Abstract
Early events leading to the establishment of hepatitis C virus (HCV) infection are not completely understood. We show that intact and dynamic microtubules play a key role in the initiation of productive HCV infection. Microtubules were required for virus entry into cells, as evidenced using virus pseudotypes presenting HCV envelope proteins on their surface. Studies carried out using the recent infectious HCV model revealed that microtubules also play an essential role in early, postfusion steps of the virus cycle. Moreover, low concentrations of vinblastin and nocodazol, microtubule-affecting drugs, and paclitaxel, which stabilizes microtubules, inhibited infection, suggesting that microtubule dynamic instability and/or treadmilling mechanisms are involved in HCV internalization and early transport. By protein chip and direct core-dependent pull-down assays, followed by mass spectrometry, we identified β- and α-tubulin as cellular partners of the HCV core protein. Surface plasmon resonance analyses confirmed that core directly binds to tubulin with high affinity via amino acids 2-117. The interaction of core with tubulin in vitro promoted its polymerization and enhanced the formation of microtubules. Immune electron microscopy showed that HCV core associates, at least temporarily, with microtubules polymerized in its presence. Studies by confocal microscopy showed a juxtaposition of core with microtubules in HCV-infected cells. In summary, we report that intact and dynamic microtubules are required for virus entry into cells and for early postfusion steps of infection. HCV may exploit a direct interaction of core with tubulin, enhancing microtubule polymerization, to establish efficient infection and promote virus transport and/or assembly in infected cells.
HCV5 infection is a major cause of chronic liver disease, which frequently progresses to cirrhosis and hepatocellular carcinoma. HCV represents a global public health problem, with 130 million people infected worldwide. There is currently no vaccine directed against HCV and the available antiviral treatments eliminate the virus in 40-80% of patients, depending on the virus genotype (for review, see Ref. 1).
HCV has a single-stranded, positive-sense RNA genome of ∼9.6 kilobases encoding a large polyprotein that is processed by both host and viral proteases to produce three structural proteins (core protein and the envelope glycoproteins E1 and E2), p7, and six nonstructural proteins, which are involved in polyprotein processing and replication of the virus genome (for review, see Ref. 2).
HCV core is a basic protein, synthesized as the most N-terminal component of the polyprotein, and is followed by the signal sequence of the E1 envelope glycoprotein (3). The polypeptide is cleaved by signal peptidase and signal peptide peptidase, resulting in the release of core from the endoplasmic reticulum membrane and its trafficking to lipid droplets (3-5). Mature core protein forms the viral nucleocapsid (6) and consists of two domains, D1 and D2. D1 lies at the protein N terminus, is composed of about 117 amino acids (aa), and is involved in RNA binding (7). D2 is relatively hydrophobic, has a length of about 55 aa, and targets HCV core to lipid droplets (8).
Microtubules (MTs) are ubiquitous cytoskeleton components that play a key role in various cellular processes relating to cell shape and division, motility, and intracellular trafficking (9). MTs are dynamic, polarized polymers composed of α/β-tubulin heterodimers that undergo alternate phases of growth and shrinkage, dependent on so-called “dynamic instability” (10). Active transport by MTs is bidirectional and involves both plus and minus end-directed motors: kinesin and dynein (11, 12).
Another mechanism of cytosolic transport on MTs, called “treadmilling” (13, 14) involves polymerization at the plus end and depolymerization at the minus end after severing of MTs by cellular katenin (15).
MTs have important functions in the life cycle of most viruses (13, 16, 17). Cytoplasmic transport on MTs provides viruses with the means to reach sites of replication or enables progeny virus to leave the infected cell. Some viruses, such as Ebola virus (18) or reovirus (19), are transported on MTs within membranous compartments, whereas other viruses like herpes simplex virus type 1 (20), murine polyoma virus (21), human cytomegalovirus (22), or adenovirus (23) interact with MT motors or MT-associated proteins to allow their transport along microtubules.
Previous studies have established that the cell cytoskeleton is involved in HCV replication, since HCV replication complexes are subjected to intracellular transport and their formation is closely linked to the dynamic organization of endoplasmic reticulum, actin filaments, and the microtubule network (24-26). In addition, intact microtubules are essential for viral morphogenesis and the secretion of progeny virus from infected cells (27). The role of microtubules in HCV cell entry and the initiation of productive HCV infection has not yet been addressed.
In this study, we provide evidence that the MT network plays a key role in HCV cell entry and postfusion steps of the virus cycle that lead to the establishment of productive HCV infection. The initial steps of the viral cycle are sensitive to MT-affecting drugs that inhibit MT formation or depolymerize or stabilize microtubules. We also show a unique property of the HCV core protein, its capacity to directly bind to tubulin and to enhance MT polymerization in vitro. Our findings suggest that HCV could exploit the MT network by polymerization-related mechanisms to productively infect its target cell. Thus, microtubules may provide a novel target for therapeutic interventions against HCV infection.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Rabbit antibodies against α- and β-tubulin were from AbCam. Fluorescein isothiocyanate-conjugated goat anti-mouse IgG (H+L) was purchased from Jackson Inc.; Alexa Fluor 568-labeled goat anti-mouse IgG(H+L), Alexa Fluor 488-labeled goat anti-rabbit IgG (H+L), and Texas Red-labeled anti-phalloidin antibodies were from Invitrogen. Control mouse and rabbit IgG were from Pierce and Santa Cruz Biotechnology, Inc., respectively.
Recombinant HCV Core Proteins—Recombinant His6-tagged HCV core proteins aa 2-169, aa 2-122, and aa 2-117 were produced in Escherichia coli and purified in native conditions on nickel-nitrilotriacetic acid-agarose and by reverse-phase high pressure liquid chromatography on a VYDAC C8 column, as previously described (28, 29). Core proteins were handled in 0.1% n-dodecyl-d-maltoside to ensure their native conformation. This detergent was present in BIAcore and tubulin polymerization experiments.
Cell Cultures—Human hepatoma Huh7 and Huh7.5 cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum and glutamine. Huh7.5 cells were supplemented with pyruvate and with 1% nonessential amino acids, antibiotics, and antifungal agents. 293T human embryo kidney cells (HEK 293T) and PLC/PRF/5 human hepatoma cells (ATCC CRL-8024) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
Infection of Huh7.5 Cells with the JFH-1 Strain of HCVcc—The JFH-1 plasmid encoding the genome of the JFH-1 strain of HCV was kindly provided by T. Wakita (30) and used to generate HCVcc. The virus was cultured as described previously (30) to obtain a virus stock of ∼1.4 × 106 focus-forming units/ml (5 × 107 IU/ml). Cells were incubated with infectious JFH-1 (40 μl containing 6 × 104 focus-forming units of virus) for 2 h at 37 °C to allow infection. After 2 h, cells were washed and incubated with complete Dulbecco's modified Eagle's medium for the indicated times at 37 °C.
To assess the effect of cytoskeleton-affecting drugs on the initiation of HCV infection, Huh7.5 cells were grown for 48 h to obtain ∼1 × 106 cells/well. The cells were incubated with complete medium containing 0.37 μm to 30 μm nocodazole, 1 nm to 30 μm vinblastin, 20 μm cytochalasin D, or 1-5 μm paclitaxel, the drugs being added before or at different time points postinfection. The amount of HCV RNA in infected cells was determined by quantitative polymerase chain reaction (qRT-PCR) 16-24 h postinfection. After treatment with drugs, cell viability was assessed by staining with trypan blue. The results were normalized by quantification of the RNA of GAPDH, the housekeeping cellular gene using the GAPDH control kit from Eurogentec.
Quantitative Real Time RT-PCR—HCV RNA was quantified by one-step real time qRT-PCR with the SuperScript® III Platinum® one-step qRT-PCR kit (Invitrogen), as described previously (31). HCV RNA was quantified relative to a reference serum, standardized with an HCV RNA quantification panel from AcroMetrix.
Cell Infection with HCVpp—Pseudoparticles were produced as described previously in human embryo kidney cells (HEK 293T) (32-34). 293T cells were co-transfected with a murine leukemia virus-based transfer vector encoding luciferase, a murine leukemia virus Gag-Pol packaging construct, and an envelope glycoprotein-expressing vector (either phCMV-E1E2, phCMV-G, or phCMV-RD114) using Exgen500 (Euromedex). The phCMV-E1E2, phCMV-G, and phCMV-RD114 expression vectors encode the HCV glycoproteins E1 and E2, the vesicular stomatitis virus G protein, and the feline endogenous virus RD114 glycoprotein, respectively. Supernatants containing pseudotyped particles were harvested 48 h after transfection, filtered through 0.45-μm pore-sized membranes, and incubated with 300 units/ml DNase I for 30 min at 37 °C to remove excess plasmid DNA (35).
To investigate the influence of vinblastin on HCVpp cell entry, confluent monolayers of PLC/PRF/5 cells were incubated for 30 min at 37 °C with 30 μm vinblastin in the culture medium and infected with 600 μl of DNase-treated HCVpp in the presence of 30 μm vinblastin. The presence of vinblastin was maintained during 3 h of cell-virus contact, and HCVpp assays were carried out as previously described (35).
HCV Replicon—Huh7 cells harboring the subgenomic JFH1 replicon were obtained and grown as previously described (36). Intracellular levels of HCV RNA were determined by qRT-PCR as described above.
Immunofluorescence Microscopy—Cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and saturated in PBS containing 0.1% Tween 20 and 1% gelatin overnight at 4 °C. Cells were then incubated with primary antibody (for 1 h at room temperature) and washed with PBS containing 0.1% Tween 20. After staining with fluorochrome-labeled secondary antibodies, the cells were mounted in Vectashield medium (Vector Laboratories) and examined under a fluorescence microscope (Zeiss Axiovert 200M) equipped with the ApoTome system.
