Abstract
p53-dependent apoptosis is modulated by the ASPP family of proteins (apoptosis-stimulating proteins of p53; also called ankyrin repeat-, Src homology 3 domain-, and Pro-rich region-containing proteins). Its three known members, ASPP1, ASPP2, and iASPP, were previously found to interact with p53, influencing the apoptotic response of cells without affecting p53-induced cell cycle arrest. More specifically, the bona fide tumor suppressors, ASPP1 and ASPP2, bind to the core domain of p53 and stimulate transcription of apoptotic genes, whereas oncogenic iASPP also binds to the p53 core domain but inhibits p53-dependent apoptosis. Although the general interaction regions are known, details of the interfaces for each p53-ASPP complex have not been evaluated. We undertook a comprehensive biophysical characterization of ASPP-p53 complex formation and mapped the binding interfaces by NMR. We found that the interaction interface on p53 for the proapoptotic protein ASPP2 is distinct from that for the antiapoptotic iASPP. ASPP2 primarily binds to the core domain of p53, whereas iASPP predominantly interacts with a linker region adjacent to the core domain. Our detailed structural analyses of the ASPP-p53 interactions provide insight into the structural basis of the differential behavior of pro- and antiapoptotic ASPP family members.
Since the discovery of p53 in 1979 (1-3), the essential function of this tumor suppressor in preventing uncontrolled cell proliferation after DNA insult has become clear; p53 induces expression of genes to initiate cell cycle arrest or apoptosis (4-7). However, the detailed numerous mechanisms by which p53 carries out its function are not fully understood. For example, control of cellular fate by p53 through cell cycle arrest rather than apoptosis (or vice versa) is still a topic of debate, and several types of p53 involvement and modulation have been described (8-12).
An early clue that these two cellular responses can be separated emerged from studies characterizing p53 mutants that were defective in their ability to induce apoptosis but not cell cycle arrest (13-15). Since these initial reports, post-translational modifications of p53 and interactions with specific cellular proteins have been found to be more closely associated with one pathway over the other. For example, phosphorylation of Ser46 (16-18) or acetylation of Lys120 in the DNA binding domain by MYST family acetyltransferases (19, 20) purportedly renders p53 more effective in activating proapoptotic programs. Proteins that bind p53 have also been implicated in promoting p53-dependent apoptosis (21, 22).
The ASPP proteins constitute a recently described new family of proteins that bind and modulate p53-dependent apoptosis (23). Their name is based on the domain organization of the proteins (ankyrin repeat, SH3,3 and proline-rich domain-containing protein) as well as their function (apoptosis-stimulating protein of p53). The founding member of the family, ASPP2, was initially identified as 53BP2 (p53-binding protein 2) in a yeast two-hybrid screen, using the p53 DNA binding core domain as bait (24). ASPP1 was identified later in a homology search (25). Functional studies revealed that p53-induced apoptosis was substantially enhanced in the presence of ASPP1 or ASPP2 (26-28), and complexes with ASPP1 or ASPP2 increased the affinity of p53 for promoters of proapoptotic genes (28, 29). Such preferential activation of apoptosis by ASPP1/2 was also observed for p63 and p73 (30). The third member of ASPP family, iASPP, was originally identified as an inhibitor of NFκB (31). Compared with ASPP1/2, iASPP inhibits p53-dependent apoptosis. Given the high sequence similarity between all three proteins, it may be possible that iASPP simply functions as a competitive inhibitor of ASPP1/2. However, iASPP specifically inhibits p53-dependent induction of proapoptotic genes (29, 32), suggesting a mechanism of inhibition that involves iASPP altering the promoter binding activity of p53.
The regions of ASPPs that interact with p53 have been mapped to the C termini, consisting of four ankyrin repeats and an SH3 domain (Fig. 1, A and B). The crystal structure of the complex between the C terminus of ASPP2 (ASPP2-CT) and the DNA binding core domain of p53 (p53-CD; Fig. 1C) reveals that the third and fourth ankyrin repeats and SH3 domain of ASPP2 contact the same p53-CD surface that interfaces with DNA (33). The crystal structure of the C-terminal region of iASPP (iASPP-CT) shows a very similar structure, and binding of iASPP-CT and ASPP2-CT to p53-CD exhibits comparable affinities (34).
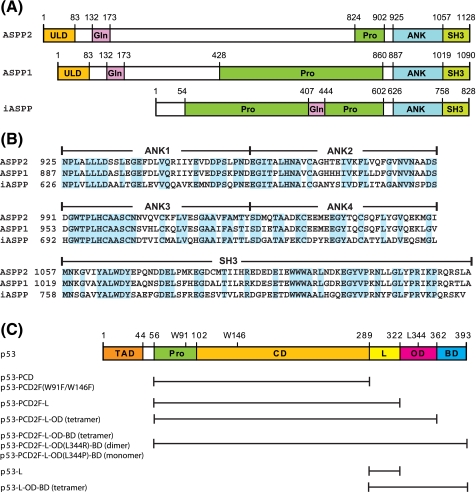
FIGURE 1.
Amino acid sequences and domain organization of ASPP and p53 proteins. A, domain organization of ASPP2, ASPP1, and iASPP. Individual domains are as follows: ubiquitin-like (ULD), glutamine-rich (Gln), proline-rich (Pro), ankyrin repeats (ANK), and Src homology 3 (SH3). B, sequence alignment of the C-terminal regions of ASPP2, ASPP1, and iASPP. Identical residues are highlighted in cyan. The domain boundaries are indicated at the top of the protein sequences. C, domain organization of p53. Individual domains are the transcription activation (TAD), the proline-rich (Pro), the DNA binding or core (CD), the linker (L), the oligomerization or tetramerization (OD), and the basic (BD) domains. All p53 protein constructs used in the present study are listed: p53-PCD (residues 56-289), p53-PCD2F (residues 56-289 with W91F and W146F), p53-PCD2F-L (residues 56-322 with W91F and W146F), p53-PCD2F-L-OD (residues 56-362 with W91F and W146F), p53-PCD2F-L-OD-BD (residues 56-393 with W91F and W146F), p53-PCD2F-L-OD(L344R)-BD (residues 56-393 with W91F, W146F, and L344R), p53-PCD2F-L-OD(L344P)-BD (residues 56-393 with W91F, W146F, and L344P), p53-L (residues 289-322), and p53-L-OD-BD (residues 289-393).
