Abstract
In cell-extracellular matrix junctions (focal adhesions), the cytoskeletal protein talin is central to the connection of integrins to the actin cytoskeleton. Talin is thought to mediate this connection via its two integrin, (at least) three actin, and several vinculin binding sites. The binding sites are cryptic in the head-to-rod autoinhibited cytoplasmic form of the protein and require (stepwise) conformational activation. This activation process, however, remains poorly understood, and there are contradictory models with respect to the determinants of adhesion site localization. Here, we report turnover rates and protein-protein interactions in a range of talin rod domain constructs varying in helix bundle structure. We conclude that several bundles of the C terminus cooperate to regulate targeting and concomitantly tailor high affinity interactions of the talin rod in cell adhesions. Intrinsic control of ligand binding activities is essential for the coordination of adhesion site function of talin.
Integrin αβ heterodimers play a central role in metazoan cell adhesion, mediating essential contacts in embryogenesis, tissue homeostasis, wound healing, vertebrate cellular immunity, and pathological processes (1). The integrins function as transmembrane receptors in cell-cell and cell-extracellular matrix adherens-type junctions mediating mechanical connection as well as transmembrane signaling. Intracellularly, integrins are connected via adaptor proteins to the actin cytoskeleton. Talin plays a central role in this process by binding to the short cytodomains of β-integrins (2) and regulating both the activity of integrin αβ heterodimers and their connection to the cytoskeleton (3-6). Genetic deletion of the TALIN 1 gene causes early embryonic lethality in mice and other organisms (7, 8). Furthermore, talin interaction with different integrins is essential for platelet activation and function (9, 10).
A key mechanism determining formation and dissolution of adhesions is the rapid exchange of cytoskeletal proteins within the intracellular adhesion complex (11). Many of the proteins involved in this complex exist in a dynamic equilibrium between two conformational states, the “inert,” soluble, cytosolic conformation and the “active,” localized, ligand-bound conformation. Exchange rates of individual proteins such as talin and vinculin are high (12-14), so that modulation of their residency times influences the fate of the adhesion site (11). Conformational regulation of ligand binding sites in talin is, thus, expected to have a decisive effect on the formation, maturation (signaling), and disassembly of the adhesion complex. Accordingly, vinculin mutants with increased half-lives confer longer residency times of talin in adhesion sites (13). Thus, although poorly understood, the regulation of ligand binding to talin is central to the control of adhesion sites dynamics and turnover.
Talin is a large 270-kDa cytoskeletal protein with an N-terminal head (amino acid residues 1-433) and an extensive C-terminal rod region (residues 434-2541). Autoregulatory interactions between these regions establish the inert cytoplasmic conformation of talin (15). The head region consists of an extended FERM domain. The FERM F3 subdomain has a phosphotyrosine binding-like-fold (IBS-1)3 that is occupied in a competitive fashion by the β-integrin cytoplasmic tail and other protein ligands (16, 17). The rod region contains ∼62 α-helices organized into a series of four- and five-helix bundles and a dimerization site (DS) at the C terminus which is formed by helix H62 (18). The rod holds one further binding site for β-integrins (IBS-2) and 11 α-helices carrying the binding motif for vinculin, so that each is a potential vinculin binding site (VBS) (19, 20). In addition, there are several binding sites for filamentous actin (F-actin, ABS), one in the head (ABS-1) (21) and at the least two in the rod (ABS-2, ABS-3) (18, 22, 23). Preliminary studies on the talin head-to-rod autoinhibited form show that the talin F3 phosphotyrosine binding domain binds to residues 1654-2344 in the talin rod (24) and suggest a role in the regulation of integrin binding.
The mechanisms of talin activation, adhesion site/membrane localization, and ligand binding remain unclear. The current model assumes that the inhibitory head-to-rod binding is released through interaction with acidic phospholipids at the membrane (25) and may involve non-integrin binding partners of the FERM domain, such as phosphatidylinositol-4-phosphate-5-kinase type I γ or layilin (15, 26). However, the basis of activation of binding sites for integrin, vinculin, and F-actin in talin remains largely elusive. The integrin binding site IBS-1 in the FERM domain is well documented by structural studies, both crystallography and NMR, and other biophysical methods (6, 16). The isolated talin head region, however, associates rather weakly with adhesion sites in intact cells, and the domains and precise molecular mechanisms mediating talin recruitment to these structures have remained controversial. The IBS-2A domain (H47-H51) in the C-terminal part of the talin rod has been investigated in more detail by localization studies of fluorescently labeled proteins and biophysical interaction measurements of isolated protein domains using surface plasmon resonance (19). Furthermore, the integrity of IBS-2A (helix H50) has been shown to be essential for the rescue of talin function in talin knock-out cells (27). At the same time, a series of deletion mutants of ABS-3/DS (H57-H62) led to the description of an independent adhesion targeting of the talin C terminus, mediated by an as-yet undetermined binding partner(s) (28).
The C-terminal part of talin rod, talinC (H47-H62), comprising the IBS-2A/B and the ABS-3/DS domain, evidently contains important binding sites and regulatory elements that convey essential aspects of talin function in cells. Elucidation of the complex interplay of binding sites in talinC requires knowledge of the positions of α-helices and the organization of the bundled domains. Based on the structural organization of talinC, we investigated localization, binding partners, and exchange kinetics of talinC domain constructs in focal adhesions (FAs). Our data provide evidence for interdependent binding site activities in IBS-2 and ABS-3, a key characteristic of talinC, allowing control of targeting to and residency times in FAs. TalinC, thus, establishes a tightly regulated connection between talin and the actin cytoskeleton.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—Vectors used for transient transfection of eukaryotic cells were modified from pEGFP-N2 and pEGFP-C2 (Invitrogen). A new multiple cloning site offering BamHI/MluI/SalI/XhoI sites was introduced by ligation of two oligonucleotides (forward, TCGATGGGATCCACGCGTGTCGACCTCGAGGT; reverse, GATCACCTCGAGGTCGACACGCGTGGATCCCA) into pEGFP plasmids via BamHI and XhoI restriction sites. Talin fragments were amplified by PCR from a mouse talin full-length cDNA clone (accession no. X56123; kindly provided by Dr. K. Yamada), cloned into pCRBlunt cloning vector (Invitrogen), sequenced, and subsequently transferred into the modified pEGFP vectors.