Confocal Microscopy—Cells were fixed for 20 min in methanol at -20 °C. Indirect immunofluorescence and 4′,6-diamidino-2-phenylindole staining were performed as described previously (8, 37). Microtubules were labeled using monoclonal anti-tubulin antibody and fluorochrome-conjugated secondary anti-mouse antibody. Core was stained with rabbit antibody, followed by fluorochrome-labeled secondary antibody. Cells were then examined with a Zeiss LSM510 META inverted confocal microscope. Images were recorded with a Plan-Apochromat ×63 lens (numerical aperture 1.4). For Z-stack analysis, 25 images were recorded at 0.14-μm intervals. For the three-dimensional reconstructions, Z-stack images were collected using optimum intervals, generating 20-25 slices/sample. Image stacks were deconvolved by three-dimensional blind deconvolution (20 iterations) using Autodeblur software (Media Cybernetics). Three-dimensional reconstructions were created with the five-dimensional viewer extension in the isosurface mode using a bin factor of 1.
Preparation of Cell Extracts from Huh7 Cells—Cell extracts from hepatoma cells were prepared as previously described (38). Briefly, 3-5 × 106 cells were detached from the surface of the flask by EDTA treatment, collected by centrifugation, washed with cold PBS, resuspended in PBS supplemented with 2% Nonidet P-40 and EDTA-free protease inhibitor mixture (Roche Applied Science), and incubated for 30 min on ice. Cell debris and nuclei were removed by centrifugation. Aliquots of supernatant were frozen and stored at -80 °C.
Protein Chip SELDI-TOF Mass Spectrometry (MS) Analyses—The IMAC3 Protein Chip Array (Ciphergen Biosystems) was incubated with 60 μl of 100 mm NiCl2 for 15 min at 20 °C. After washing, core protein (2 μg in 50 μl of PBS) was spotted onto the chip, incubated for 50 min at 20 °C, and washed in 0.1% Triton X-100 in PBS. We then spotted 30-60 μg of cell extracts in the same buffer onto the chip, and incubated it for 1 h at 20 °C. The chip was washed with PBS containing 5 mm HEPES, and energy-absorbing molecule solution was loaded onto the sample and retained on the chip surface. An unrelated His6-tagged protein or PBS was used as a negative control. The Ciphergen protein chip reader (PBS II model) was used to analyze the arrays, and the data were processed with Protein Chip software version 3.2.1.
Pull-down Assays—Pull-down experiments using IMAC (BIOSEPRA)-cellulose beads were employed to identify putative cellular partners of the HCV core protein present in cell extracts. His6-tagged core proteins were immobilized on Ni2+-coated cellulose beads. Beads were then incubated with 50-70 μg of cell extracts for 20-120 min at 4 or at 20 °C, washed in a buffer containing 0.1%, Triton X-100 and 5 mm HEPES in PBS, suspended in 30 μl of Laemmli buffer, and analyzed by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue.
Peptide Mass Mapping and Bioinformatic Analyses—Matrix-associated laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) was used to identify proteins of interest isolated by pull-down assays followed by SDS-PAGE separation, using the procedure originally described by Shevchenko et al. (39). Briefly, protein bands stained with Coomassie Blue were excised from the gel, washed, reduced, S-alkylated, and digested with sequencing grade porcine trypsin (Promega) overnight at 37 °C. The resulting peptides were extracted and desalted using ZipTips (Millipore). Peptide mass was analyzed with an 0.5-μl tryptic digest mixture, containing 0.5 μl of saturated cyanohydroxycinnamic acid (Sigma). The results were analyzed by MALDI-TOF MS (Voyager DE STR; Applied Biosystems). Proteins were identified by searches of the Swiss-Prot data base, using a local copy of the MS-FIT 3.2 part of the PROTEIN PROSPECTOR package.
Surface Plasmon Resonance (SPR) Analyses—Real time binding experiments were carried out with a BIAcore 3000 biosensor system (BIAcore, Uppsala, Sweden). All experiments were performed at 25 °C, at a flow rate of 20 μl/min. The eluent consisted of 10 mm HEPES, pH 7.4, 150 mm NaCl, 50 mm EDTA, and 0.005% P20 surfactant. The dispenser buffer consisted of 10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, and 0.005% P20. Core proteins, aa 2-169 or aa 2-117, were immobilized via their C-terminal His6 tag residues on the surface of an nitrilotriacetic acid sensor chip, previously activated with 500 mm NiCl2. Bovine tubulin (at concentrations of 50, 100, 250, 500, and 1,000 nm) was injected in dispenser buffer, pH 7.4. Changes in surface concentration resulting from the interaction of tubulin with surface-fixed core protein were detected as an optical phenomenon affecting the SPR signal, expressed in RU, where 1 RU corresponds to an immobilized protein concentration of 1 pg/mm2. The results obtained for a blank, produced by injecting tubulin into a mock sensorchip, were subtracted from each sensogram.
Tubulin Polymerization Assay—To investigate the effect of the core protein on tubulin polymerization, we used the fluorescence-based in vitro tubulin polymerization assay kit from Cytoskeleton (Denver, CO). This assay is based on fluorescence enhancement due to the incorporation of a fluorescent reporter into MTs as polymerization occurs. The assay was carried out using highly purified tubulin (99% pure). HCV core proteins were used in a concentration range of 5-200 μg/ml. The reading parameters in SAFAS SP-2000 were set as follows. Fluorescence wavelengths were 360 nm for excitation and 420 nm for emission, and kinetics were of one reading/min at 37 °C.
Electron Microscopy—The effect of core on MT formation in vitro was controlled by electron microscopy. The polymerization assays were carried out as described above in the presence of 20 μg/ml core protein aa 1-117 or in the absence of core and in the presence or absence of MT-affecting drugs (3 μm nocodazol or 3 μm paclitaxel). Microtubules were then fixed with 2.5% glutaraldehyde and stained with 4% uranyl acetate. Grids were examined in a Jeol electron microscope 1200 EX at 80 kV at a magnification of ×120,000.
RESULTS
Initiation of HCV Infection Requires the Intact MT Network—The JFH1 strain of HCV (2a genotype) generates infectious virus particles in the hepatoma cell line Huh7.5, which are released from cells and are capable of establishing productive infection in naive Huh7.5 cells. To determine whether the cytoskeleton plays a role in HCV cell entry and the initiation of the HCV life cycle, the effects of MT-affecting drugs vinblastin and nocodazole and the actin filament-depolymerizing drug cytochalasin D were investigated using this HCVcc infection model.
Huh7.5 cells were incubated with drugs for 30 min before and 2 h during infection with HCVcc. The efficiency of infection in treated versus nontreated cells was determined by measuring the levels of HCV RNA produced in cells cultured in the medium without drugs 16 h postinfection by qRT-PCR. Since these drugs could have a toxic effect, cell viability of drug-treated and nontreated cells was compared after trypan blue staining. In addition, the results obtained by qRT-PCR were normalized by comparative quantification of GAPDH RNA.
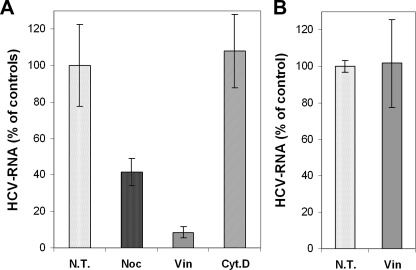
Nocodazole or vinblastin resulted in a substantial reduction of HCV RNA levels as compared with untreated cells (Fig. 1A). These findings contrasted with those obtained in the presence of the actin filament-depolymerizing drug cytochalasin D, which did not show any significant effect on HCV RNA levels. In control experiments carried out at 4 °C (when endocytosis is blocked), we excluded the effect of MT-affecting drugs on virus attachment (Fig. 1B). These results indicated that MTs but not actin filaments are required for the initiation of HCV infection.
FIGURE 1.
Intact microtubule network is required for the establishment of productive HCV infection. A, Huh7.5 cells were pretreated for 30 min with 20 μm nocodazol, 30 μm vinblastin, or 20 μm cytochalasin D. Infection of Huh7.5 cells with JFH1 HCV strain was subsequently carried out for 2 h in the presence of these compounds, and then cells were washed and cultured for 16 h in the absence of drugs. The levels of infection were determined by quantification of HCV RNA by qRT-PCR. The results were normalized by quantification of RNA corresponding to GAPDH, the housekeeping gene, and presented as a percentage of HCV RNA as compared with nontreated cells (100%). Dotted bar, control, untreated cells (N.T.). B, control experiments to show that vinblastin does not affect HCV binding to cells. Huh7.5 cells were incubated with cold medium containing 20 μm vinblastin for 30 min and then incubated with JFH1 for 2 h at 4 °C. Cells were washed, and virus bound to the cells in the presence or absence of vinblastin (N.T.) was quantified by qRT-PCR.
Intact Microtubule Network Is Required for HCV Cell Entry—The observed inhibition of cell infection by MT-affecting drugs could involve virus cell entry (from virus binding at the plasma membrane until the release of the nucleocapsid from the endocytic vesicle) or rather later steps following fusion and liberation of the viral nucleocapsid. To investigate whether microtubules are involved in early endocytic events occurring between virus attachment at the plasma membrane and fusion mediated by the E1E2 envelope glycoproteins, we used pseudoviral particles presenting the HCV envelope glycoproteins on a retroviral core (HCVpp model) (32-34). Hepatoma cells were pretreated with vinblastin for 30 min before infection, and the drug was maintained during cell infection with HCVpp. HCVpp cell entry was measured using a PCR-based assay, which quantifies the amount of retroviral genome after retrotranscription that occurs following capsid delivery to the cytoplasm of the target cell (35).