Given the opposing functional outcomes of ASPP2 and iASPP interactions, we investigated whether the detailed molecular interactions of these two proteins with p53 are distinct. Here, we report on biochemical and structural studies of ASPP2-CT and iASPP-CT with various constructs of p53 (Fig. 1C). ASPP-CT binding to p53 that contains the Pro-rich and DNA-binding core domains (p53-PCD) was followed by isothermal titration calorimetry (ITC), fluorescence, and NMR methods. Our results show that both ASPP2-CT and iASPP-CT utilize very similar interaction surfaces to contact p53-PCD. However, in stark contrast to previous reports, we found that ASPP2-CT binds to p53-PCD much more tightly than iASPP-CT (∼60-fold). Extension of these binding studies to several p53 constructs (Fig. 1C) revealed that iASPP-CT and ASPP2-CT exhibit different affinities, suggesting that the molecular interaction determinants are unique to the individual complexes. Although the core domain of p53 is the primary binding target for ASPP2-CT, we found that the p53 linker region is the highest affinity target for iASPP-CT. Therefore, different regions of p53 appear to contribute to the proapopototic and antiapoptotic complexes, respectively, allowing modulation of the functional outcome. Detailed knowledge of the differential interfaces in the ASPP2-p53 and iASPP-p53 complexes will aid in the design of small therapeutic molecules that specifically target either of the two interactions.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—cDNAs encoding human p53 and ASPP2 were purchased from Invitrogen. Constructs coding for residues 56-289 (p53-PCD), 56-322 (p53-PCD-L), 56-362 (p53-PCD-L-OD), 289-322 (p53-L), 289-393 (p53-L-OD-BD), and 56-393 (p53-PCD-L-OD-BD) of p53 (Fig. 1C) were amplified and subcloned into pET21a (EMD Chemicals, Inc., San Diego, CA) using NdeI and XhoI sites. The final protein products included an N-terminal Met and Leu-Glu-His6 at their C termini. Gene constructs coding for residues 289-322 of p53 (p53-L), 925-1128 of ASPP2 (ASPP2-CT), and 623-828 of iASPP (iASPP-CT) were amplified and subcloned into pET32a (EMD Chemicals) using EcoRI and XhoI sites. A TEV protease recognition sequence (ENLYFQS) was created between the BamHI and EcoRI sites in pET32, at the C terminus of thioredoxin. Final ASPP proteins, after TEV protease cleavage, contained Ser-Glu-Phe and Leu-Glu-His6 at their N termini and C termini, respectively. p53-L only possessed the three extra residues (Ser-Glu-Phe) at the N terminus. All p53 mutant constructs, p53-PCD-W91F/W146F, p53-PCD-L-OD-W91F/W146F, p53-PCD-L-OD-BD-W91F/W146F, p53-PCD-L-OD-BD-W91F/W146F/L344R, and p53-PCD-L-OD-BD-W91F/W146F/L344P (Fig. 1C), were created using QuikChange site-directed mutagenesis kits (Stratagene, La Jolla, CA). Nucleotide sequences were verified for the entire coding regions of all constructs.
Proteins were expressed in Escherichia coli Rosetta 2 (DE3), cultured in Luria-Bertani medium, using 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for induction and growth at 18 °C for 16 h. Soluble forms of His-tagged proteins were first purified using 5 ml of Ni2+-nitrilotriacetic acid columns, and aggregated material was removed by gel filtration column chromatography using Hi-Load Superdex200 16/60 (GE Healthcare) equilibrated with a buffer containing 25 mm sodium phosphate, pH 7.5, 50 mm NaCl, 1 mm dithiothreitol, and 0.02% sodium azide. The thioredoxin portions of the fusion proteins were cleaved from p53-L, ASPP2-CT, and iASPP-CT by incubating with TEV protease (ratio of 20:1) overnight at 4 °C. The expression plasmid PRK709 for TEV protease was obtained from Addgene Inc. (Cambridge, MA), and TEV protease was purified as described by Nallamsetty et al. (35). All final purifications of p53 proteins were performed over a Hi-Trap SP column (GE Healthcare) at pH 6.5 using a 0-1 m NaCl gradient. ASPP2-CT and iASPP-CT were purified over a Hi-Trap QP column (GE Healthcare) at pH 7.5 and a 0-1 m NaCl gradient. Buffer exchange was carried out using Amicon concentrators (Millipore, Billerica, MA), and proteins were stored at 4 °C in solution or as lyophilized powders at -80 °C. The p53-L peptides were purified over a C4 reverse phase column using an acetonitrile (5-80%) gradient with 0.1% formic acid. For isotopic labeling, proteins were expressed in modified minimal medium using 15NH4Cl and [U-13C6]glucose or [U-13C6, 2H7]-glucose as sole nitrogen and/or carbon sources, respectively. For deuterium labeling, growth media were prepared with 99.9% 2H2O. U-13C,15N,2H-Labeled proteins with selective Val/Leu-methyl and Phe/Tyr-12C,1H-protonation were prepared as described for the isotopic labeling above, except that U-13C5,15N,2H2(2,3)-labeled valine and/or unlabeled tyrosine and phenylalanine were added prior to induction (36, 37). The molecular masses of all purified proteins were confirmed by mass spectrometry, and isotope labeling efficiency was estimated to be greater than 98% for all labeled proteins. Protein concentrations were determined using theoretical extinction coefficients based on amino acid sequences using the ExPASy Proteomics Server (available on the World Wide Web). The concentration of p53-L was determined by amino acid analysis.
ITC—Typically, p53 protein or ASPP2-CT, at concentrations of 20-40 μm, stirred at 307 rpm in the sample cell, were titrated with aliquots of 240-400 μm solutions of ASPP2-CT or p53 at 12 °C in the calorimeter (MicroCal Inc., Northampton, MA). Titrations were carried out in 25 mm sodium phosphate, pH 7.5, 1 mm dithiothreitol, and 0.02% sodium azide with varying amounts of NaCl (50-150 mm). In some titrations, 25 mm sodium phosphate, pH 7.5, was replaced by 25 mm Tris-HCl, pH 7.5. All samples were extensively buffer-exchanged using Amicon concentrators (Millipore) prior to the titrations. 10-μl aliquots were used for injections (the first injection was 3 μl) at 250-s intervals (initial delay, 60 s). Titrations into the buffers were used for base-line corrections. Data analyses were performed using Origin 7 software (OriginLab Corp., Northampton, MA).
NMR Spectroscopy—All NMR experiments were conducted at 17 °C using protein samples in 25 mm sodium phosphate buffer, pH 7.5, 50 or 150 mm NaCl, 1 mm dithiothreitol, and 0.02% sodium azide, using Bruker Avance 900-, 800-, 700-, and 600-MHz spectrometers, equipped with 5-mm, triple resonance and z axis gradient cryoprobes. For backbone chemical shift assignments of ASPP2-CT and iASPP-CT, two-dimensional 1H-15N HSQC, and three-dimensional HNCACB and HN(CO)CACB experiments were performed on U-13C,15N,2H-labeled proteins or U-13C,15N,2H-labeled ones with selective Val/Ile/Leu-13C1H3 - and Phe/Tyr-12C,1H-protonation. Binding site mapping and ligand titration studies were conducted using 1H-15N TROSY-HSQC experiments on U-15N,2H- or U-13C,15N,2H-labeled samples after adding aliquots of unlabeled partner protein. A total of 5-8 1H-15N TROSY-HSQC spectra were acquired for each titration. Titration curves were plotted for five TROSY-HSQC resonances that exhibited reasonable shifts and no peak overlap or broadening. Dissociation constants were calculated by nonlinear best fitting the 1HN titration curves using KaleidaGraph (Synergy Software, Reading, PA), averaging over the five curves. All spectra were processed with NMRPipe (38) and analyzed using NMRDraw and NMRView (39).