Protein Purification and Analysis of Interactions—The cDNAs encoding various murine talin rod domains were subcloned into the expression vector pET23A-T7 (29). This vector encodes an N-terminal T7 tag and a C-terminal His tag for easy protein purification using affinity chromatography. The vinculin head (Vh), residues 1-716, D1-D3 wild type, and the D1-D3 A50I mutant were cloned into the pGEX-4T vector (GE Healthcare) encoding an N-terminal glutathione S-transferase tag (20). After expression in BL21 Codon Plus, His-tagged talin constructs were purified on nickel-nitrilotriacetic acid-agarose (Qiagen) and glutathione S-transferase-tagged vinculin constructs on glutathione-Sepharose 4 Fast Flow (Amersham Biosciences) according to the manufacturer's protocols. Protein interaction was tested in a pulldown assay. Magnetic protein G beads (25 μl, Invitrogen) preloaded with T7-antibody (1 μl, Novagen) were used to analyze binding of talin constructs to Vh D1-D3 or Vh D1-D3 A50I (100 μl in PBS, final concentration, 0.8 μm each). After incubation at 4 °C overnight, protein G beads were added for 2 h, and bound protein complexes were washed extensively in PBS, 0.5% Triton X-100. Beads were boiled in 20 μl of SDS sample buffer, and 5 μl of eluates were used for immunoblots. Negative controls without talin were treated in parallel. 4 μl of each protein interaction mix were kept and used as input control (total). Talin and vinculin constructs were detected with anti-T7 tag and hVin-1 antibody (Sigma), respectively, and secondary horseradish peroxidase anti-mouse Ig antibody (Dianova). Spot peptide analysis of Vh D1-D3 was performed using 50 nm recombinant protein as described (20).
Cell Culture—C2C12 myoblasts, mouse embryonal fibroblasts (MEFs) derived from vinculin knock-out mice (30), MEF-Vcl null, and wild type MEF (controls) derived from littermates, MEF-wt, were cultured in Dulbecco's modified Eagle's medium (PAA Laboratories GmbH), 10% fetal calf serum (Sigma), and 2 mm l-glutamine (PAA Laboratories GmbH). Subconfluent cells were split and seeded onto fibronectin (25 μg/ml)-coated glass coverslips. On the following day myoblast cells were transfected with talin pEGFP constructs using Nanofectin transfection reagent (PAA) according to the manufacturer's instructions. To confirm protein size and integrity, expression of GFP-tagged talin constructs was controlled by immunoblot. Protein levels in transiently transfected populations ranged between 0.5- and 1.4-fold of talinC expression (data not shown).
Immunofluorescence Microscopy—For immunostaining, the culture medium was removed 24 h after transfection, and cells were fixed directly in prewarmed 4% paraformaldehyde, PBS, washed in PBS, and permeabilized with 0.5% Triton X-100, PBS. Cells were stained with an antibody raised against vinculin (hVin-1, Sigma) and with phalloidin-Alexa Fluor 568 (Invitrogen). The goat anti-mouse-Alexa Fluor 568 conjugate (Invitrogen) was used as the secondary antibody. Micrographs were taken with a ZEISS Axiovert 200M inverted fluorescence microscope using the AxioVision Software (Version 4.6) and further processed in Photoshop CS2 (Adobe). To avoid bias in the interpretation of adhesion site targeting of talin constructs, three members of the laboratory assigned categories of “none,” “weak,” or “strong” to cells in a blinded fashion yielding identical results. In addition, obvious changes in the occurrence, position, or morphology of adhesion sites as well as the observation of protein aggregates were documented and reported together with the assignment.
Fluorescence Recovery after Photobleaching (FRAP)—Transfected C2C12 cells grown on fibronectin-coated coverslips were observed in an open heating chamber (Warner Instruments) mounted on an Olympus double scan-headed FluoView1000 confocal microscope equipped with a Plan-Apo 100×/1.45 oil objective and controlled by FV10-ASW software (Olympus). A 488-nm argon laser and 405-nm diode laser were used for enhanced GFP excitation and bleaching, respectively. The two scan heads allowed simultaneous bleaching and image acquisition. FRAP movies (one frame per 1.107 s) were taken as follows. Bleaching of enhanced GFP fluorescence in focal adhesions, which was done by a fast circular movement (“tornado tool”, Olympus software) of the 405-nm laser beam (∼1-2 frames long), was initiated 5 frames after the movie start, and recovery of fluorescence was recorded over time as indicated. For FRAP analysis, only peripheral focal adhesions of cells expressing low levels of GFP-tagged talin constructs and sufficient signal-to-noise ratio were selected. Expression of talin constructs used in this analysis did not affect occurrence, position, or morphology of adhesion sites unless otherwise indicated. Based on immunoblots and fluorescence-activated cell sorter analysis, the expression of talin constructs in FRAP experiments was estimated to range from equal to moderately increased (2-3-fold overexpression) compared with endogenous talin.