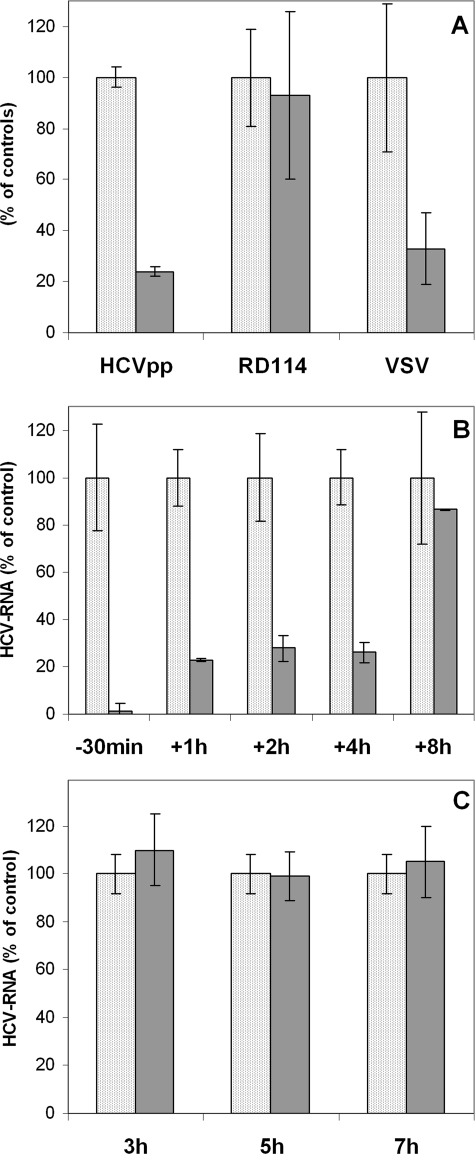
Pseudotyped particles harboring VSV-G or RD114 glycoproteins on their surface were employed as controls to exclude a nonspecific effect of the drugs or their influence on the fusion process. As shown in Fig. 2A, HCVpp cell entry was significantly reduced in the presence of vinblastin. The drug equally reduced cell entry of control pseudotypes VSV-Gpp, which also require MT-dependent endocytic transport. In contrast, cell entry of RD114pp was not affected, as expected for a virus known to enter the cell by direct fusion at the plasma membrane. Hence, we conclude that early endocytic events of HCV cell entry occurring between virus attachment at the plasma membrane and fusion mediated by the E1E2 envelope glycoproteins require an intact MT network.
FIGURE 2.
Intact microtubules are required for HCV entry into cells and early postentry steps of infection. A, vinblastin inhibits HCVpp cell entry. Cells were infected with pseudoviral particles expressing on their surface either the HCV envelope proteins (HCVpp) or particles harboring VSV-G or RD114 glycoproteins in the presence (gray bar) or absence of vinblastin (dotted bar). Cells were preincubated with 30 μm vinblastin for 30 min and infected with pseudoviral particles in the presence of the drug, which was maintained for 3 h. Results are expressed as percentage of virus cell entry relative to murine leukemia virus cDNA copies (normalized by quantification of the DNA from the albumin gene) in the presence or absence of the drug. RD114pp cell entry is not affected by vinblastin, as expected for a virus that does not require endosomal transport and enters the cell by direct fusion at the plasma membrane. B, vinblastin inhibits early, postfusion steps. Cells were incubated with the JFH1 strain of HCV for 30 min at 4 °C to bind the virus, and then unbound virus was removed by washing, and cells were incubated for 2 h at 37 °C. Vinblastin (10 μm) was applied at different time points: 30 min before infection (-30min) or 1 h (+1h), 2 h (+2h), 4 h (+4h), or 8 h (+8h) postinfection. Cells were then cultured in the presence of the compound, and the levels of intracellular RNA were determined by qRT-PCR 16 h postinfection. The results were normalized by the determination of RNA corresponding to GAPDH and presented as a percentage of HCV RNA as compared with nontreated cells grown in the same experimental conditions (100%). C, vinblastin does not inhibit the subgenomic JFH1 replicon. Huh7 cells expressing JFH1 subgenomic HCV replicon were treated with 30 μm vinblastin for 3, 5, and 7 h. Intracellular HCV RNA was then quantified by qRT-PCR. Results obtained were normalized by quantification of RNA corresponding to GAPDH and presented as a percentage of HCV RNA as compared with nontreated cells. Vinblastin-treated cells are shown as a gray bar, and untreated cells are shown as a dotted bar.
Intact Microtubule Network Is Also Required for Early “Postentry” Steps—In addition to their role in virus entry into cells from virus binding until the fusion mediated by virus envelope proteins, which can be analyzed using the HCVpp model, functional MTs may play an important role at postentry steps, after fusion and release of the virus nucleocapsid into the cytosol. These steps cannot be investigated using the HCVpp model, which is based on retrovirus core and thus does not contain HCV core protein.
We therefore investigated, using the HCVcc JFH1 model, whether MT-affecting compounds can inhibit early postentry steps after virus fusion, which is considered to be completed by 2 h after infection. MT-affecting drugs were applied at different time points before or after infection. The cells were thus treated either with 10 μm vinblastin for 30 min 30 min before infection or infected with HCV and grown for 1, 2, 4, or 8 h and then treated with the drug. HCV RNA levels were quantified by RT-PCR after 16 h in drug-treated cells, compared with nontreated cells. As shown in Fig. 2B, a significant decrease in the production of the virus was observed when vinblastin was applied up to 4 h postinfection. Vinblastin applied 8 h postinfection had no effect on abundance of intracellular HCV RNA, measured after 16 h.
To exclude the influence of the drug on virus replication, control experiments were carried out using a subgenomic replicon derived from the same JFH1 virus strain (36). The application of 30 μm vinblastin over a similar period of time (3-7 h) had no visible effect on intracellular HCV RNA levels as measured by qRT-PCR (Fig. 2C). Thus, treatment with the drug before or at the beginning of infection, substantially reduced levels of virus produced and concerned early virus transport rather than replication process. These data confirmed that the intact microtubule network is needed for early events of the HCV cell cycle, which happen until 8 h after infection. These events most probably involve virus cell entry, nucleocapsid release, and early transport, subsequent to virus fusion.
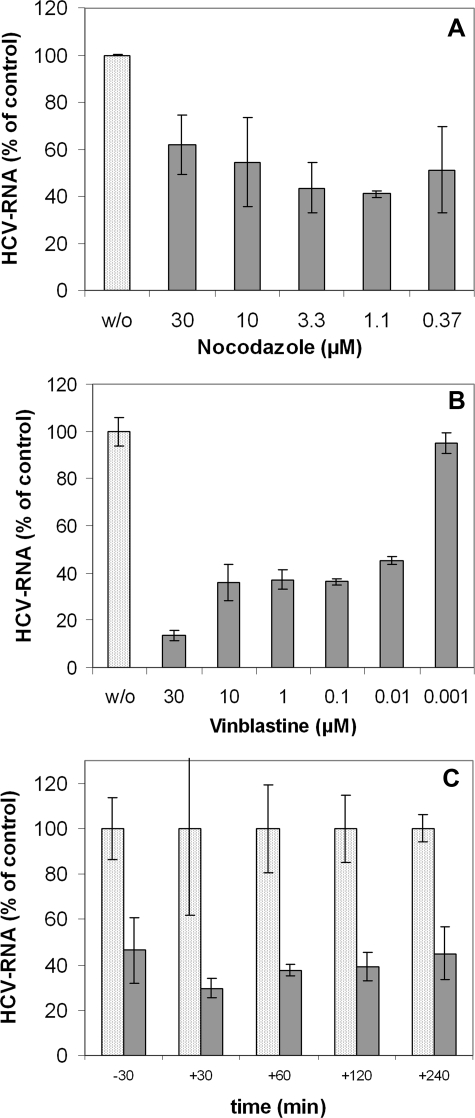
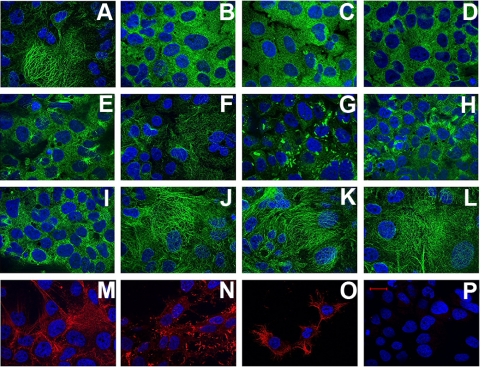
Dynamic Microtubules Are Required for the Initiation of HCV Infection—The effect of MT-affecting drugs is concentration-dependent, thus the concentration of compounds that have inhibitory effects on infection may reflect different mechanisms involved in virus interaction with MTs (40). We therefore investigated the influence of various concentrations of vinblastin and nocodazole on establishment of productive HCVcc infection. As shown in Fig. 3A, nocodazole used in a large range of concentrations (0.37 μm to 30 μm) had a substantial effect on the initiation of HCV infection. These concentrations of nocodazole induced visible changes in MT structure in accordance with the disassembly of tubulin polymers (Fig. 4, B-E), whereas lower concentrations of the compound (0.37 μm) (Fig. 4F) did not induce perceptible changes in microtubule structure but still impaired virus infection. The concentrations used are known to suppress MT dynamic instability (41).