Fluorescence Spectroscopy—Fluorescence titrations were carried out using a QuantaMaster Spectrofluorometer (Photon Technology International, Inc., Birmingham, NJ). Typically, solutions contained 1-2 μm ASPP2-CT or iASPP-CT in 25 mm sodium phosphate, pH 7.5, 1 mm dithiothreitol, 0.02% sodium azide, 50-150 mm NaCl, and varying amounts of nonfluorescent p53 proteins (0-8 μm). Trp fluorescence was excited at 295 nm, and emission spectra were recorded by scanning from 300 to 380 nm with a step size of 1 nm and integration of 1 s. Trp fluorescence experiments were carried out at 12 °C, the temperature of the emission maximum. Any background fluorescence of p53 was subtracted from the fluorescence of ASPPCT-p53 mixtures at the emission maximum. The net increase of Trp fluorescence was plotted against the concentration of added p53 proteins, and the binding isotherms were nonlinear best fitted using Prism (GraphPad Software, La Jolla, CA) to extract dissociation constants.
RESULTS AND DISCUSSION
Binding of ASPP2-CT and iASPP-CT to p53-PCD; Affinities and Binding Site Mapping by NMR—The interactions between the C-terminal domains of ASPP2 and iASPP (ASPP2-CT (residues 925-1128) and iASPP-CT (residues 623-828)) and p53 that comprised the Pro-rich and core domains of p53 (p53-PCD; residues 56-289) were investigated by NMR chemical shift mapping. Backbone resonance assignments of ASPP2-CT and iASPP-CT based on three-dimensional HNCACB and HN(CO)CACB triple resonance experiments were essentially complete for ASPP2-CT (188 of 195 nonproline residues) and almost complete for iASPP-CT (168 of 193 nonproline residues). The binding sites for p53 on ASPP2-CT (Fig. 2, A and B) and iASPP-CT (Fig. 2, D and E) were determined using 1H-15N TROSY-HSQC spectroscopy following the amide resonances upon the addition of increasing amounts of p53-PCD. For ASPP2-CT, both fast (Thr1028, Trp993, and Met1021, each exhibiting small chemical shift changes upon binding) and intermediate-slow (Val1109, Ser1024, and Tyr1108, each exhibiting larger chemical shift changes upon binding) exchange was observed (insets in Fig. 2A). In contrast, most of the iASPP-CT resonances exhibited fast exchange upon p53-PCD binding (inset in Fig. 2D), indicating that the binding of iASPP-CT to p53-PCD is weaker than that of ASPP2-CT. Identification of the bound positions was straightforward for all resonances in the fast exchange regime but was difficult for those exhibiting intermediate-slow exchange; ASPP2-CT resonances exhibiting intermediate-slow exchange were frequently too broad to be observed at the mid-point of titration (insets in Fig. 2A; yellow). In these cases, the chemical shift differences between the free (black) and the first titration point (green) and between the last two titration points (cyan and red) were carefully investigated. Even if changes were very small, this approach allowed us to identify the direction of the shifts and to unambiguously distinguish bound from free frequencies for most residues in both ASPP-CT (Fig. 2B) and iASPP-CT (Fig. 2E).
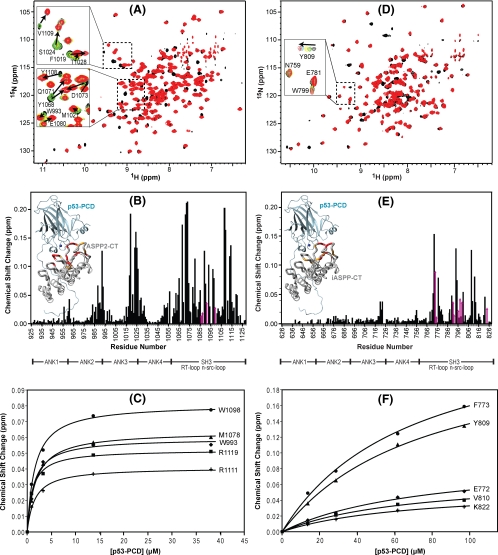
FIGURE 2.
Chemical shift mapping of the interaction between p53-PCD and ASPP2-CT and iASPP-CT. Superposition of the 1H-15N TROSY-HSQC spectra of U-15N,2H-labeled ASPP2-CT (A) and 15N,2H-labeled iASPP-CT (D) without (black) and in the presence (red) of unlabeled p53-PCD. Two selected regions are expanded, and individual resonances along the titration are shown in the inset (increasing amounts of p53-PCD are shown with the colors black → green → yellow → cyan → red). B and E, magnitude of the chemical shift changes versus residue number for ASPP2-CT and iASPP, respectively. The chemical shift change (Δδ) is calculated using the square root of ΔδHN2 + (ΔδN × 0.1)2, with ΔδHN and ΔδN representing the 1HN and 15N chemical shift differences, respectively, between free ASPP-CT (black spectrum in A and D) and the final mixture (red spectrum in A and D). Changes for residues whose resonances broaden and/or are difficult to follow through the entire titration are estimated from earlier titration points and indicated with magenta arrows. Structural mapping of the p53 binding site on ASPP2-CT and iASPP-CT is provided in the insets. Domain boundaries for ASPP2-CT and iASPP-CT are indicated below the residue numbers. The complex structures of ASPP2-CT·p53-PCD and iASPP-CT·p53-PCD were modeled based on the ASPP2-CT·p53-CD crystal structure (Protein Data Bank code 1YCS) by comparative modeling using MODELLER (46), and residues are colored according to the magnitude of their associated chemical shift changes: red, Δδ > (Δδaverage + 2 × S.D.); orange,(Δδaverage + 2 × S.D.) >Δδ > (Δδaverage + 1 × S.D.). C and F, titration curves for selected ASPP2-CT and iASPP-CT 1HN resonances, respectively.
Several resonances in the ASPP2-CT spectrum exhibited significant spectral perturbations upon p53-PCD binding (Fig. 2B); for 23 residues, chemical shift changes larger than the mean plus one S.D. (0.068 ppm) are noted. Structural mapping of the affected residues by NMR reveal them to be clustered in the SH3 domain and in the loops between ankyrin repeats 2 and 3 and repeats 3 and 4. These NMR results are in good agreement with the x-ray model of the ASPP2-CT·p53-CD complex (33). Extraction of a dissociation constant from the titration curves of selected ASPP2-CT resonances (Fig. 2C) yielded very similar values, with an average Kd of 1.3 ± 0.2 μm (also see Table 1).
TABLE 1.