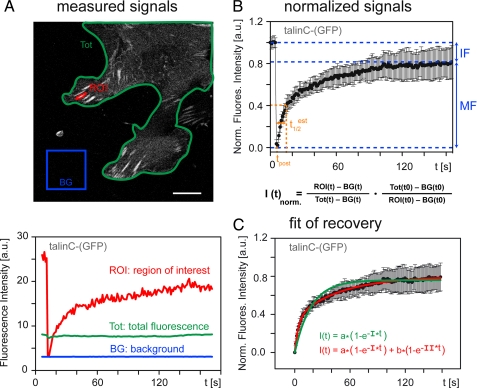
Image Processing and Statistical Analysis—FRAP movies were analyzed using ImageJ software (National Institutes of Health, rsb.info.nih.gov, Version 1.38). Movies showing clear focus drifts or focal adhesion site growth or dissolution were discarded. Regions of interest were defined, with one including the photobleached focal adhesion, and the average intensity was determined for all frames of the movie. Measured signals were corrected for background fluorescence and acquisition photobleaching and normalized as described (31). Graph plots and statistical analyses were carried out using SigmaPlot 10 (Systat). Mono- and biexponential regression functions were used for the calculation of half-life times of recovery and protein mobility distributions of enhanced GFP-tagged talin fragments in focal adhesion sites.
RESULTS
FA Targeting of Talin—In cells, talin displays a prominent adhesion site localization assumed to represent the active protein and a large diffuse cytoplasmic pool reflecting the conformation of the protein “inactive” in ligand binding. Here, we employed GFP-tagged talin constructs to examine essential properties of the domains (Fig. 1, A and B). The isolated head region (residues 1-439), which binds to and activates integrins but cannot provide the mechanical link to the actin cytoskeleton in vivo (32), did not localize discernibly to adhesion sites. Thus, ligand interactions of the talin rod appear to be required for adhesion site localization of the active protein. This was supported by the pronounced FA localization observed for the talin rod (residues 434-2541). However, the precise ligand interactions responsible for FA targeting of the rod remained unclear. Therefore, we generated three new constructs corresponding to proposed functional units of the rod which considered known or predicted boundaries of helical bundles (20). These extended helical (H) bundles comprise (i) H1-H15, with multiple VBS helices and constitutive vinculin binding (33), (ii) H15-H50, with VBS helices, an actin binding site (ABS-2), and part of an integrin binding site (IBS-2A), and (iii) H47-H62, also with VBS helices, the complete IBS-2 and the ABS-3, the latter two of which combine an F-actin (ABS) with the integrin binding site IBS-2 (19, 22). F-actin binding as such was not expected to promote specific FA-localization, as subcellular positioning of ABS domains is not restricted to focal adhesions and leads to general actin filament decoration. All three talin rod constructs targeted to FAs, albeit with different efficiency (Fig. 1, A and B), indicating that active binding sites for either vinculin or β-integrins suffice to mediate some sort of localization. Consistently, FA targeting of a core IBS-2A four-helix bundle (H47-50, the proposed minimal integrin binding site) has been reported by Kieffer and co-workers (19). However, a construct introduced by McCann and co-workers (28), comprising N-terminal VBS helices (H1-H15), the ABS-2, and a partial IBS-2 domain (Fig. 1A, residues 434-2197), did not localize, suggesting the presence of autoinhibitory elements and/or structural requirements for ligand binding. Indeed, helix bundle boundaries in IBS-2B of the latter construct were not considered, so that this construct terminated in two “unsaturated” amphipathic helices H52-H53 (2140-2193), which may well negatively influence the IBS-2A bundle (H47-H51) and interfere with FA targeting. In contrast, recruitment of talin rod constructs comprising the complete C-terminal domain arrangement, IBS-2 and ABS-3 (H47-H62), termed talinC, was extremely pronounced and appeared to be more efficient than any of the other constructs tested either here or by others (19, 28) (Fig. 1, A and B). For this reason we decided to focus our further analyses investigating the mechanism(s) of FA localization on talinC as the key player in connecting talin to the cytoskeleton.
FIGURE 1.
FA localization of talin domain constructs. A, ligand binding sites for F-actin (ABS), β-integrins (IBS), vinculin (VBS), and the dimerization site (DS) of talin are shown. Constructs are indicated (bars), numbers refer to amino acid residues of the N/C terminus, and the relative strength of FA targeting is summarized (strong, weak, or none). Constructs used by others (19, 28), which we have re-analyzed for the purposes of comparison, are indicated (light gray bars). B, representative fluorescence images of GFP-tagged talin variants co-stained for vinculin to visualize FAs (red). Construct information is given as α-helices number (H) of the talin rod. Insets showing enlarged views of vinculin (v, top) or talin variants (t, middle) in FAs and the merged image (m, bottom) refer to selected regions (frames). Size bar, 20 μm (inset, 8 μm).