FIGURE 3.
Dynamic instability and/or treadmilling mechanisms are required for the initiation of HCV infection. Huh7.5 cells were pretreated with the indicated concentrations of nocodazol (A) or vinblastin (B) for 30 min. Infection of cells with JFH1 was subsequently carried out for 2 h in the presence of these compounds, and then cells were washed and cultured for 16 h in the absence of the drugs. C, influence of paclitaxel on the initiation of HCV infection. Cells were incubated with the JFH1 strain of HCV for 30 min at 4 °C to bind the virus, and then cells were washed and incubated further for 2 h at 37 °C. Paclitaxel (2 μm) was applied at different time points: either 30 min before infection (-30) or 30 min (+30), 1 h (+60), 2 h (+120), or 4 h (+240) postinfection. Cells were then washed and cultured in the presence of the compound up to 16 h postinfection. The levels of infection (in A-C) were determined by the quantification of HCV RNA by qRT-PCR. The results were normalized by determination of RNA corresponding to GAPDH, the housekeeping gene, and presented as percentage of HCV RNA as compared with nontreated cells grown in the same experimental conditions (100%).
FIGURE 4.
Effect of drugs on the cell cytoskeleton assessed by immunofluorescence microscopy. Untreated Huh 7.5 cells (A) and cells treated for 2 h with different concentrations of nocodazol (30 μm (B), 10 μm (C), 3.3 μm (D), 1.1 μm (E), and 0.37 μm (F)) are shown as are Huh7.5 cells treated with various concentrations of vinblastin: 30 μm (G), 10 μm (H), 1 μm (I), 100 nm (J), 10 nm (K), and 1 nm (L). Staining was with rabbit anti-α-tubulin antibody followed by Alexa 488-labeled anti-rabbit IgG. Cells treated with 20 μm cytochalasin D (N) or nontreated cells (M) were stained with Texas Red-labeled anti-phalloidin antibody. O, cells stained with Oregon Green 488 taxol (Tubulin Tracker). Control cells stained with a nonrelated antibody, followed by Alexa 488-labeled anti-rabbit IgG, are shown in P. The nucleus is stained with 4′,6-diamidino-2-phenylindole. Bar, 20 μm.
Depending on its concentration, vinblastin is known to induce different effects on the structure and function of microtubules (40). In accordance with this notion, we observed that vinblastin in concentrations of 10-30 μm caused the aggregation of MTs in paracrystalline arrays (Fig. 4, G and H), whereas 1 μm vinblastin induced changes suggesting inhibition of MT assembly (Fig. 4I). For nanomolar concentrations of vinblastin, the changes in MT structure were not perceptible (Fig. 4, J-L); however, at these concentrations, vinblastin is known to suppresses MT dynamic instability and treadmilling mechanisms (20, 42). As shown in Fig. 3B, vinblastin concentrations as low as 10 nm to 100 nm still severely impaired HCV infection. These observations indicated that not only an intact but also a dynamic MT network is required to establish a robust HCV infection. These data suggest that MT tread-milling and/or dynamic instability mechanisms are involved in early steps of the virus cell cycle.
This conclusion was further supported by experiments carried out in the presence of paclitaxel (taxol), the MT-stabilizing drug known to block MT treadmilling mechanisms and/or their dynamic instability (20, 42). Paclitaxel, when applied at different time points either 0.5 h before infection or 0.5, 1, 2, or 4 h postinfection, induced substantial inhibitory effects on the efficiency of HCV infection (Fig. 3C). We conclude, therefore, that also postentry steps of the HCV cell cycle, which take place 2-8 h postinfection, require an intact and dynamic MT network. Since virus traffic during internalization and/or transport cannot take place on stabilized microtubules, our findings suggest that treadmilling and/or MT dynamic instability mechanisms might be involved in intracellular transport of the virus nucleocapsid.
Identification of α- and β-Chains of Tubulin as Core Protein Cellular Partners—The above findings raised the possibility that the HCV core protein could interact with microtubules or microtubule-related proteins during early transport of virus nucleocapsid. Thus, we searched for cellular partners that directly bind to HCV nucleocapsid protein and that could be related to MTs, by protein chip technology and pull-down assays, followed by mass spectrometry (Fig. 5).
FIGURE 5.
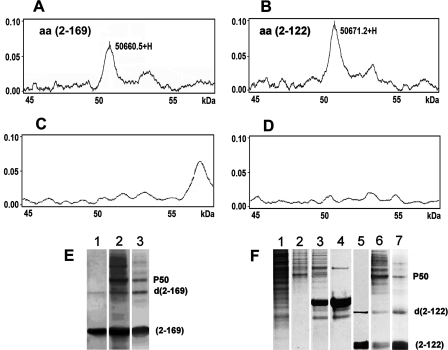
Identification of a 50-kDa cellular component interacting with HCV core protein. A-D, protein chip analyses of cellular components binding to the HCV core protein. A SELDI-TOF mass spectrometry profile of HCV core interactions with cell extracts of Huh7 cells is shown, in the region of 45-60 kDa with a peak of 50.6605 kDa corresponding to a molecule that binds to full-length core protein aa 2-169 (A) and a 50.6712-kDa molecule that binds to the truncated core protein aa 2-122 (B). The corresponding profiles obtained with a control, unrelated, His-tagged protein coated on protein chips and incubated with the same cell extracts (C) and the profile obtained for core aa 2-169-coated chips incubated with PBS instead of the cell extract (D) are shown. E and F, identification of a 50-kDa component from Huh7 cells interacting with the HCV core proteins in pull-down assays. Analyses were carried out by SDS-PAGE and followed by Coomassie Blue staining. E, lane 1, core protein aa 2-169 coated on beads; lane 2, core protein aa 2-169 interacting with cell extract analyzed in nonstringent conditions (10 μl of IMAC beads, 70 μl of cell extract, and incubation for 2 h at room temperature); lane 3, the same as lane 2 in stringent conditions (10 μl of beads, 50 μl of cell extract, and incubation carried out for 20 min at 4 °C). F, lane 1, total cell extract of Huh7 cells; lane 2, Huh7 cell extract interacting with noncoated IMAC beads; lane 3, unrelated protein (His-tagged GFP)-coated beads analyzed with Huh7 cell extract; lane 4, His-tagged GFP coated on beads; lane 5, core protein aa 2-122-coated beads alone; lane 6, core aa 2-122 and cell extract analyzed in nonstringent conditions; lane 7, core protein aa 2-122 and cell extract analyzed in stringent conditions. P50 is a protein band identified by pull-down assays. d(2-169) and d(2-122) correspond to dimers of core proteins aa 2-169 and aa 2-122, respectively.
Protein chip technology is based on capturing proteins on protein chip arrays followed by analysis on a SELDI-TOF mass spectrometer (protein chip reader). Core proteins aa 2-169 (corresponding to domains D1 + D2) and aa 2-122 (corresponding to domain D1), harboring a C-terminal histidine tag, were immobilized on IMAC3 chips, and their interactions with proteins in extracts prepared from Huh7 cells were analyzed. Profiles obtained with immobilized core proteins (Fig. 5, A and B) were compared with corresponding controls: protein chips coated with control His6-GFP protein, analyzed with extract from hepatoma cells (Fig. 5C), and core protein-coated chips incubated with PBS instead of cell extracts (Fig. 5D).
In protein chip analyses, ∼12 peaks relating to cell proteins bound to core were observed in the region corresponding to molecular masses from 11 to 60 kDa. A major peak of 50 kDa was identified in this region for both core proteins aa 2-169 and aa 2-122. This peak was not detected by protein chip analyses carried out with control, unrelated protein (Fig. 5C) or uncoated chips (Fig. 5D). Thus, the 50-kDa component was considered as a cellular partner interacting with HCV core.
Pull-down assays were then carried out with the IMAC system, also with two HCV core proteins aa 2-169 and aa 2-122 as a bait. In these experiments, different stringency conditions were used to confirm the specificity of protein/protein interactions (e.g. differing the amounts of cell extracts and beads and various incubation times and temperatures along with washing steps). In addition, the following series of controls was performed: (i) immobilized core proteins were analyzed to verify whether bait proteins were successfully captured to the beads; (ii) uncoated beads were used to identify cellular proteins, which could nonspecifically bind to the beads; (iii) beads were coated with a nonrelevant protein (purified His6-tagged GFP).
As shown in Fig. 5E, under low stringency conditions, a major 50 kDa (P50) band was detected in SDS-PAGE, corresponding to the cellular component that bound to both core proteins aa 2-169 (composed of domains D1 + D2) and aa 2-122 (composed of domain D1). Other cellular proteins, which associated with core-coated beads, progressively disappeared under more stringent conditions (Fig. 5F). Thus, the 50 kDa band corresponded to the 50 kDa component detected by protein chip assays.