Summary of dissociation constants for ASPP2-CT and iASPP-CT binding to various constructs of p53 determined by NMR, ITC, or fluorescence
The quaternary states of the p53 constructs that contain the oligomerization domain are indicated: monomer (m), dimer (d), and tetramer (t). NMR data were recorded at 17 °C, whereas the ITC and fluorescence data were collected at 12 °C.
| p53 variants | ASPP variants | Kd (NMR) | Kd (ITC) | Kd (fluorescence) |
|---|---|---|---|---|
| μm | μm | μm | ||
| p53-PCD | ASPP2-CT | 1.3 ± 0.2a | 1.5 ± 0.1a | |
| p53-PCD | iASPP-CT | 76.7 ± 10.0a | —b | |
| p53-PCD2F | ASPP2-CT | 2.2 ± 0.6c | 2.3 ± 0.3a | 2.8 ± 0.6a |
| p53-PCD2F | iASPP-CT | 107.5 ± 12.1c | —b | —b |
| p53-PCD2F-L | ASPP2-CT | 2.2 ± 0.9c | 3.0 ± 0.5c | |
| p53-PCD2F-L | iASPP-CT | 11.1 ± 1.7c | 7.7 ± 5.9c | |
| p53-PCD2F-L-OD (t) | ASPP2-CT | 2.7 ± 0.6c | ||
| p53-PCD2F-L-OD (t) | iASPP-CT | 7.6 ± 1.9c | ||
| p53-PCD2F-L-OD-BD (t) | ASPP2-CT | 1.7 ± 0.2, 33 ± 8c,d | 1.9 ± 0.3c | |
| p53-PCD2F-L-OD-BD (t) | iASPP-CT | —e | 4.6 ± 1.2c | |
| p53-PCD2F-L-OD(L344R)-BD (d) | ASPP2-CT | 1.9 ± 0.2, 29 ± 7c,d | 1.7 ± 0.3c | |
| p53-PCD2F-L-OD(L344R)-BD (d) | iASPP-CT | —e | 3.3 ± 1.5c | |
| p53-PCD2F-L-OD(L344P)-BD (m) | ASPP2-CT | 1.5 ± 0.7c | 1.4 ± 0.3, 18 ± 5c,d | 1.6 ± 0.2c |
| p53-PCD2F-L-OD(L344P)-BD (m) | iASPP-CT | 4.3 ± 1.1c | —e | 3.0 ± 0.6c |
| p53-L | ASPP2-CT | 39.7 ± 4.8c | ||
| p53-L | iASPP-CT | 15.9 ± 2.0c | ||
| p53-L-OD-BD (t) | ASPP2-CT | 38.9 ± 5.0c | ||
| p53-L-OD-BD (t) | iASPP-CT | 17.8 ± 0.9c |
Kd values were obtained in the presence of 50 mm NaCl.
ITC and fluorescence experiments were not performed for these pairs due to weak interaction.
Kd values were obtained in the presence of 150 mm NaCl.
The titration curve was best fitted to two sequential binding events.
ITC data using iASPP-CT were not analyzed, because the sample precipitated during the experiment.
The effects of p53-PCD binding, as seen in the spectral data of iASPP-CT, were similar to those observed for ASPP2-CT in some ways but distinct in others. As in ASPP2-CT, resonances of residues residing in the SH3 domain and the junction between ankyrin repeats 3 and 4 of iASPP-CT exhibited significant perturbations upon interaction with p53-PCD (Fig. 2E), and 18 residues experienced chemical shift changes larger than the overall mean value plus 1 S.D. (0.031 ppm; Fig. 2E). However, all perturbations were much smaller than those observed for ASPP2-CT, and the large effects that were observed for residues residing between ankyrin repeats 3 and 4 of ASPP2-CT were significantly smaller in iASPP-CT. In addition, no effects were observed for residues connecting ankyrin repeats 2 and 3 in iASPP-CT. The NMR binding site mapping data clearly show that the interface in the iASPP-CT·p53-PCD complex is different from that in the ASPP2·p53-PCD complex; iASPP-CT mainly uses the SH3 domain to interact with p53-PCD, whereas ASPP2-CT uses both SH3 and ankyrin repeats. The results of our NMR binding experiments provide the first structural data on the p53/iASPP interface. The dissociation constant extracted from the NMR titration curves of selected iASPP-CT residues (Fig. 2F) for the iASPP-CT·p53-PCD complex was Kd = 76.7 ± 10.0 μm. This value reveals that the affinity of iASPP-CT to p53-PCD is ∼60-fold weaker than that of ASPP2-CT to p53-PCD.
Our binding constants are in sharp contrast to previous reports that stated similar Kd values of 20-30 nm for both ASPP2-CT and iASPP-CT binding to the core domain of p53 (p53-CD), as determined by surface plasmon resonance or enzyme-linked immunosorbent assay (33, 34). This discrepancy cannot be easily explained based on the use of different lengths of proteins. The previously employed ASPP2-CT and iASPP-CT protein constructs are very similar to those used here, with ASPP2-CT and iASPP-CT comprising residues 905-1128 and 625-828 (34) compared with 925-1128 and 623-828, respectively. It seems unlikely that the minor N-terminal variations have any impact on the interaction, since our NMR data clearly show that no interaction with p53 at the N termini of ASPP2-CT or iASPP-CT occurs (Fig. 2, B and E). However, it cannot be ruled out that the discrepancies might result from the use of different techniques for the binding studies.
Tidow et al. (40) determined a Kd of 2.2 μm for ASPP2-CT (residues 891-1128) binding to the p53-CD that lacked the Prorich region (residues 94-310) by ITC. Here, we determined a Kd of 1.3 ± 0.2 μm with a p53 construct that contained the additional Pro-rich sequence (residues 56-93) at the N terminus of the p53 core domain and missed several amino acids at the C terminus. The fact that essentially the same binding constants were determined for the ASPP2-CT interaction, irrespective of the absence or presence of the Pro-rich region in the p53 core domain, indicates that this region does not contribute to ASPP2-CT binding in a significant manner.
Interaction of ASPP2-CT with p53 Fluorescence and ITC Studies—p53 typically functions as a dimer of dimers, with oligomerization achieved primarily via its oligomerization domain (OD). Although p53 dimers and tetramers have been characterized by NMR (41, 42), NMR structural studies of ASPP2-CT in complex with oligomeric p53 are fraught with difficulties, given the large molecular mass of the tetramer. We therefore explored whether other biophysical techniques could reliably determine p53/ASPP binding affinities when NMR methodologies became unfeasible. As a first step, we compared the binding affinities obtained using NMR (described above) with those obtained by fluorescence spectroscopy and ITC (described here) for identical proteins.
Based on our NMR structural mapping of the interaction interfaces between ASPP2-CT or iASPP-CT and p53-PCD, we determined that a stretch of three Trp residues (1097-1099 of ASPP2 and 798-800 of iASPP) resides in the p53 binding site. These unique Trp residues are conserved in all three ASPP proteins (Fig. 1B). We therefore decided to exploit Trp fluorescence as a read-out for any conformational changes that take place upon binding to p53. Initial analyses revealed that indeed an increase in fluorescence occurs after the addition of p53-PCD to ASPP2-CT (after subtraction of background Trp fluorescence from p53-PCD alone; data not shown). In order to eliminate background p53-PCD fluorescence, the two Trp residues in p53-PCD were changed to Phe (W91F and W146F). NMR characterization of this mutant (p53-PCD2F) confirmed that its global structure was essentially identical to wild-type p53-PCD (data not shown). In addition, the binding affinity of ASPP2-CT for p53-PCD2F, measured by NMR, was comparable with that for wild-type p53-PCD (2.2 ± 0.6 μm versus 1.3 ± 0.2 μm, respectively; Table 1). Similarly, the binding affinity of iASPP-CT to p53-PCD2F was 107.5 ± 12.1 μm, comparable with the Kd value for p53-PCD, 76.7 ± 10.0 μm (Table 1). Based on this equivalence of p53-PCD2F and p53-PCD, we used nonfluorescent p53-PCD2F for titrations with ASPP2-CT or iASPP-CT, determining the affinities by fluorescence. Changes were seen in the emission maximum, which shifted from 339 to 334 nm, and in intensity, with a ∼16% increase in fluorescence at 334 nm upon the addition of p53-PCD2F (Fig. S1A). Using this fluorescence assay, the dissociation constant for the ASPP2-CT·p53-PCD2F complex was determined to be 2.8 ± 0.6 μm (Table 1). The very weak interaction between iASPP-CT and p53-PCD2F (Kd ∼100 μm as determined by NMR) did not allow the equivalent fluorescence experiment to be performed.