Domain Structure and Binding Sites of TalinC—The domain structure of talinC has recently been solved by crystal structure and NMR analysis. TalinC consists of three five-helix bundles (Fig. 2A) and a C-terminal dimerization site (18).4 The first two helical bundles (H47-H56), termed IBS-2A and IBS-2B, share a common helix with a distinct kink at its center and contain an integrin binding site, which has not yet been precisely mapped. The third helical bundle domain, ABS-3 (H57-H61), is functionally linked to a C-terminal helix responsible for dimerization (H62, DS). F-actin binding to ABS-3/DS is negatively regulated by helix H57 and strictly depends on dimerization (18) (the ABS-3/DS actin binding module is homologous to that in the yeast protein Sla2p, the huntingtin-interacting protein HIP1, and the related protein HIP1R; the module has also been referred to as (I/L)WEQ motif or THATCH domain (23, 34)). A range of domain constructs designed to respect helix bundle domains is shown in Fig. 2B. Consistent with results from Kieffer and co-workers (19, 27), IBS-2A/B constructs did localize to FAs, albeit with a strong background of cytoplasmic and perinuclear protein (Fig. 2, B and C). The minimal requirement for localization is a four-helix bundle construct (H47-H50) from IBS-2A (19), and integrin binding to either IBS-2A/B or IBS-2A conveys weak constitutive FA targeting. Furthermore, in agreement with McCann and co-workers (28), the ABS-3/DS construct (H57-H62) was efficiently expressed but did not localize to adhesion sites or to F-actin (data not shown). However, removal of helix H57 strongly enhances actin binding of ABS-3/DS (18), and McCann and co-workers (28), when analyzing the H57 deletion construct H58-H62, suggested that the (I/L)WEQ motif (ABS-3/DS) was responsible for talin localization to adhesion sites. Our results confirmed strong FA targeting of talin H58-H62, although our transfected cells frequently displayed aberrantly large and curved adhesion sites, consistent with a strong and de-regulated interaction with F-actin. This suggested a potential requirement for H57 to control ABS-3/DS activity (supplementary Fig. S1). The addition of a partial IBS-2A/B domain to ABS-3/DS, as in talin H50-H62 but not of IBS-2B alone (H52-H62), was sufficient to restore adhesion site localization (Fig. 2, B and C). We concluded that in cells, one of the proposed binding partners of helix H50, vinculin or β-integrin (20, 27), may be required to support activation of ABS-3/DS. Notably in this respect, FAs were not adversely affected in talin H50-H62-transfected cells. The pronounced adhesion site localization of this construct appeared to be counterbalanced by the presence of helix H57 in the ABS-3 five-helix bundle (H57-H61) (18, 23, 35). Thus, our localization analysis confirmed the presence of two FA targeting modules in talinC; IBS-2A/B, allowing weak constitutive localization, and ABS-3/DS, which upon displacement (“activation”) or removal of helix H57 mediated strong targeting.
FIGURE 2.
Structure and FA localization of talinC domain constructs. A, top view of talinC and ribbon model of helix bundle structures for the ABS-3/DS domain (18).4 VBS helices H50 and H58 are highlighted in blue. B, scheme and localization observed for talinC domain constructs (see Fig. 1A); asterisks indicate an altered FA morphology, and ag indicates the observation of protein aggregates in cells. For talin H47-H56, some strongly stained FAs were observed. Size bar, 20 μm (inset, 8 μm). C, representative fluorescence images of GFP-tagged talinC constructs co-stained for vinculin (see Fig. 1B).
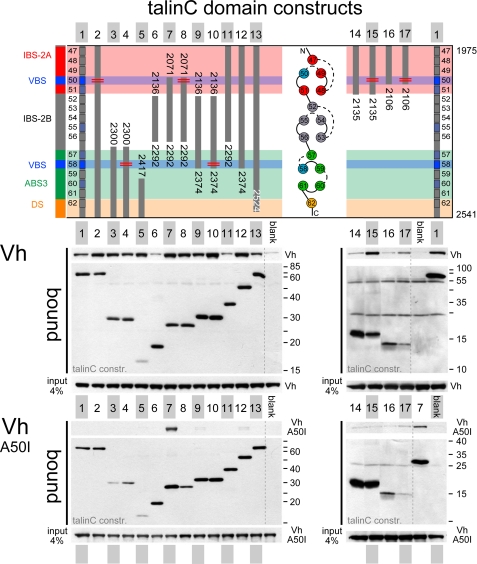
Vinculin Head Interactions with TalinC—Talin rod contains multiple VBS helices that bind activated vinculin with nanomolar affinity, provided the hydrophobic helix surfaces required for the interaction are accessible. As we have reported previously, talinC helices H50 and H58 show the highest affinity for vinculin, whereas helices H51, H55, H57, and H60 may serve as additional sites of interaction (see Fig. 3) (20). We addressed the issue of vinculin binding to talinC helical bundles in pulldown experiments using domains D1-D3 of Vh as a constitutive high affinity binding partner for talin VBS helices. To control for affinity and specificity of the interaction, we used as the negative control the Ala-50 to Ile mutant of vinculin head (Vh A50I), the binding of which is strongly reduced because of stabilization of a four-helix bundle in vinculin head D1 (36). Fig. 3 shows a representative pulldown experiment (n = 3 repeats). When testing talinC variants with intact five-helix bundles, we first observed constitutive strong binding of Vh to ABS-3/DS, whereas interactions with IBS-2A and -2B and IBS-2A/B were not clearly distinguishable from the background (Fig. 3 constructs 3, 6, 11, and 14). TalinC and a C-terminal deletion mutant, residues 1975-2524, bound rather weakly, suggesting reduced VBS accessibility in the entire domain arrangement (Fig. 3, 1 and 13). None of the above-mentioned talinC constructs bound to Vh A50I. Second, all constructs containing truncated helical bundles (including “unsaturated” helices H50 or H58) displayed strong interactions as expected, and even the very short H60-H62 construct supported Vh binding (Fig. 3, constructs 5, 7, 9, and 12). Remarkably, talin H50-H56 bound strongly to Vh A50I, suggesting that it may activate vinculin (head), a capacity that has been previously attributed to VBS peptides and accessible VBS helices (37). The interaction of talin H50-H56 was specific, as limited digest of the complex with vinculin head revealed protection of helix H50, and furthermore, a lower affinity variant, H50-H56:H50* (V2087A/L2091A), did not bind to Vh A50I (Fig. 3, constructs 7 and 8); supplemental Fig. S2). Third, mutations introduced into helices H50 and H58, which were designed to suppress vinculin interaction while leaving the talinC five-helix bundles intact, showed reduced binding in a spot peptide assay (supplemental Fig. S2) but did not abolish interactions with the vinculin head in pulldown assays (Fig. 3, constructs 2, 4, 8, 10, 15, and 17). As detailed below, Vh binding is critical for the interpretation of adhesion site dynamics of talinC:H50* mutants. Taken together, our analysis of interactions between purified proteins demonstrated that binding of activated vinculin (or vinculin head) is a constitutive property of the ABS-3/DS domain and cryptic for IBS-2A/B. The high affinity site of helix H50 appears to require the conformational rearrangement of helices in the IBS-2A five-helix bundle.