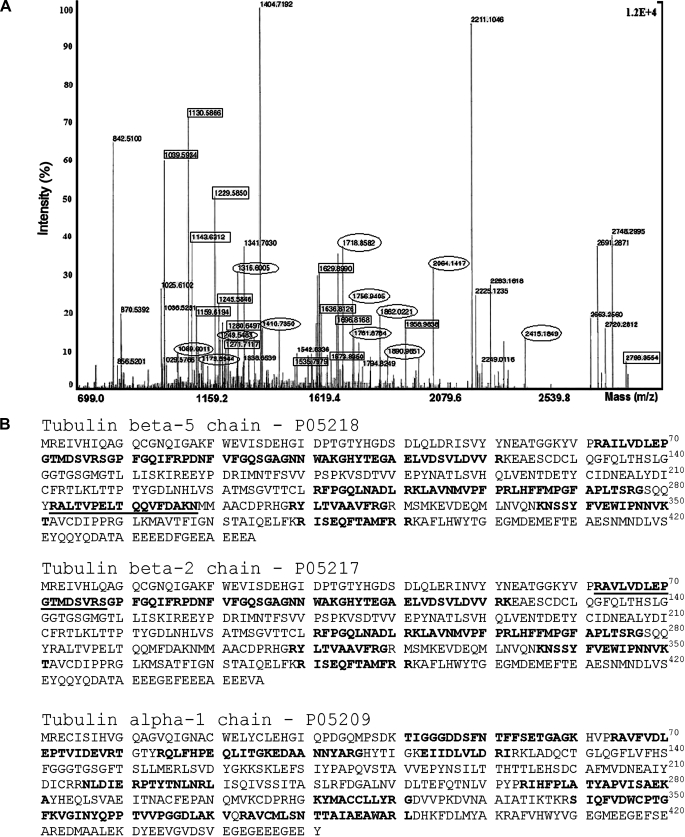
To identify the 50-kDa component that interacted with HCV core protein, the protein band detected by SDS-PAGE was digested with proteolytic enzymes, and the resulting peptides were analyzed by mass spectrometry (MALDI-TOF MS). The human β-tubulin 5-chain and β-tubulin 2-chain were identified as the most probable candidates for HCV core protein cellular partners, with 31 and 28% sequence coverage, respectively (Table 1). In addition, the human α-tubulin 3 and 1 isotypes were identified with 33% sequence coverage and a significant MOWSE score. These MOWSE scores strongly suggest that β- and α-tubulin chains are cellular partners that interact specifically with HCV core protein. Since β- and α-tubulins usually associate with each other, it is likely that the HCV core interacts with α/β-tubulin heterodimers.
TABLE 1.
Proteins identified by mass spectrometry
| Rank | MOWSE score | Accession number (Swiss-Prot) | Number of peptides | Masses matched (%) | Protein mass | Sequence coverage | Identified protein |
|---|---|---|---|---|---|---|---|
| kDa | % | ||||||
| 1a | 1.42e+010 | P05218 | 15 | 15 of 74 (20%) | 49.7 | 31 | Human β-tubulin 5-chain |
| 2b | 2.36e+009 | P07437 | 14 | 14 of 74 (18%) | 49.8 | 28 | Human β-tubulin 1-chain |
| 2.36e+009 | P05217 | 14 | 14 of 74 (18%) | 49.8 | 28 | Human β-tubulin 2-chain | |
| 2.36e+009 | P68371 | 14 | 14 of 74 (18%) | 49.8 | 28 | Human β-tubulin 2-chain | |
| 3c | 1.22e+009 | Q71U36 | 12 | 12 of 59 (20%) | 50.1 | 33 | Human α-tubulin 3-chain |
| 1.22e+009 | P05209 | 12 | 12 of 59 (20%) | 50.2 | 33 | Human α-tubulin 1-chain | |
| 1.22e+009 | P68363 | 12 | 12 of 59 (20%) | 50.2 | 33 | Human α-tubulin ubiquitous or (1B) chain |
The highest MOWSE score indicates that β-tubulin (5-chain) is the most probable candidate for core interacting protein.
The second candidate is β-tubulin (2-chain).
The third candidate is α-tubulin (3- and/or 1-chain, or ubiquitous chain of α-tubulin). These results are indicative of the presence of both β- and α-tubulin in the sample analyzed; β-tubulin 5- and 2-chains and α-tubulin 3/1-chains are the most probable candidates for the core protein cellular partners.
In mammals, six α-tubulin and seven β-tubulin genes have been described (43). In liver cells, α-tubulin isotypes, such as α1A/1B and α4A+Y, and α1C and β isotypes, such as β-I, β-II, β-Vb, and β-V, are the most prevalent (44). Tubulin isotypes are defined by 15-20 amino acid residues located at the C terminus of the tubulin chain (called the isotype-defining region). Peptides matching β-tubulin allowed the identification of tubulin β5 and β2 as the most relevant β-tubulin isotypes, preferentially reacting with HCV core protein. However, none of the peptides that matched α-tubulin targeted the C-terminal isotype-defining region, which rendered impossible the discrimination between chains α1 and α3 of α-tubulin (Fig. 6B). We also analyzed by mass spectrometry the 50-kDa protein that bound to uncoated beads (shown in Fig. 5F, lane 2). This protein was identified as human elongation factor 1-α1 (accession number NP68104) with a 105 lower MOWSE score as compared with the human β- and α-tubulins. Since the elongation factor is a highly expressed protein, we suggest that it is a nonspecific component, which binds to cellulose beads.
FIGURE 6.
Identification ofα- andβ-tubulin as HCV core-interacting proteins by MALDI-TOF analyses. To identify the 50-kDa component interacting with core, the protein band detected by SDS-PAGE was digested with proteolytic enzymes, the resulting peptides were analyzed by mass spectrometry (MALDI-TOF MS), and results were compared with the Swiss-Prot data bank. A, MALDI-TOF mass spectrogram of 50-kDa protein. Circles and rectangles indicate peptides matched with β-tubulin and α-tubulin, respectively. B, amino acid sequences identified by mass spectrometry. In boldface type are mass matches with the human β5-tubulin and β2-tubulin chains and the human α-1 and/or 3 tubulin chains. Underlined are sequences allowing isotype identification.
We conclude from this analysis that α- and β-chains of tubulin interact with core proteins composed either of D1 + D2 or D1 alone. These findings suggest a key role of D1 in such interactions.
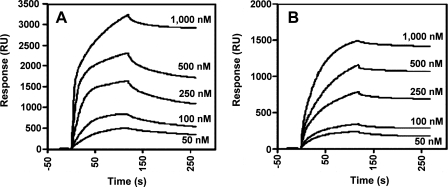
The HCV Core Protein Interacts with α/β-Tubulin in Vitro and in Vivo—Analyses by SPR were carried out to investigate whether core protein could directly bind to tubulin. A high purity preparation of bovine tubulin was used, which has almost complete sequence homology with human tubulin (45) and is a heterodimer composed of one α and one β chain, each with a molecular mass of 55 kDa. The molar equivalent of tubulin is typically defined as a heterodimer with a molecular mass of 110 kDa. For SPR studies, core proteins were immobilized on the nitrilotriacetic acid sensor chip by their C-terminal His residues (via Ni2+), and various concentrations of tubulin were injected. As shown in Fig. 7, a direct and dose-dependent binding of tubulin to both forms of core protein was detected. The kinetic constants for core proteins aa 2-169 and aa 2-117 were determined, using various tubulin concentrations. Apparent dissociation constants (Kd(app)) were 100 nm for core protein aa 2-169 and 75 nm for core protein aa 2-117 (Fig. 7, A and B, respectively). Thus, HCV core binds directly to α/β-tubulin with high affinity, in the nanomolar range, and the interaction site is located in the N-terminal D1 domain of the protein.
FIGURE 7.
Analysis of the interactions between HCV core and tubulin in vitro by SPR (BIA-core). Binding of tubulin to N-His-tagged recombinant HCV core proteins aa 2-169 (D1 + D2) (A) and aa 2-117 (D1) (B) immobilized on Ni2+ sensor chips. Increasing concentrations of purified tubulin were injected. Results are expressed in resonance units (RU).
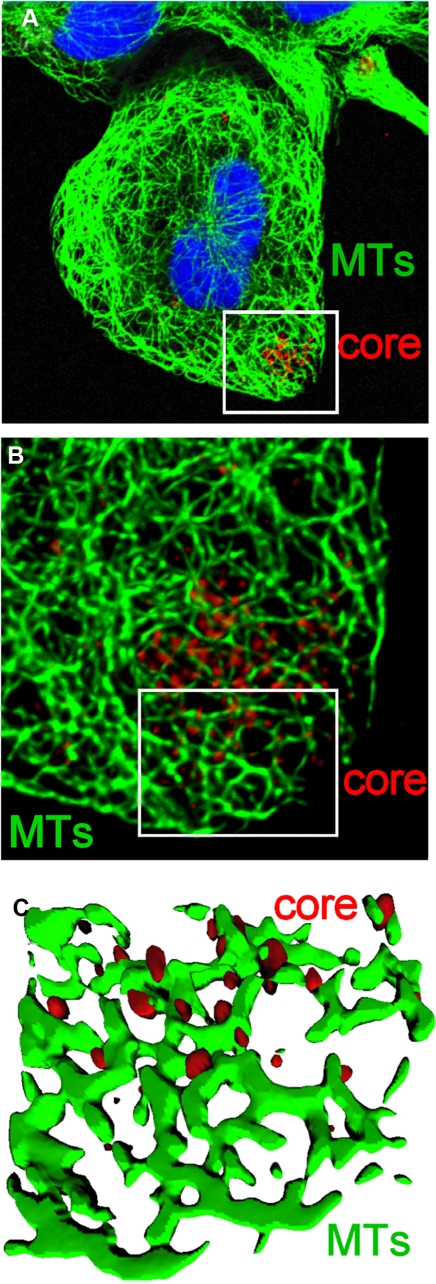
Next we investigated whether HCV core can interact with tubulin and/or MTs in HCV-infected cells. Studies carried out by confocal microscopy on the JFH1 model revealed a juxtaposition of core protein with MTs in HCV-infected cells at early phase of infection (Fig. 8). These studies suggested that HCV core may also interact with MTs in HCV-infected cells.