As a third methodology, we employed ITC experiments to extract binding constants for the ASPP-CT interaction with various p53-PCD domain constructs. Titration of ASPP2-CT with p53-PCD (Fig. S1B) yielded a Kd of 1.5 ± 0.1 μm, again in excellent agreement with the NMR titration (Table 1). In addition, using p53-PCD2F for the titration resulted in a Kd very similar to the corresponding NMR value (2.3 ± 0.3 versus 2.2 ± 0.6 μm; see Table 1). Incidentally, iASPP-CT could not be studied in equivalent experiments, since the very weak interaction does not produce sufficient heat for detection during titration.
Together, the above results demonstrate unambiguously that NMR, fluorescence spectroscopy, and ITC binding between ASPP2-CT and the p53 constructs produce essentially identical dissociation constants and that the binding behavior of the tryptophanless p53-PCD mutant (p53-PCD2F) toward ASPPs is similar to that of wild-type p53-PCD. Therefore, any of these three complementary techniques can be used to characterize the binding of various p53 constructs to ASPPs, making a wide range of molecular sizes and affinities accessible.
The Linker Region between the Core and Oligomerization Domains of p53 Contributes to ASPP Interaction—The fact that substantially weaker affinity was found for iASPP-CT interacting with p53-PCD challenges a simple competitive inhibition model, in which iASPP is proposed to disrupt the ASPP2-p53 complex. For iASPP to efficiently displace ASPP2 from p53, its level of expression would have to be ∼100-fold higher than that of ASPP2. Alternatively, additional p53 domains not present in the core constructs studied here or previously may be required for complex formation. To test this hypothesis, we prepared a variety of p53 proteins that extend from the core domain toward the C terminus. In particular, the OD and basic domain (BD) were thought to be important in this context, since avidity contributions in oligomers and the known binding potential of the basic region could influence any interaction. All p53 proteins that were tested for binding to ASPP-CT are summarized in Fig. 1C: p53-PCD2F-L (residues 56-322 with W91F and W146F); p53-PCD2F-L-OD (residues 56-362 with W91F and W146F); p53-PCD2F-L-OD-BD (residues 56-393 with W91F and W146F); p53-PCD2F-L-OD(L344R)-BD (residues 56-393 with W91F, W146F, and L344R); and p53-PCD2F-L-OD(L344P)-BD (residues 56-393 with W91F, W146F, and L344P). Constructs that contain Leu344 substitutions were created to control p53 oligomerization, since Leu344 is a highly conserved residue in p53 that is critical for tetramer formation (43, 44). Specifically, the L344R mutation allows for p53 dimerization but not tetramerization, whereas L344P abolishes oligomerization altogether, rendering p53 monomeric (43, 44). We confirmed, by multiangle light scattering (Fig. S2), that the L344R and L344P mutants of p53-PCD2F-L-OD-BD exist as dimer and monomer, respectively, compared with the tetrameric wild-type construct.
Binding affinities of ASPP2-CT or iASPP-CT to the various p53 constructs were measured by Trp fluorescence, and the resulting data are summarized in Table 1. For ASPP2-CT, the addition of the linker, OD, and BD to the p53 core domain did not significantly alter the binding affinity that was observed for the core domain alone (Kd values varying from 3.0 to 1.6 μm; Fig. 3A and Table 1). This finding is consistent with NMR titration experiments that yield a Kd of 1.5 ± 0.7 μm when monomeric p53-PCD2F-L-OD(L344P)-BD was used (Table 1). In sharp contrast to the results with ASPP2CT, iASPP-CT binding was clearly influenced by the inclusion of additional p53 sequences. Although for p53-PCD2F, the binding was too weak to be detectable by ITC or Trp fluorescence, p53 constructs with the linker or other C-terminal domains caused significant changes in Trp fluorescence (Fig. 3B). For the p53-PCD2F-L construct, a dissociation constant of 7.7 ± 5.9 μm was determined (Table 1). This finding suggests that the linker region of p53 provides additional interaction energy for iASPP-CT binding. NMR titration experiments with p53-PCD2F-L also support this conclusion; a Kd of 11.1 ± 1.7 μm was determined for iASPP-CT, compared with the much larger value of 108 ± 12 μm for the p53-PCD2F construct (Table 1). This has an important functional consequence and implies that for a full-length tetrameric p53, iASPP (Kd = 4.6 ± 1.2 μm) may be able to act as an effective competitive inhibitor of ASPP2 (Kd = 1.9 ± 0.3 μm).
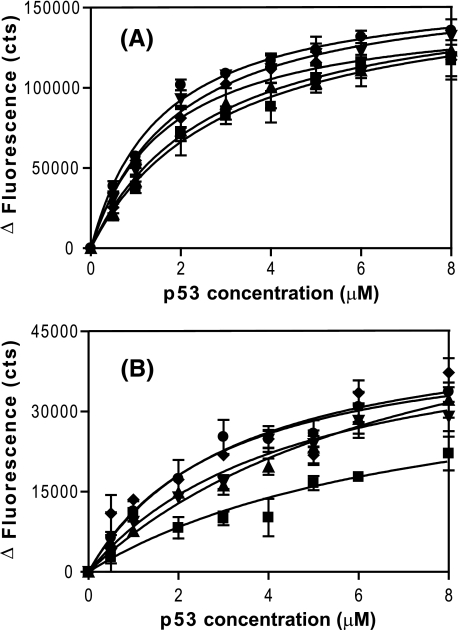
FIGURE 3.
Trp fluorescence binding curves for ASPP2-CT and iASPP-CT to various p53 constructs. Net changes of fluorescence at the emission maximum of either ASPP2-CT (A) or iASPP-CT (B) are plotted against the concentrations of different p53 constructs: p53-PCD2F-L (▪), p53-PCD2F-L-OD (▴), p53-PCD2F-L-OD-BD (▾), p53-PCD2F-L-OD(L344R)-BD (♦), and p53-PCD2F-L-OD(L344P)-BD (•). The data were fitted using a single-site binding site isotherm. The resulting dissociation constants for ASPP2-CT and p53-PCD2F-L, p53-PCD2F-L-OD, p53-PCD2F-L-OD-BD, p53-PCD2F-L-OD(L344R)-BD, and p53-PCD2F-L-OD(L344P)-BD are 3.0 ± 0.5, 2.7 ± 0.6, 1.9 ± 0.3, 1.7 ± 0.3, and 1.6 ± 0.2 μm, respectively. Equivalent dissociation constants for iASPP-CT are 7.7 ± 5.9, 7.6 ± 1.9, 4.6 ± 1.2, 3.3 ± 1.5, and 3.0 ± 0.6 μm, respectively. The errors in the iASPP-CT titration experiment are larger, since the net Trp fluorescence change was about 25% of the change for ASPP2-CT. All dissociation constants are summarized in Table 1.