FIGURE 3.
Accessibility of VBS helices in talinC. In a pulldown assay, the interaction of recombinant Vh with T7-tagged talin domain constructs was determined using αT7 antibody to immobilize purified talin fragments on magnetic beads. The Ala-50 to Ile mutant of vinculin head (Vh A50I) was used as specificity control. Note the increased stability of a four-helix bundle in the Vh A50I mutant limits interactions to binding partners that expose very high affinity VBS helices. 1, H47-62; 2, H47-62:V2087A, L2091A; 3, H57-62; 4, H57-62:V2360A, I2352A; 5, H60-62; 6, H52-56; 7, H50-56; 8, H50-56:V2087A, L2091A; 9, H52-5; 10, H52-58:V2360A, I2352A; 11, H47-56; 12, H47-58; 13, H47-62 (amino acids 2524); 14, H47-51; 15, H47-51:V2087A, L2091A; 16, H47-50; 17, H47-50:V2087A, L2091A; blank, no partner added.
Dynamics of Talin in Adhesion Sites—FRAP experiments allow determination of release kinetics (koff) and half-life times of fluorescently labeled proteins in adhesion sites. Protein kinetics depend on the equilibrium of bound to free (cytosolic) protein, and dissociation of the bleached protein from adhesions is the rate-limiting step in recovery of the fluorescence signal (38, 39). To establish the method in transiently transfected C2C12 myoblasts, we employed GFP-tagged talin and talinC constructs using a double scan-headed confocal microscope for simultaneous bleaching and image acquisition. Analysis of fluorescence recovery required a biexponential fit for both talinC and talin, indicating the presence of two distinct kinetic pools in adhesion sites (Fig. 4). Subsequent analyses of other talin constructs as well as of vinculin confirmed these two pools, which had been described previously for vinculin (14). The biexponential fit of our experimental data provided two half-life times (t½) of typically 1-3 s for t½(I) and >10 s to 1 min for t½(II), with both well below 10-15 min, the approximate half-life of adhesion sites in resting cells (11). In addition to the mobile fraction, there was a variable amount of immobile protein (IF) in FAs, which was determined from the percentage of fluorescence recovery (Fig. 4B). This third pool, comprised 10-20% of the protein in FAs, was not accessible to kinetic analysis and most likely reflected protein states with very slow turnover and/or partially unfolded immobile protein. Exchange rates of the fast pool t½(I) were distinct from diffusion, representing protein in a tethered state, which can lead to binding or immediate release. Here, we focus our analysis on the slow pool t½(II) of bound protein and describe half-life times together with the percentage of FA-localized protein in this fraction.
FIGURE 4.
FRAP analysis of GFP-tagged talinC in FAs. A, for the regions indicated, fluorescence intensities were monitored over time, and mean gray values were determined and plotted. Size bar, 10 μm. B, intensity values corrected for background and acquisition photobleaching were normalized (see the formula). Averaged curves (>10 adhesions, several cells) provided the percentage of mobile (MF) and immobile (IF) fraction of talin molecules in FAs. C, regression analysis comparing mono- and biexponential fit of fluorescence recovery revealed two kinetic pools for talinC in FAs, which represent tethered (half-life times, t½(I) = 2.8 ± 0.5 s) and bound protein (t½(II) = 33.5 ± 3.6 s), respectively. a.u., arbitrary units.
In C2C12 cells, the half-life time t½(II) of talin was determined to be 45.3 ± 2.2 s, consistent with published observations for talin in other cell lines (13). In comparison, talinC showed a reduced half-life time t½ (II) of 33.5 ± 3.6 s with a higher fraction of the protein in the tethered state, indicating the presence of cryptic binding sites (supplemental Fig. S3). To address the involvement of vinculin in talin rod kinetics, we also studied talin rod constructs by FRAP analysis in mouse embryonic fibroblasts deficient for vinculin (MEF-Vcl null) and their controls (MEF-wt) (30) (Table 1). We found vinculin to significantly decrease half-life times of talin and some talinC domain constructs in FAs (for details see below).
TABLE 1.
Kinetics of talin domain constructs in FAs
The half-life times of talin fragments were determined by FRAP analysis in C2C12 cells, vinculin-deficient mouse embryonic fibroblasts, MEF-Vcl null, and their control cells, MEF-wt. Construct description indicates the helices (H) and mutations (*) in helices H50, V2087A/L2091A and in H57, L2323A. The kinetic values were calculated from a biexponential fit of FRAP curves (n > 10) (further explanations are shown under “Results”). The table indicates the half-life times t1/2(II) and the percentages of protein (II) in the bound fraction as well as the sum of bound and immobile fractions (II + IF) for each construct.