FIGURE 8.
Juxtaposition of core in HCVcc-infected cells. A, Huh7.5 cells were inoculated with medium from cells electroporated with JFH-1 RNA. Cells were stained with anti-core and anti-tubulin antibody followed by fluorochrome-labeled secondary antibodies 24 h postinoculation. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole. B and C, Z-stack analysis of the zoomed area (white box in A). B shows a maximal intensity projection of a Z-series generated from a stack of 20 images. To remove the blurring effect due to Z-stack acquisition, the Z-series was deconvolved using a blind deconvolution protocol. The image in C is a three-dimensional reconstruction of the zoomed area (indicated by the white box). Microtubules (in green) and core protein (in red) are represented as solid isosurfaces.
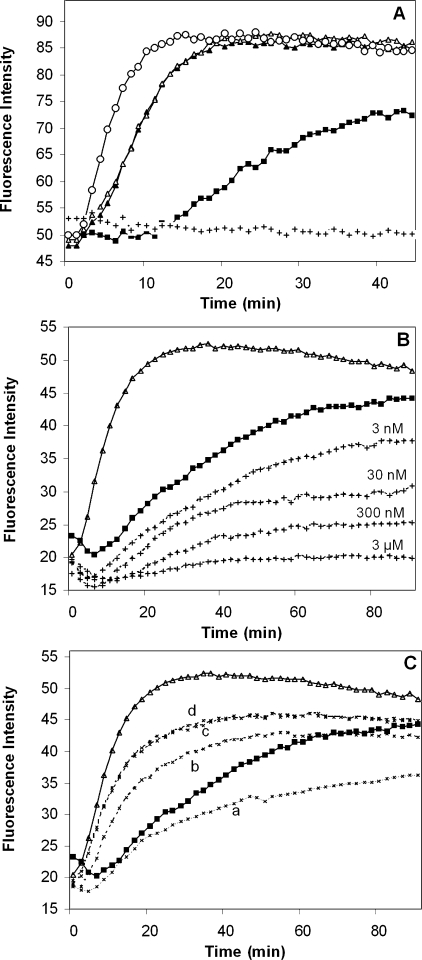
HCV Core Protein Enhances Tubulin Polymerization in Vitro—To determine whether the binding of the HCV core protein to tubulin via D1 affects tubulin polymerization, we conducted in vitro tubulin polymerization analyses. This assay is based on fluorescence enhancement due to the incorporation of a fluorescent reporter to MTs as polymerization occurs, generating a polymerization curve that represents the three phases of microtubule formation: nucleation, growth, and steady state equilibrium. Compounds that interact with tubulin could alter one or more of these phases of polymerization. The addition of either of the two core proteins aa 2-169 or aa 2-117 to a tubulin polymerization reaction significantly increased the rate of microtubule polymerization to a similar extent as the antimitotic drug paclitaxel, which eliminates the nucleation phase and enhances the growth phase (Fig. 9A). Adding vinblastin decreased the polymerization rate and caused significant reduction in polymer mass. Various concentrations of vinblastin induced a dose-dependent decrease of microtubule mass in vitro (Fig. 9B).
FIGURE 9.
The HCV core protein promotes polymerization of tubulin in vitro. A, polymerization of tubulin was carried out in the absence of core protein (filled squares) or in the presence of core protein aa 2-169 (open triangles) or aa 2-117 (filled triangles) at a concentration of 20 μg/ml. Polymerization in the presence of vinblastin (3 μm) (crosses) or paclitaxel (3 μm) (open circles) is shown. B, to investigate the influence of various concentrations of vinblastin on tubulin polymerization in vitro, the drug was added to the standard polymerization mixture at a concentration range from 3 nm to 3 μm. The effect of the drug was monitored by a fluorescence assay as a function of time (crosses). Tubulin polymerization in the absence of drug (filled squares) and in the presence of HCV core protein aa 2-122 (open triangles) is shown. C, the HCV core protein and vinblastin were simultaneously added to the polymerization mixture. Tubulin polymerization in the absence of core (filled squares) and in the presence of core aa (2-122) (open triangles) was assessed. Curves (crosses) represent polymerization of tubulin in the presence of core protein aa 2-122 and decreasing concentrations of vinblastin (3 μm, 300 nm, 30 nm, and 3 nm, designated as a-d, respectively), when both compounds were added simultaneously.
We examined further whether the HCV core could promote tubulin polymerization in the presence of vinblastin. In this series of experiments, different concentrations of vinblastin and a constant concentration of HCV core were used. The polymerization process was monitored by an in vitro polymerization assay in comparison with standard polymerization conditions. As shown in Fig. 9C, in the absence of vinblastin, HCV core aa 2-122 significantly increased tubulin polymerization. Vinblastin in a concentration range from 3 nm to 3 μm decreased the core-mediated enhancement of MT formation in a dose-dependent manner. Nevertheless, in the presence of core, the effect of the drug was attenuated, as compared with that induced in the absence of core. These observations are consistent with the potent effect of the HCV core protein on tubulin polymerization and the role of the N-terminal D1 domain of core in this process.
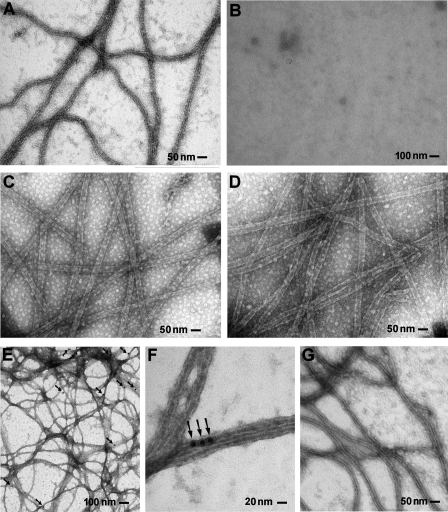
HCV Core Protein Remains Associated with Polymerized Microtubules—To ascertain that, indeed, the HCV core protein enhances tubulin polymerization and induces formation of microtubules, MTs formed in vitro in the presence of core were analyzed by electron microscopy. This series of experiments demonstrated that MTs formed in the presence of core (shown in Fig. 10D) were very similar to those formed in the presence of paclitaxel (Fig. 10C), which also enhances tubulin polymerization. Strikingly, MTs polymerized in the presence of core bound anti-core antibodies, as ascertained by immune electron microscopy. These observations provided evidence that the HCV core enhances MT polymerization in the absence of other co-factors (such as MT-associated proteins) and associates, at least temporarily, with MTs (Fig. 10, E and F).
FIGURE 10.
HCV core protein promotes tubulin polymerization and associates with MTs. A-D, control experiments to ascertain that MT polymerizes in the presence of core protein. A, microtubules formed in vitro in standard conditions, when tubulin (at a concentration of 2 mg/ml) was incubated in polymerization buffer for 10 min at 37 °C in the absence of drugs and HCV core protein. B, microtubules are not observed when Nocodazol (3 μm) is added to the MT polymerization mixture; C, microtubules polymerized in the presence of 5 μm paclitaxel, a drug that enhances polymerization and stabilizes microtubules; D, microtubules formed in the presence of HCV core protein aa 2-117 in the polymerization mixture at a concentration of 20 μg/ml. Samples were coated on Formvar grids, fixed with 2% glutaraldehyde, washed, and stained with 4% uranyl acetate. E-G, immune electron microscopy analyses of microtubules polymerized in vitro in the presence of core protein. Microtubules formed in the presence of core protein aa 2-117 (E and F) and in the presence of paclitaxel as negative control (G). Samples were fixed with 2% glutaraldehyde, washed, and stained with 4% uranyl acetate. Arrows, colloidal gold beads.
DISCUSSION
HCV is considered to enter the cell by clathrin-mediated endocytosis and fusion between the viral envelope and endosome membranes that occurs upon acidification of the endosomal compartment (35, 46). Several cell surface molecules have been identified as HCV receptors (CD-81, scavenger receptor SR-BI, Claudin-1, and occludin) (for a review, see Ref. 47); however, the mechanisms of virus traffic during cell entry and early postentry steps remain unknown.
In this report using the JFH1 in vitro model, which reproduces a complete virus cell cycle, and drugs affecting main cytoskeleton components, we provide evidence that intact and dynamic MTs play a key role in the early steps of the virus cycle leading to the establishment of productive HCV infection. Indeed, drugs that inhibit tubulin polymerization (vinblastin) or disrupt (nocodazol) or stabilize (paclitaxel) MTs inhibit also early steps of infection. In contrast, cytochalasin D, an inhibitor of actin polymerization and thus actin-dependent cellular transport, had no direct effect on the initiation of HCV infection.
Using virus pseudotypes (HCVpp) as a model for studies of HCV cell entry (32-35), we demonstrate that the first steps of virus internalization, from its attachment to the cell surface until fusion of the viral envelope glycoproteins within an endosomal compartment, depend on the intact MT network. Indeed, vinblastin, which blocked HCVpp infection, equally reduced cell entry of other pseudotype particles (VSV-Gpp), which require MT-dependent endocytic transport.
Our further studies carried out on the HCVcc model demonstrated also that early postentry steps of the virus cycle, after fusion of the virus envelope, which is estimated to be completed 2 h after the initiation of infection, require functional MTs. Indeed, a significant decrease in the production of the virus was observed when vinblastin was applied at various time points up to 8 h postinfection. These data showed that an intact microtubule network is needed for early events of the HCV cell cycle. These events most probably also concern virus nucleocapsid release and early transport, subsequent to virus fusion.