Significantly, neither ASPP2-CT nor iASPP-CT binding to p53 was affected by the oligomerization state of p53 (Table 1 and Fig. 3). For ASPP2-CT, both monomeric and dimeric p53 mutant proteins were found to bind ASPP2-CT essentially the same (Kd = 1.6 ± 0.2 and 1.7 ± 0.3 μm, respectively), indistinguishable from the tetrameric protein (Kd = 1.9 ± 0.3 μm). iASPP-CT also exhibited similar binding constants for interaction with the monomeric, dimeric, and tetrameric forms of p53-PCD2F-L-OD-BD (Kd values are 3.0 ± 0.6, 3.3 ± 1.5, and 4.6 ± 1.2 μm, respectively; Table 1 and Fig. 3B), demonstrating that the quaternary state of p53 does not significantly affect the interaction. NMR titration experiments with p53-PCD2F-L-OD(L344P)-BD gave a comparable Kd value (4.3 ± 1.1 μm; Fig. S3). Taken together, the above results clearly demonstrate that oligomerization does not enhance the binding affinity in either p53-ASPP2 or p53-iASPP complex formation.
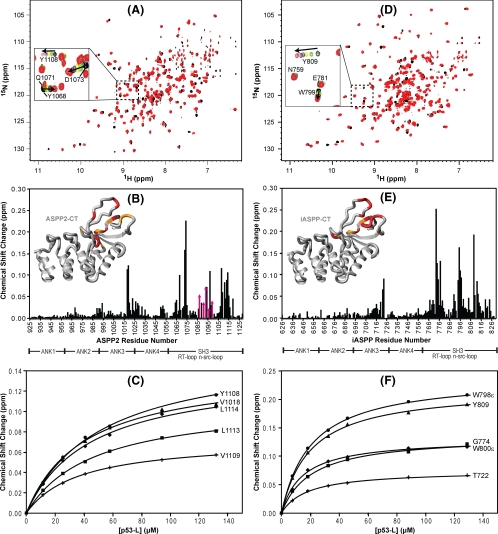
The p53 Linker Region Binds to the Same Site on ASPPs as the p53 Core Domain—Since our binding data indicated that the linker region of p53 provides additional energy in the iASPP-CT interaction, we prepared a peptide comprising only the linker residues 289-322 (p53-L) and investigated its binding to iASPP-CT and ASPP2-CT by 1H-15N TROSY-HSQC NMR (Fig. 4). We observed interaction for both ASPP2-CT (Fig. 4, A-C) and iASPP-CT (Fig. 4, D-F) and noted that resonances of residues in the SH3 domain and the loop between ankyrin repeats 3 and 4 were affected. Indeed, the region affected by p53-L binding is very similar to that affected by the core domain (p53-PCD) (Fig. 2). It seems that the interaction of the linker peptide with iASPP-CT is slightly stronger (Kd = 15.9 ± 2.0 μm) than that with ASPP2-CT (39.7 ± 4.8 μm), contrasting with the data for the p53 core domain. Interestingly, the Kd value for the p53-L·ASPP2-CT complex (39.7 μm) is very similar to the second Kd value obtained from the ITC data for the p53-PCD2F-L-OD-BD·ASPP2-CT complex (33 μm; Table 1 and Fig. S4), suggesting that the p53 does in fact have two binding sites for ASPP2-CT; one is strong and located on the core domain (Kd ∼1-2 μm), and the other is weak on the linker region (Kd ∼40 μm). Furthermore, this secondary event was not dependent on the oligomerization state of p53 (Table 1). For the iASPP-CT/p53 interaction, a tighter binding site occurs with the linker region (Kd ∼16 μm) compared with the core domain (Kd ∼100 μm), whereas the opposite is true for ASPP2-CT. Furthermore, a tetrameric p53 construct that contains the linker, oligomerization, and basic domains without the core domain (p53-L-OD-BD; residues 289-393) results in Kd values of 38.9 ± 5.0 and 17.8 ± 0.9 μm for ASPP2-CT and iASPP-CT, respectively, as determined by NMR. These affinities are essentially the same as those obtained for the linker peptide alone, confirming that the oligomerization and basic domains of p53 do not contribute to binding.
FIGURE 4.
Chemical shift mapping of the interaction between ASPP2-CT and iASPP-CT and the p53 linker peptide. A and D, superposition of the 1H-15N TROSY-HSQC spectra of U-15N,2H-labeled ASPP2-CT (A) and 15N,2H-labeled iASPP-CT (D) in the absence (black) and presence (red) of unlabeled p53-L. A selected region is expanded in each spectrum, and individual resonances along the titration are shown in the inset (increasing amounts of p53-L are shown with the colors black → green → yellow → blue → burgundy → cyan → red). B and E, magnitude of the chemical shift changes versus residue number for ASPP2-CT and iASPP, respectively. Domain boundaries for ASPP2-CT and iASPP-CT are indicated below the residue numbers. Structural mapping of the p53 binding site on ASPP2-CT and iASPP-CT is provided in the insets. The insets show the crystal structures of ASPP2-CT and iASPP-CT (Protein Data Bank codes 1YCS and 2VGE), where residues are colored according to the magnitude of their associated chemical shift changes: red, Δδ > (Δδaverage + 2 × S.D.); orange,(Δδaverage + 2 × S.D.) >Δδ > (Δδaverage + 1 × S.D.). C and F, titration curves for selected ASPP2-CT and iASPP-CT 1HN resonances, respectively.
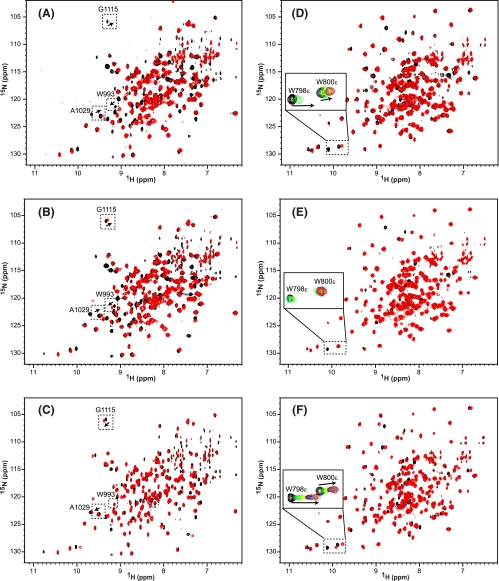
Given all of the above data, the question arose which region of p53, the core domain or linker region, provides the dominant interface in the interactions with ASPP2-CT and iASPP-CT. Careful analysis of the NMR titration data for p53-PCD2F-L, p53-PCD2F, and p53-L (Fig. 5) allowed us to answer this question. Very similar spectral perturbations in ASPP2-CT (Fig. 5, A-C) and iASPP-CT (Fig. 5, D-F) were noted for binding to all three constructs due to overlapping interfaces. Still, some subtle differences in the perturbation patterns could be detected. For example, both p53-PCD2F-L (Fig. 5A) and p53-PCD2F (Fig. 5B) produced similar peak movements for Trp993, Ala1029, and Gly1115 in the ASPP2-CT spectrum, whereas the addition of p53-L was distinct (Fig. 5C), indicating that the core domain of p53-PCD2F-L rather than the linker dominates in interactions with ASPP2. The opposite result was seen in the iASPP-CT spectrum; p53-PCD2F-L induced the same changes as p53-L, distinct from p53-PCD2F (note the changes in Trp798 and Trp800 resonances in Fig. 5, D-F). Therefore, our NMR data indicate that iASPP-CT interacts predominantly with the linker region in p53 and not the core domain. Differential engagement of distinct p53 domains by ASPP2 and iASPP is consistent with the different binding affinities that were found: ASPP2-CT binds to p53-PCD2F (Kd = 2 μm) about 20-fold more strongly than p53-L (Kd = 40 μm), whereas iASPP-CT binds to p53-L (Kd = 16 μm) about 7-fold more strongly than to p53-PCD2F (Kd = 108 μm). Further support for this distinct difference in binding mode for ASPP2 compared with iASPP is provided by our data indicating that essentially the same affinity is observed for p53-PCD2F-L (Kd = 2.2 μm) and p53-PCD2F (Kd = 2.2 μm) to ASPP2-CT, whereas for iASPP-CT, p53-PCD2F-L (Kd = 11 μm) and p53-L (Kd = 16 μm) exhibit similar binding affinity. Taken together, our data suggest that iASPP should be an efficient inhibitor of ASPP2 binding to p53. However, in contrast to simple, competitive inhibition by iASPP, which would involve the same interfaces as seen in the p53-ASPP2 complex, iASPP engages the linker region of p53; this interaction is weaker in ASPP2 compared with iASPP.