|
Protein
|
C2C12
|
MEF-wt
|
MEF-Vcl null
|
|||
|---|---|---|---|---|---|---|
| t½ (S.D.) | II (II + IF) | t½ (S.D.) | II (II + IF) | t½ (S.D.) | II (II + IF) | |
| s | % | s | % | s | % | |
| (GFP)-talin | 45 ± 2 | 60 (81) | 62 ± 4 | 70 (80) | 116 ± 18 | 51 (67) |
| TalinC-(GFP) | 33 ± 4 | 46 (63) | 25 ± 4 | 32 (54) | 19 ± 2 | 45 (60) |
| TalinC:H50*-(GFP) | 8 ± 2 | 45 (69) | 10 ± 2 | 32 (47) | 12 ± 4 | 30 (37) |
| TalinC:R2526G-(GFP) | 10 ± 2 | 32 (65) | ND | ND | ||
| Talin H47-H56-(GFP) | 6 ± 1 | 32 (70) | ND | 12 ± 3 | 28 (42) | |
| Talin H47-H51-(GFP) | 11 ± 3 | 27 (39) | ND | No localizationa | ||
| Talin H47-H50-(GFP) | 10 ± 1 | 62 (79) | 18 ± 4 | 48 (50) | 20 ± 5 | 48 (54) |
| (GFP)-talin H47-H58 | 47 ± 11 | 31 (73) | ND | 31 ± 3 | 40 (60) | |
| Talin H58-H62-(GFP) | 51 ± 1 | 68 (100)b | 54 ± 1 | 84 (100)b | 55 ± 13 | 81 (84) |
| Talin H57*-H62-(GFP) | 36 ± 4 | 70 (84) | ND | ND | ||
| Talin H50-H62-(GFP) | 36 ± 3 | 69 (86) | 26 ± 3 | 64 (72) | 44 ± 7 | 41 (46) |
Very weak localization; the signal/noise ratio was not sufficient for FRAP analysis.
Mono-exponential fit used.
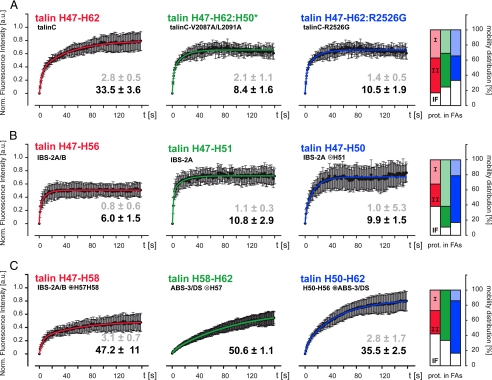
Effects of IBS-2A/B and ABS-3/DS on TalinC Kinetics—We addressed the influence of IBS-2A/B and ABS-3/DS by FRAP analysis of point mutants. The mutant constructs were designed to inhibit integrin/vinculin or F-actin binding, respectively, without affecting the helical bundle structure of talinC subdomains. The conservative Leu-2087/Val-2091 to Ala mutation (L2087A/V2091A) was introduced in talinC with the intention of removing the VBS motif of helix H50 (H50*) (20), which concomitantly ablates β-integrin binding as previously reported by others (19, 27). As discussed above, Vh binding was actually sustained in the H50* mutant despite a reduction of affinity (Fig. 3), and because of the abolishment of the β-integrin interaction, this construct allowed us to address consequences of integrin binding. The Arg-2526 to Gly mutation (R2526G) of talinC results in a monomeric protein that does not bind to the actin cytoskeleton, as dimerization is a prerequisite for the ABS-3/DS interaction with F-actin (18, 40). Both talinC constructs localized to FAs, and compared with wild type talinC, mutations caused a dramatic drop in t½(II) of bound protein from 33.5 ± 3.6 s to 8.4 ± 1.6 s and 10.5 ± 1.9 s, respectively, for the H50* and R2526G variant of talinC (Fig. 5A, Table 1). Thus, FRAP analysis of theses variants revealed a loss of ligand binding site activity in FAs. Moreover, t½(II) of talinC-H50* was also reduced sharply as compared with talinC in MEF-Vcl null cells, confirming a strong effect of β-integrin on talinC kinetics even in the absence of vinculin (Table 1). Hence, both the IBS-2A/B and ABS-3/DS domains cooperate and contribute significantly to kinetics and binding affinity of talinC in FAs, revealing critical interactions of talin rod with β-integrin and F-actin. The involvement of vinculin, although apparent in the short half-life t½(II) of talinC in MEF-Vcl null cells, is more complex and requires additional analysis.
FIGURE 5.
FRAP analysis of talinC variants in adhesion sites. In C2C12 cells, residency times and mobility distribution of talinC and domain constructs thereof were determined using biexponential fit of normalized FRAP recovery curves. For each construct, half-life times (± S.D.) for tethered t½(I) (gray) and bound t½(II) (black) protein fractions are indicated together with the distribution between kinetic pools of tethered (I, light color), bound (II, dark color) and immobile protein (IF, white) in FAs. A, talinC kinetics depend on both β-integrin binding to IBS-2A/B and F-actin binding to ABS-3/DS. B, integrin binding to IBS-2A and IBS-2A/B is inhibited. C, high affinity interactions of talinC require co-stimulation of several binding partners (see “Results” for further information). a.u., arbitrary units.