The effect of MT-affecting drugs is concentration-dependent and thus may reflect the mechanisms involved in virus interaction with MTs (40, 41). We observed the substantial effect of vinblastin and nocodazol on the initiation of HCV infection, when these drugs were used even in low micromolar and nanomolar concentrations. Indeed, in micromolar concentrations, nocodazole induces depolymerization of MTs and tubulin aggregation. Nocodazol at low concentrations suppresses the association and dissociation rates of tubulin, stabilizing the MT dynamics, but does not alter the equilibrium between a MT polymer and soluble tubulin. Equally, nanomolar concentrations of vinblastin do not induce perceptible changes in MT structure; however, at these concentrations, vinblastin is known to suppress MT dynamic instability and treadmilling mechanisms. Paclitaxel, an MT-stabilizing drug, also had a significant inhibitory effect on the establishment of productive HCV infection. Altogether, these findings suggested that early virus transport depends on MT dynamic instability and/or treadmilling mechanisms and thus may involve the interaction of virus core protein with microtubules.
Indeed, our observations using confocal microscopy evidenced juxtaposition of core with microtubules in HCV-infected cells. These data corroborated with our in vitro findings, which identified α- and β-tubulin chains as cellular proteins directly binding HCV core and evidenced direct interactions between the HCV core protein and tubulin. SPR (BIAcore) studies permitted the identification of the N-terminal D1 as a part of the core protein that directly binds to α/β-tubulin heterodimers with high affinity (in the nanomolar range). We equally provided evidence that the direct interaction of D1 of core with tubulin, without any mediators (such as MT-associated proteins), promotes tubulin polymerization in vitro. HCV core displayed an effect opposite to that of vinblastin, a drug that inhibits MT polymerization. When applied in the presence of vinblastin, core attenuated the inhibitory effect of the drug on MT polymerization. Our observations by electron microscopy confirmed that microtubules formed in the presence of core are very similar to those formed in the presence of paclitaxel, a drug known to enhance microtubule polymerization (48). Most importantly, immune electron microscopy analyses revealed that core protein remained associated with microtubules formed in vitro in its presence.
Viral genome-protein complexes, viruses, and subviral particles can be transported within the host cell cytoplasm during cell entry, from the plasma membrane to the site of viral replication, and during viral assembly and egress to the plasma membrane for their release into the extracellular milieu (13, 16). The association of viral proteins with microtubule components has been reported for several viruses: vaccinia virus (49), herpes simplex virus (50), murine coronavirus (51), pseudorabies virus (52), vesicular stomatitis virus (53), rotavirus (54), and M protein of Sendai virus (55). During infection, some viruses are transported on MTs within membranous vesicles (18, 19), whereas others use mechanisms driven by microtubule motors for transport of their capsids or subviral particles on MTs interacting either with microtubule motors (20-23) or with microtubule-associated proteins (56).
Only three proteins have been shown up to now to directly interact with tubulin and to enhance tubulin polymerization in vitro: human immunodeficiency virus Tat protein (57), tobacco mosaic virus movement protein (58), and Ebola virus matrix protein VP40 (59). Nevertheless, the exact mechanisms and role of these interactions in virus transport remain to be elucidated.
Transport of molecules along MTs can be achieved by using MT-dependent motors, such as kinesins and dyneins (11, 12, 60). These motors use energy derived from ATP hydrolysis to move cargo, but motor-driven processes are not related to polymerization and depolymerization kinetics (14). Another mechanism of cytosolic transport on MTs, called “treadmilling” (13, 14) involves polymerization at the plus end and depolymerization at the minus end after severing of MTs by cellular katenin (15). Thus, intracellular transport of the HCV core, similarly to the tobacco virus encoded movement protein or the human immunodeficiency virus Tat protein, which also enhance MT polymerization (57, 58), can be mediated by mechanisms related to tubulin polymerization.
HCV-RNA replication takes place in the cytoplasm in membrane-associated replication complexes designated as membranous webs (61). The cytoskeleton components (microtubules and actin filaments) provide tracks for the movement of replication complexes (24-26). Two types of HCV replication complexes have recently been reported; large structures, with restricted mobility appeared 24 h postinfection, and small replication complexes were formed 12-24 h postinfection and moved in a MT-dependent manner (26). These studies estimated that the formation of replication complexes starts about 12 h after the initiation of HCVcc infection, thus later than the early events analyzed in our study.
HCV morphogenesis and the secretion of progeny virus also require a functional microtubule network (27). HCV core protein probably recruits nonstructural proteins and replication complexes to lipid droplet-associated membranes, and this process is directly connected to the virus assembly pathway in HCV-infected cells (62). In HCV-infected cells, newly synthesized core protein loads lipid droplets and progressively coats the entire organelle, replacing adipocyte differentiation-related protein (37). Core protein-coated lipid droplets are transported on microtubules in a dynein-dependent manner, 48-72 h postinfection, to the perinuclear area, where virus morphogenesis takes place (27). The above quoted studies focused either on the formation and transport of HCV replication complexes or on virus morphogenesis and release but did not address the role of microtubules in HCV cell entry and early transport. Our study provides the first evidence that a microtubule network that is both intact and dynamic plays a key role in virus internalization, leading to the establishment of infection. Although many viruses use the microtubule network for virus transport at various steps of their life cycle, our study suggests that the establishment of productive HCV infection requires a dynamic process driven by microtubule polymerization. Such mechanisms have been reported for a few viruses, such as duck hepatitis B virus or HHP-8 (63, 64). Indeed, for these viruses, early steps of infection subsequent to the virus cell entry depend on both intact microtubules and their dynamic turnover. Although this point might require further studies, our observations by electron microscopy favor the hypothesis that similarly to the tobacco virus X encoded movement protein (58) or the human immunodeficiency virus Tat protein (57), HCV core could integrate into the microtubule lattice to exploit assembly dynamics and/or treadmilling mechanisms for transport of virus nucleocapsid in infected cells.
Another possible function of the HCV core protein could be the regulation of dynein- and kinesin-based motions. Since bidirectional motion is widespread and direction of the microtubule-dependent transport depends on the balance between plus end and minus end directed motion, the cargo-based regulation of motor-MT interactions may be a key mechanism that is used to control cargo transport (11, 12, 60). Proteins showing the ability to bind to MTs independently of motors could play a crucial role in such regulation (12). In accordance with this notion, direct binding of core to tubulin and activation of microtubule polymerization could alter the balance of dynein- and kinesin-based motions. Indeed, the regulation of motor complexes by virus capsid proteins has been suggested (65), but the exact mechanism of such regulations has not yet been elucidated.
Our findings show a novel and potentially important property of the core protein and suggest that HCV might exploit the MT lattice for transport of the virus nucleocapsid by polymerization-related mechanisms. Such interactions could also play a role at later steps of the viral cycle, during viral morphogenesis and secretion. Improving our understanding of the consequences of core-MT interactions might facilitate the development of new therapeutic approaches targeted to inhibit HCV infection. Indeed, molecules modifying MT dynamics are already an integral part of anti-cancer treatment (66).
Acknowledgments
We thank Y. Rouille for valuable discussion, T. Wakita for providing the JFH-1 HCVcc clone, and J. F. Delagneau for ACAP-27 monoclonal antibody. We express our gratitude to J. D'Alayer for help in protein chip and pull-down assays and E. Perret in immunofluorescence microscopy. We are grateful to K. M. Kean for a critical review of the manuscript.
This work was supported in part by grants from the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS) and Action Concerté de l'Institut Pasteur.
Footnotes
The abbreviations used are: HCV, hepatitis C virus; HCVcc, cell culture-produced hepatitis C virus; MT, microtubule; SPR, surface plasmon resonance; HCVpp, pseudoparticles harboring envelope glycoproteins of hepatitis C virus; GADPH, glyceraldehyde-3-phosphate dehydrogenase; SELDI, surface-enhanced laser desorption ionization; MALDI, matrix-assisted laser desorption ionization; TOF, time of flight; aa, amino acids; RT, reverse transcription; qRT, quantitative reverse transcription; PBS, phosphate-buffered saline; MS, mass spectrometry; MOWSE, molecular weight search.