FIGURE 5.
Chemical shift mapping of the binding of different p53 constructs to ASPP2-CT and iASPP-CT. A-F, superposition of 1H-15N TROSY-HSQC spectra of U-15N,2H- or 15N,13C,2H-labeled ASPP2-CT and U-15N,2H- or 15N,13C,2H-labeled iASPP-CT in the absence (black) and presence (red) of unlabeled p53-PCD2F-L (A and D), p53-PCD2F (B and E), and p53-L (C and F). A-C, select resonances of ASPP2-CT are highlighted in dashed boxes and labeled with residue name and number. D-F, titrations of selected iASPP-CT resonances are displayed in the enlarged boxes, and the progression in the titration is shown by black → green → yellow → blue → burgundy → cyan → red contour colors.
Conclusion—Utilizing ITC, fluorescence and NMR, we determined the binding affinities and interaction sites between the C-terminal domains of ASPP2 and iASPP and various constructs of p53, spanning from the Pro-rich domain to the C-terminal basic domain. We discovered that each of these proteins interact with p53 through two distinct primary interfaces, with both contributing to binding. Our data indicate that the C terminus of ASPP2 primarily engages the DNA binding core domain of p53 in its complexes, while the C terminus of iASPP preferentially binds to the linker region adjacent to the p53 core domain. Our results suggest a mechanism for how iASPP can inhibit the ASPP2-p53 interaction: iASPP can engage the linker region of p53, even in the presence of bound ASPP2.
The detailed molecular mechanism for how ASPP2 specifically enhances DNA binding activity of p53 at the promoter sites of pro-apoptotic genes is not yet known. Since the DNA binding surface of the p53 core domain is also utilized in the interaction with ASPP (33, 40), secondary binding interfaces of p53 would be needed in any ASPP-p53-DNA ternary complex. The identification of a secondary ASPP binding site on p53 involving the linker region may explain the molecular mechanism of ASPP modulation of the proapoptotic activity of p53. In addition, ASPP2 might recruit additional factors using its N terminus. Irrespectively, a simple competition model of iASPP versus ASPP2 for p53 interaction is not sufficient to explain selective inhibition by iASPP of the transcriptional activity of p53 on proapoptotic genes (29, 32). It is indeed possible that iASPP interferes with the proapoptotic activity of p53 by engaging the linker region and thereby alters the precise positioning of the DNA core binding domain(s) at the promoters of apoptotic genes.
Many p53-interacting cellular factors which regulate the activity of p53 have been identified (5), and all regions of p53, such as the transcriptional activation domain, Pro-rich region, DNA binding core domain, oligomerization domain, and basic domain, except the linker, have been identified to be involved in interactions with cellular factors. Here, we provide the first report that the linker region of p53 can provide a molecular interface for ASPP family members. Our extensive binding data for the interaction between ASPP proteins and p53 allow us to advance a mechanistic explanation for ASPP2 acting as an activator of p53-dependent apoptosis and iASPP operating as an inhibitor. Therefore, our results open the way for designing small molecules or peptides that directly target the iASPP-p53 interface, thereby curtailing the oncogenic function of iASPP while preserving the proapoptotic activity of ASPP2 in tumors that express wild-type p53 and overexpress iASPP (29, 32, 45).
Supplementary Material
Acknowledgments
We thank Drs. Teresa Brosenitsch, Charles J. DiComo, and Carol Prives for critical reading of the manuscript and Jason Concel for expert technical assistance.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: SH3, Src homology 3; ASPP2-CT, the C terminus of ASPP2; p53-CD, the DNA-binding core domain of p53; iASPP-CT, the C terminus of iASPP; p53-PCD, the Pro-rich and DNA-binding core domains of p53; p53-PCD2F, p53-PCD with W91F and W146F; p53-PCD2F-L, p53-PCD2F with linker; p53-PCD2F-L-OD, p53-PCD2F-L with oligomerization domain; p53-PCD2F-L-OD-BD, p53-PCD2F-L-OD with basic domain; p53-L, p53 linker between CD and OD; p53-L-OD-BD, p53 linker, oligomerization and basic domain; TEV, tobacco etch virus; ITC, isothermal titration calorimetry; HSQC, heteronuclear single quantum correlation; TROSY, transverse relaxation optimized spectroscopy; OD, oligomerization domain; BD, basic domain.