Bundle Stability and Ligand Binding of the Talin IBS-2—In vitro, protein binding assays using purified IBS-2 constructs have revealed a cryptic high affinity interaction of H50 with vinculin head. In addition, analysis of β-integrin binding indicated that IBS-2A/B bound with similar strength as talinC, whereas IBS-2A and -2B individually showed little or no interaction.4 In cells, we found FA interaction of IBS-2A/B to be very short-lived, with a t½(II) of 6.0 ± 1.5 s and 11.9 ± 2.8 s for C2C12 and MEF-Vcl null, respectively (Fig. 5B, Table 1), suggesting rather weak interactions. Consistently, localization was faint and, in MEF-Vcl null cells, 58% of the FA-localized protein was tethered. The remarkably high immobile fraction (38%) of IBS-2A/B in C2C12 cells may indicate the presence of long-lived interactions in a different conformation. The IBS-2A domain localizes very weakly in MEF-Vcl null cells and showed a short t½(II) of 10.8 ± 2.9sinC2C12 cells, with 61% of protein tethered (Fig. 5B, Table 1). These measurements do not indicate strong interactions of the IBS-2A five-helix bundle with either of its ligands, vinculin or β-integrin. To address the potential of the respective binding sites, we generated talin H47-H50, the proposed minimal IBS-2 (19), which lacks the C-terminal helix, most likely destabilizing the protein fold. For talin H47-H50, we determined a residency time t½(II) of 9.9 ± 1.5sinC2C12 cells together with a high percentage (62%) of bound protein (Fig. 5B, Table 1). Comparison to MEF-Vcl null cells, however, was more informative; in contrast to IBS-2A (H47-H51), the four-helix bundle clearly localized in vinculin deficient cells, with a half-life time t½(II) of 20.0 ± 4.9 s, much higher than that of IBS-2A/B. The fact that enhanced integrin binding was not observed in either C2C12 or MEF-wt cells suggests a negative influence of vinculin on residency times. In these cells t½(II) of talin H47-H50 was low and clearly reduced compared with that of talinC, a difference not seen in vinculin-deficient MEF-Vcl null cells (Table 1). Thus, protein-protein interactions, domain targeting, and kinetic analysis in FAs consistently demonstrated inhibition of both vinculin and β-integrin binding to the IBS-2A/B 10-helix bundle, strongly indicating a requirement for the activation of cryptic ligand binding sites via conformational regulation of this domain.
Activation of Binding Sites in TalinC—Targeting and kinetic studies using talinC subfragments indicated weak constitutive binding of the integrin binding IBS-2A/B domain (H47-H56) and an inability of the ABS-3/DS domain (H57-H62) to localize because of the inhibition of F-actin binding. Hence, enhanced targeting and adhesion site kinetics observed for talinC (H47-H62) strongly suggested activation of one or both domain folds in FAs. We, therefore, designed new constructs that should report the activation of the different binding sites and potential interaction between the two domains. C-terminal extension of IBS-2A/B by two helices resulted in a stable protein construct H47-H58 that localized to FAs without adversely affecting their morphology (Fig. 2C). Although helix H58 contains a high affinity VBS (20), neither helices H57 nor H58 is involved in F-actin binding (18). Talin H47-H58 displayed long half-life times t½(II) of 47.2 ± 10.6 s and 31.2 ± 3.1 s in C2C12 and MEF-Vcl null cells, respectively, reflecting a strong induction of β-integrin binding to IBS-2A/B. In both cell lines, t½(II) was several fold (8/2.5) longer for talin H47-H58 than for IBS-2A/B and exceeded half-life times of talinC. Furthermore, the long residency time of talin H47-H58 in MEF-Vcl null cells proved that vinculin was dispensable in this activation process (Fig. 5C, Table 1), consistent with the idea that helix bundle arrangement may be critical to the induction of integrin binding. In a similar manner, bundle stability appears to control F-actin binding to ABS-3. After removal of helix H57 from ABS-3/DS, the H58-H62 talin construct targeted effectively, leading to deformed FAs and actin bundles (supplemental Fig. S1), but even in morphologically normal FAs, t½ was very long at 50.6 ± 1.1sinC2C12 cells and around a minute in both MEF cell lines. In addition, the percentage of bound protein (II + IF = 84-100%) exceeded values of talinC and was at the top end of all constructs in our analysis. In contrast, a functionally conservative Leu-2323 to Ala mutation in helix H57, which enhanced in vitro actin binding (18) and destabilized the ABS-3 five-helix bundle, also restored ABS-3/DS domain targeting without adverse effects on cells. This talin variant as well as the N-terminal extended talin construct H50-H62 showed adhesion site kinetics t½(II) of 35.5 ± 2.5 s and 25.8 ± 3.5 s, similar to those of talinC in C2C12 and MEF-wt cells, respectively, but a high fraction of bound protein in FAs. Moreover, talin H50-H62 targeted in MEF-Vcl null cells with an enhanced residency time t½(II) of 43.9 ± 6.7 s, revealing the effect of integrin binding in the absence of vinculin competition for helix H50. Thus, kinetic measurements using ABS-3/DS domain constructs in vinculin-deficient cells suggested that integrin binding to helix H50 of IBS-2A could induce F-actin binding to ABS-3/DS via the rearrangement of bundled helices, relieving inhibition by helix H57. Furthermore, competition by vinculin for H50 reduced half-life times of talin H50-H62 in C2C12 and MEF-wt cells, and consistent with the extreme residency times t½(II) of 115.5 ± 27.8 s observed for full-length talin in MEF-Vcl null cells (Table 1), vinculin appears to modulate talin binding kinetics in FAs. Hence, the analysis of talinC domain constructs harboring properties of both targeting modules IBS-2A/B and ABS-3/DS strongly suggests that binding site activity of talinC is induced in adhesion sites by costimulation of several binding partners.
DISCUSSION
The localization and time-resolved analyses of talin domain constructs outlined herein confirm that talinC mediates efficient FA targeting of the talin rod. Localization depends on two binding modules, the IBS-2A/B and the ABS-3/DS domain. Both modules contribute to the kinetic properties of talinC in FAs and require activation of their high affinity binding sites for β-integrin and F-actin, respectively. The residency times of talinC in FAs are modulated by vinculin, which appears to be involved in both the process of activation and inactivation. Based on protein-protein interactions and kinetic analyses, we propose a sequence of events for FA localization and release of talinC from adhesion sites, as illustrated in Fig. 6. Targeting of talinC dimers is initiated by constitutive, low affinity integrin binding of IBS-2. In FAs, ABS-3 can establish low affinity interaction with F-actin and also encounters activated vinculin, which binds to constitutive VBS helices. The induction of high affinity interactions with integrins or F-actin then requires conformational rearrangement of helix bundles in IBS-2A/B and ABS-3/DS domains. The activation process can be supported by other factors, including vinculin binding to further VBS helices and mechanical stress resulting from a coupling of integrins to the actin cytoskeleton. Furthermore, reorganization of the IBS-2A five-helix bundle for integrin binding may allow competitive interaction of vinculin with helix H50, the high affinity VBS in IBS-2A and interaction site for β-integrins (18, 27, 41). Subsequently, displacement of β-integrins from IBS-2A/B could initiate the release of talinC and limit its residency time in adhesion sites.
