubMed] [Google Scholar]
- 12.Amarilio, R., Ramachandran, S., Sabanay, H., and Lev, S. (2005) J. Biol. Chem. 280 5934-5944 [DOI] [PubMed] [Google Scholar]
- 13.Kagiwada, S., Hosaka, K., Murata, M., Nikawa, J., and Takatsuki, A. (1998) J. Bacteriol. 180 1700-1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura, Y., Hayashi, M., Inada, H., and Tanaka, T. (1999) Biochem. Biophys. Res. Commun. 254 21-26 [DOI] [PubMed] [Google Scholar]
- 15.Weir, M. L., Klip, A., and Trimble, W. S. (1998) Biochem. J. 333 247-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skehel, P. A., Fabian-Fine, R., and Kandel, E. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1101-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soussan, L., Burakov, D., Daniels, M. P., Toister-Achituv, M., Porat, A., Yarden, Y., and Elazar, Z. (1999) J. Cell Biol. 146 301-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapierre, L. A., Tuma, P. L., Navarre, J., Goldenring, J. R., and Anderson, J. M. (1999) J. Cell Sci. 112 3723-3732 [DOI] [PubMed] [Google Scholar]
- 19.Pennetta, G., Hiesinger, P. R., Fabian-Fine, R., Meinertzhagen, I. A., and Bellen, H. J. (2002) Neuron 35 291-306 [DOI] [PubMed] [Google Scholar]
- 20.Foster, L. J., and Klip, A. (2000) Am. J. Physiol. 279 C877-C890 [DOI] [PubMed] [Google Scholar]
- 21.Shirane, M., and Nakayama, K. I. (2003) Nat. Cell Biol. 5 28-37 [DOI] [PubMed] [Google Scholar]
- 22.Natsume, T., Yamauchi, Y., Nakayama, H., Shinkawa, T., Yanagida, M., Takahashi, N., and Isobe, T. (2002) Anal. Chem. 74 4725-4733 [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa, T., Shirane, M., Iemura, S., Natsume, T., and Nakayama, K. I. (2007) Genes Cells 12 709-719 [DOI] [PubMed] [Google Scholar]
- 24.Davies, T. H., and Sanchez, E. R. (2005) Int. J. Biochem. Cell Biol. 37 42-47 [DOI] [PubMed] [Google Scholar]
- 25.Wyles, J. P., McMaster, C. R., and Ridgway, N. D. (2002) J. Biol. Chem. 277 29908-29918 [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, S. E., Brickner, J. H., Reilein, A. R., Fenn, T. D., Walter, P., and Brunger, A. T. (2005) Structure 13 1035-1045 [DOI] [PubMed] [Google Scholar]
- 27.Pelham, H. R. (1996) Cell Struct. Funct. 21 413-419 [DOI] [PubMed] [Google Scholar]
- 28.Loewen, C. J., and Levine, T. P. (2005) J. Biol. Chem. 280 14097-14104 [DOI] [PubMed] [Google Scholar]
- 29.Freedman, R. B., Hirst, T. R., and Tuite, M. F. (1994) Trends Biochem. Sci. 19 331-336 [DOI] [PubMed] [Google Scholar]
- 30.Favaloro, V., Spasic, M., Schwappach, B., and Dobberstein, B. (2008) J. Cell Sci. 121 1832-1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teuling, E., Ahmed, S., Haasdijk, E., Demmers, J., Steinmetz, M. O., Akhmanova, A., Jaarsma, D., and Hoogenraad, C. C. (2007) J. Neurosci. 27 9801-9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takenawa, T., and Itoh, T. (2006) IUBMB Life 58 296-303 [DOI] [PubMed] [Google Scholar]
- 33.Nishimura, A. L., Mitne-Neto, M., Silva, H. C., Richieri-Costa, A., Middleton, S., Cascio, D., Kok, F., Oliveira, J. R., Gillingwater, T., Webb, J., Skehel, P., and Zatz, M. (2004) Am. J. Hum. Genet. 75 822-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanekura, K., Nishimoto, I., Aiso, S., and Matsuoka, M. (2006) J. Biol. Chem. 281 30223-30233 [DOI] [PubMed] [Google Scholar]
- 35.Mitne-Neto, M., Ramos, C. R., Pimenta, D. C., Luz, J. S., Nishimura, A. L., Gonzales, F. A., Oliveira, C. C., and Zatz, M. (2007) Protein Expression Purif. 55 139-146 [DOI] [PubMed] [Google Scholar]