References
- 1.Pawlotsky, J. M. (2004) Trends Microbiol. 12 96-102 [DOI] [PubMed] [Google Scholar]
- 2.Penin, F., Dubuisson, J., Rey, F. A., Moradpour, D., and Pawlotsky, J. M. (2004) Hepatology 39 5-19 [DOI] [PubMed] [Google Scholar]
- 3.McLauchlan, J., Lemberg, M. K., Hope, G., and Martoglio, B. (2002) EMBO J. 21 3980-3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hope, R. G., and McLauchlan, J. (2000) J. Gen. Virol. 81 1913-1925 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki, R., Sakamoto, S., Tsutsumi, T., Rikimaru, A., Tanaka, K., Shimoike, T., Moriishi, K., Iwasaki, T., Mizumoto, K., Matsuura, Y., Miyamura, T., and Suzuki, T. (2005) J. Virol. 79 1271-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi, K., Kishimoto, S., Yoshizawa, H., Okamoto, H., Yoshikawa, A., and Mishiro, S. (1992) Virology 191 431-434 [DOI] [PubMed] [Google Scholar]
- 7.Fan, Z., Yang, Q. R., Twu, J. S., and Sherker, A. H. (1999) J. Med. Virol. 59 131-134 [PubMed] [Google Scholar]
- 8.Boulant, S., Montserret, R., Hope, R. G., Ratinier, M., Targett-Adams, P., Lavergne, J. P., Penin, F., and McLauchlan, J. (2006) J. Biol. Chem. 281 22236-22247 [DOI] [PubMed] [Google Scholar]
- 9.Nogales, E. (2000) Annu. Rev. Biochem. 69 277-302 [DOI] [PubMed] [Google Scholar]
- 10.Heald, R., and Nogales, E. (2002) J. Cell Sci. 115 3-4 [DOI] [PubMed] [Google Scholar]
- 11.Welte, M. A. (2004) Curr. Biol. 14 525-537 [DOI] [PubMed] [Google Scholar]
- 12.Gross, S. P., Vershinin, M., and Shubeita, G. T. (2007) Curr. Biol. 17 478-486 [DOI] [PubMed] [Google Scholar]
- 13.Sodeik, B. (2000) Trends Microbiol. 8 465-472 [DOI] [PubMed] [Google Scholar]
- 14.Rodionov, V. I., and Borisy, G. G. (1997) Science 275 215-218 [DOI] [PubMed] [Google Scholar]
- 15.Quarmby, L. (2000) J. Cell Sci. 113 2821-2827 [DOI] [PubMed] [Google Scholar]
- 16.Dohner, K., Nagel, C. H., and Sodeik, B. (2005) Trends Microbiol. 13 320-327 [DOI] [PubMed] [Google Scholar]
- 17.Greber, U. F., and Way, M. (2006) Cell 124 741-754 [DOI] [PubMed] [Google Scholar]
- 18.Yonezawa, A., Cavrois, M., and Greene, W. C. (2005) J. Virol. 79 918-926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgi, A., Mottola-Hartshorn, C., Warner, A., Fields, B., and Chen, L. B. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6579-6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sodeik, B., Ebersold, M. W., and Helenius, A. (1997) J. Cell Biol. 136 1007-1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanjuan, N., Porras, A., and Otero, J. (2003) Virology 313 105-116 [DOI] [PubMed] [Google Scholar]
- 22.Ogawa-Goto, K., Tanaka, K., Gibson, W., Moriishi, E., Miura, Y., Kurata, T., Irie, S., and Sata, T. (2003) J. Virol. 77 8541-8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suomalainen, M., Nakano, M. Y., Keller, S., Boucke, K., Stidwill, R. P., and Greber, U. F. (1999) J. Cell Biol. 144 657-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, C. K., Jeng, K. S., Machida, K., and Lai, M. M. (2008) J. Virol. 82 8838-8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. M., Gretton, S. N., McLauchlan, J., and Targett-Adams, P. (2007) J. Gen. Virol. 88 470-475 [DOI] [PubMed] [Google Scholar]
- 26.Wolk, B., Buchele, B., Moradpour, D., and Rice, C. M. (2008) J. Virol. 82 10519-10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boulant, S., Douglas, M. W., Moody, L., Budkowska, A., Targett-Adams, P., and McLauchlan, J. (2008) Traffic 9 1268-1282 [DOI] [PubMed] [Google Scholar]
- 28.Maillard, P., Lavergne, J. P., Siberil, S., Faure, G., Roohvand, F., Petres, S., Teillaud, J. L., and Budkowska, A. (2004) J. Biol. Chem. 279 2430-2437 [DOI] [PubMed] [Google Scholar]
- 29.Boulant, S., Becchi, M., Penin, F., and Lavergne, J. P. (2003) J. Biol. Chem. 278 45785-45792 [DOI] [PubMed] [Google Scholar]
- 30.Wakita, T., Pietschmann, T., Kato, T., Date, T., Miyamoto, M., Zhao, Z., Murthy, K., Habermann, A., Krausslich, H. G., Mizokami, M., Bartenschlager, R., and Liang, T. J. (2005) Nat. Med. 11 791-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maillard, P., Huby, T., Andreo, U., Moreau, M., Chapman, J., and Budkowska, A. (2006) FASEB J. 20 735-737 [DOI] [PubMed] [Google Scholar]
- 32.Bartosch, B., Dubuisson, J., and Cosset, F. L. (2003) J. Exp. Med. 197 633-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu, M., Zhang, J., Flint, M., Logvinoff, C., Cheng-Mayer, C., Rice, C. M., and McKeating, J. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7271-7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummer, H. E., Maerz, A., and Poumbourios, P. (2003) FEBS Lett. 546 385-390 [DOI] [PubMed] [Google Scholar]
- 35.Blanchard, E., Belouzard, S., Goueslain, L., Wakita, T., Dubuisson, J., Wychowski, C., and Rouille, Y. (2006) J. Virol. 80 6964-6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Targett-Adams, P., and McLauchlan, J. (2005) J. Gen. Virol. 86 3075-3080 [DOI] [PubMed] [Google Scholar]
- 37.Boulant, S., Targett-Adams, P., and McLauchlan, J. (2007) J. Gen. Virol. 88 2204-2213 [DOI] [PubMed] [Google Scholar]
- 38.Luby-Phelps, K. (2000) Int. Rev. Cytol. 192 189-221 [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850-858 [DOI] [PubMed] [Google Scholar]
- 40.Jordan, M. A., and Wilson, L. (1998) Methods Enzymol. 298 252-276 [DOI] [PubMed] [Google Scholar]
- 41.Jordan, M. A., Thrower, D., and Wilson, L. (1992) J. Cell Sci. 102 401-416 [DOI] [PubMed] [Google Scholar]
- 42.Mabit, H., Nakano, M. Y., Prank, U., Saam, B., Dohner, K., Sodeik, B., and Greber, U. F. (2002) J. Virol. 76 9962-9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luduena, R. F. (1998) Int. Rev. Cytol. 178 207-275 [DOI] [PubMed] [Google Scholar]
- 44.Miller, L. M., Menthena, A., Chatterjee, C., Verdier-Pinard, P., Novikoff, P. M., Horwitz, S. B., and Angeletti, R. H. (2008) Biochemistry 47 7572-7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan, K. F. (1988) Annu. Rev. Cell Biol. 4 687-716 [DOI] [PubMed] [Google Scholar]
- 46.Meertens, L., Bertaux, C., and Dragic, T. (2006) J. Virol. 80 11571-11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burlone, M., and Budkowska, A. (2009) J. Gen. Virol. 90 in press [DOI] [PubMed]
- 48.Mamber, S. W., Mikkilineni, A. B., Pack, E. J., Rosser, M. P., Wong, H., Ueda, Y., and Forenza, S. (1995) J. Pharmacol. Exp. Ther. 274 877-883 [PubMed] [Google Scholar]
- 49.Ploubidou, A., Moreau, V., Ashman, K., Reckmann, I., Gonzalez, C., and Way, M. (2000) EMBO J. 19 3932-3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott, G., and O'Hare, P. (1998) J. Virol. 72 6448-6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalicharran, K., and Dales, S. (1996) Trends Microbiol. 4 264-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaelin, K., Dezelee, S., Masse, M. J., Bras, F., and Flamand, A. (2000) J. Virol. 74 474-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melki, R., Gaudin, Y., and Blondel, D. (1994) Virology 202 339-347 [DOI] [PubMed] [Google Scholar]
- 54.Nejmeddine, M., Trugnan, G., Sapin, C., Kohli, E., Svensson, L., Lopez, S., and Cohen, J. (2000) J. Virol. 74 3313-3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogino, T., Iwama, M., Ohsawa, Y., and Mizumoto, K. (2003) Biochem. Biophys. Res. Commun. 311 283-293 [DOI] [PubMed] [Google Scholar]
- 56.de Soultrait, V. R., Caumont, A., Durrens, P., Calmels, C., Parissi, V., Recordon, P., Bon, E., Desjobert, C., Tarrago-Litvak, L., and Fournier, M. (2002) Biochim. Biophys. Acta 1575 40-48 [DOI] [PubMed] [Google Scholar]
- 57.de Mareuil, J., Carre, M., Barbier, P., Campbell, G. R., Lancelot, S., Opi, S., Esquieu, D., Watkins, J. D., Prevot, C., Braguer, D., Peyrot, V., and Loret, E. P. (2005) Retrovirology 2 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyko, V., Ferralli, J., and Heinlein, M. (2000) Plant J. 22 315-325 [DOI] [PubMed] [Google Scholar]
- 59.Ruthel, G., Demmin, G. L., Kallstrom, G., Javid, M. P., Badie, S. S., Will, A. B., Nelle, T., Schokman, R., Nguyen, T. L., Carra, J. H., Bavari, S., and Aman, M. J. (2005) J. Virol. 79 4709-4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schliwa, M., and Woehlke, G. (2003) Nature 422 759-765 [DOI] [PubMed] [Google Scholar]
- 61.Gosert, R., Egger, D., Lohmann, V., Bartenschlager, R., Blum, H. E., Bienz, K., and Moradpour, D. (2003) J. Virol. 77 5487-5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyanari, Y., Atsuzawa, K., Usuda, N., Watashi, K., Hishiki, T., Zayas, M., Bartenschlager, R., Wakita, T., Hijikata, M., and Shimotohno, K. (2007) Nat. Cell Biol. 9 1089-1097 [DOI] [PubMed] [Google Scholar]
- 63.Funk, A., Mhamdi, M., Lin, L., Will, H., and Sirma, H. (2004) J. Virol. 78 8289-8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naranatt, P. P., Krishnan, H. H., Smith, M. S., and Chandran, B. (2005) J. Virol. 79 1191-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ploubidou, A., and Way, M. (2001) Curr. Opin. Cell Biol. 13 97-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drukman, S., and Kavallaris, M. (2002) Int. J. Oncol. 21 621-628 [PubMed] [Google Scholar]