References
- 1.Lane, D. P., and Crawford, L. V. (1979) Nature 278 261-263 [DOI] [PubMed] [Google Scholar]
- 2.Linzer, D. I., and Levine, A. J. (1979) Cell 17 43-52 [DOI] [PubMed] [Google Scholar]
- 3.Linzer, D. I., Maltzman, W., and Levine, A. J. (1979) Virology 98 308-318 [DOI] [PubMed] [Google Scholar]
- 4.Vousden, K. H. (2000) Cell 103 691-694 [DOI] [PubMed] [Google Scholar]
- 5.Vousden, K. H., and Lu, X. (2002) Nat. Rev. Cancer 2 594-604 [DOI] [PubMed] [Google Scholar]
- 6.Lu, X. (2005) Curr. Opin. Genet. Dev. 15 27-33 [DOI] [PubMed] [Google Scholar]
- 7.Olivier, M., Hussain, S. P., Caron de Fromentel, C., Hainaut, P., and Harris, C. C. (2004) IARC Sci. Publ. 157 247-270 [PubMed] [Google Scholar]
- 8.Balint, E. E., and Vousden, K. H. (2001) Br. J. Cancer 85 1813-1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sionov, R. V., and Haupt, Y. (1999) Oncogene 18 6145-6157 [DOI] [PubMed] [Google Scholar]
- 10.Haupt, S., Berger, M., Goldberg, Z., and Haupt, Y. (2003) J. Cell Sci. 116 4077-4085 [DOI] [PubMed] [Google Scholar]
- 11.Liebermann, D. A., Hoffman, B., and Vesely, D. (2007) Cell Cycle 6 166-170 [DOI] [PubMed] [Google Scholar]
- 12.Pietsch, E. C., Sykes, S. M., McMahon, S. B., and Murphy, M. E. (2008) Oncogene 27 6507-6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig, R. L., Bates, S., and Vousden, K. H. (1996) Mol. Cell Biol. 16 4952-4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowan, S., Ludwig, R. L., Haupt, Y., Bates, S., Lu, X., Oren, M., and Vousden, K. H. (1996) EMBO J. 15 827-838 [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan, K. M., and Vousden, K. H. (1998) Mol. Cell Biol. 18 3692-3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oda, K., Arakawa, H., Tanaka, T., Matsuda, K., Tanikawa, C., Mori, T., Nishimori, H., Tamai, K., Tokino, T., Nakamura, Y., and Taya, Y. (2000) Cell 102 849-862 [DOI] [PubMed] [Google Scholar]
- 17.D'Orazi, G., Cecchinelli, B., Bruno, T., Manni, I., Higashimoto, Y., Saito, S., Gostissa, M., Coen, S., Marchetti, A., Del Sal, G., Piaggio, G., Fanciulli, M., Appella, E., and Soddu, S. (2002) Nat. Cell Biol. 4 11-19 [DOI] [PubMed] [Google Scholar]
- 18.Di Stefano, V., Rinaldo, C., Sacchi, A., Soddu, S., and D'Orazi, G. (2004) Exp. Cell Res. 293 311-320 [DOI] [PubMed] [Google Scholar]
- 19.Sykes, S. M., Mellert, H. S., Holbert, M. A., Li, K., Marmorstein, R., Lane, W. S., and McMahon, S. B. (2006) Mol. Cell 24 841-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Y., Luo, J., Zhang, W., and Gu, W. (2006) Mol. Cell 24 827-839 [DOI] [PubMed] [Google Scholar]
- 21.Flores, E. R., Tsai, K. Y., Crowley, D., Sengupta, S., Yang, A., McKeon, F., and Jacks, T. (2002) Nature 416 560-564 [DOI] [PubMed] [Google Scholar]
- 22.Shikama, N., Lee, C. W., France, S., Delavaine, L., Lyon, J., Krstic-Demonacos, M., and La Thangue, N. B. (1999) Mol. Cell 4 365-376 [DOI] [PubMed] [Google Scholar]
- 23.Trigiante, G., and Lu, X. (2006) Nat. Rev. Cancer 6 217-226 [DOI] [PubMed] [Google Scholar]
- 24.Iwabuchi, K., Bartel, P. L., Li, B., Marraccino, R., and Fields, S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6098-6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagase, T., Ishikawa, K., Suyama, M., Kikuno, R., Hirosawa, M., Miyajima, N., Tanaka, A., Kotani, H., Nomura, N., and Ohara, O. (1998) DNA Res. 5 355-364 [DOI] [PubMed] [Google Scholar]
- 26.Lopez, C. D., Ao, Y., Rohde, L. H., Perez, T. D., O'Connor, D. J., Lu, X., Ford, J. M., and Naumovski, L. (2000) Mol. Cell Biol. 20 8018-8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ao, Y., Rohde, L. H., and Naumovski, L. (2001) Oncogene 20 2720-2725 [DOI] [PubMed] [Google Scholar]
- 28.Samuels-Lev, Y., O'Connor, D. J., Bergamaschi, D., Trigiante, G., Hsieh, J. K., Zhong, S., Campargue, I., Naumovski, L., Crook, T., and Lu, X. (2001) Mol. Cell 8 781-794 [DOI] [PubMed] [Google Scholar]
- 29.Bergamaschi, D., Samuels, Y., Sullivan, A., Zvelebil, M., Breyssens, H., Bisso, A., Del Sal, G., Syed, N., Smith, P., Gasco, M., Crook, T., and Lu, X. (2006) Nat. Genet. 38 1133-1141 [DOI] [PubMed] [Google Scholar]
- 30.Bergamaschi, D., Samuels, Y., Jin, B., Duraisingham, S., Crook, T., and Lu, X. (2004) Mol. Cell Biol. 24 1341-1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, J. P., Hori, M., Sanda, T., and Okamoto, T. (1999) J. Biol. Chem. 274 15662-15670 [DOI] [PubMed] [Google Scholar]
- 32.Bergamaschi, D., Samuels, Y., O'Neil, N. J., Trigiante, G., Crook, T., Hsieh, J. K., O'Connor, D. J., Zhong, S., Campargue, I., Tomlinson, M. L., Kuwabara, P. E., and Lu, X. (2003) Nat. Genet. 33 162-167 [DOI] [PubMed] [Google Scholar]
- 33.Gorina, S., and Pavletich, N. P. (1996) Science 274 1001-1005 [DOI] [PubMed] [Google Scholar]
- 34.Robinson, R. A., Lu, X., Jones, E. Y., and Siebold, C. (2008) Structure 16 259-268 [DOI] [PubMed] [Google Scholar]
- 35.Nallamsetty, S., Kapust, R. B., Tozser, J., Cherry, S., Tropea, J. E., Copel- and, T. D., and Waugh, D. S. (2004) Protein Expression Purif. 38 108-115 [DOI] [PubMed] [Google Scholar]
- 36.Rosen, M. K., Gardner, K. H., Willis, R. C., Parris, W. E., Pawson, T., and Kay, L. E. (1996) J. Mol. Biol. 263 627-636 [DOI] [PubMed] [Google Scholar]
- 37.Smith, B. O., Ito, Y, Raine, A., Ben-Tovim, L., Nietlispach, D., Broadhurst, W., Terada, T., Kelly, M., Oschkinat, H., Shibata, T., Yokoyama, S., and Laue, E. D. (1996) J. Biomol. NMR 8 360-368 [DOI] [PubMed] [Google Scholar]
- 38.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 39.Johnson, B. A. (2004) Methods Mol. Biol. 278 313-352 [DOI] [PubMed] [Google Scholar]
- 40.Tidow, H., Veprintsev, D. B., Freund, S. M., and Fersht, A. R. (2006) J. Biol. Chem. 281 32526-32533 [DOI] [PubMed] [Google Scholar]
- 41.Ayed, A., Mulder, F. A., Yi, G. S., Lu, Y., Kay, L. E., and Arrowsmith, C. H. (2001) Nat. Struct. Biol. 8 756-760 [DOI] [PubMed] [Google Scholar]
- 42.Veprintsev, D. B., Freund, S. M., Andreeva, A., Rutledge, S. E., Tidow, H., Canadillas, J. M., Blair, C. M., and Fersht, A. R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2115-2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davison, T. S., Yin, P., Nie, E., Kay, C., and Arrowsmith, C. H. (1998) Oncogene 17 651-656 [DOI] [PubMed] [Google Scholar]
- 44.Davison, T. S., Nie, X., Ma, W., Lin, Y., Kay, C., Benchimol, S., and Arrowsmith, C. H. (2001) J. Mol. Biol. 307 605-617 [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z. J., Zhang, Y., Zhang, X. B., and Yang, X. (2004) Leukemia 18 880. [DOI] [PubMed] [Google Scholar]
- 46.Sali, A., and Blundell, T. L. (1993) J. Mol. Biol. 234 779-815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.