FIGURE 6.
Activation model of ligand binding in the talin C terminus. FA targeting of talinC (1) is induced by low affinity binding of IBS-2A/B and ABS-3 to their respective ligands (black arrows). Activation of high affinity states in talinC (2) is reversible (bold arrows), depends on additional factors such as mechanical stress and vinculin (blue lines, blue arrow), and is abolished if low affinity ligand binding to either domain is inhibited (red).
Impact of C-terminal Binding Sites on Talin Function—In a Drosophila knock-out model, talin head has been shown to be essential for the regulation of integrin-mediated adhesion to extracellular matrix but not for linkage to the cytoskeleton. A talin construct with defective integrin binding to the head F3 subdomain could still maintain the connection of integrins to the cytoskeleton (32). In addition, selective mutations in talinC highlight the crucial role of the talin rod C terminus in the regulation of talin function. Inhibition of dimerization by a point mutation in the dimerization site blocks talin localization and cellular function (18, 40), whereas a mutation in helix H50 of IBS-2 that inhibits integrin binding of the C terminus also inactivates talin so that the mutant full-length protein cannot rescue adhesion-site formation in talin-deficient cells (27). Thus, the C-terminal region of the talin rod appears to provide the essential link between talin and the cytoskeleton.
Regulation of Binding Sites—Although in talin, actin and integrin binding are not unique features of ABS-3 and IBS-2, their inactivation is not compensated for by other binding sites, indicating tight regulation of full-length talin in a fashion dependent on talinC. Consistent with this, biochemical analysis of talin has been hampered over many years by the fact that ligand binding sites in talin could only be characterized in suitable domain fragments (42, 43). In particular, vinculin binding, requiring exposure of VBS helices, is strongly inhibited in most domains of the talin rod (20, 33). Similarly, integrin and F-actin binding sites have also only recently been elucidated in more detail. We propose that the tight regulation of high affinity binding sites is an essential feature of talin rod that is critical to the cellular control of protein dynamics in the cell adhesion complex. Uncontrolled or deregulated ligand binding activities are detrimental to cell motility and function. The consequences of regulation defects are apparent in partial domain folds like talin H58-H62, which induces disturbed architecture and dynamics of adhesion sites in transfected cells because of its inherently deregulated F-actin binding. Similarly, constitutively active vinculin (vinculin head) has been proposed to occupy high affinity binding sites, blocking inactivation of talin in the adhesion complex and subsequently adhesion site turnover. Vinculin head-expressing cells are characterized by a strong increase in number, size, and overall area of adhesion sites (44). In contrast, physiological activation of vinculin requires combinatorial input from several ligand partners and is readily reversible (36, 45). Therefore, constitutive exposure of high affinity VBS helices by the talin rod such as helix 50 of IBS-2A would have the potential to directly activate vinculin and seriously disturb protein kinetics in adhesion sites. Both vinculin and actin binding require intrinsic counterbalance as elucidated for talinC.
The regulation of talin cytoskeletal associations depends on binding site activities in the talin C terminus, which can subsequently trigger further binding sites. The two domains IBS-2A/B and ABS-3/DS demonstrate properties that would allow formation of a cytoskeletal connection in a regulated fashion, limiting both timing and localization of talin rod interaction. We conclude that talinC by acting as the targeting module in talin rod mediates transient attachment of integrins to the actin cytoskeleton. This connection is controlled by conformational regulation of binding sites in the IBS-2A/B and the ABS-3/DS domain involving reversible interaction with vinculin.
Acknowledgments
We are grateful to Dr. Kurt Anderson, Dr. David R. Critchley, and all members of the cell migration group of the IZKF Leipzig for stimulating discussion and to Dr. Thomas Riemer for support with the evaluation of FRAP data. We thank Gabriele Oehme for excellent technical support, Sabine Schnorr for her contribution to protein pulldown experiments, and Dr. Ken Yamada for talin cDNA.
This work was supported by Deutsche Forschungsgemeinschaft Grants Zi545/3 (SPP1150) and Zi545/5-1 (to W. H. Z.) and Ro2414/1-1 (to K. R.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-3.
Footnotes
The abbreviations used are: IBS, integrin binding site; DS, dimerization site; VBS, vinculin binding site; Vh, vinculin head; MEF mouse embryonal fibroblasts; FRAP, fluorescence recovery after photobleaching; ABS, actin binding site; H, α-helix; FA, focal adhesion; GFP, green fluorescent protein; IF, immobile fraction; F-actin, filamentous actin; talinC, talin residues 1975-2541 (H47-H62).
Gingras, A. R., Ziegler, W. H., Bobkov, A. A., Joyce, M. G., Fasci, D., Himmel, M., Rothemund, S., Ritter, A., Grossmann, J. G., Patel, B., Bate, N., Goult, B. T., Emsley, J., Barsukov, I. L., Roberts, G. C., Liddington, R. C., Ginsberg, M. H., and Critchley, D. R. (2009) J. Biol. Chem. 284, 8866-8876.
References
- 1.Hynes, R. (2002) Cell 110 673-687 [DOI] [PubMed] [Google Scholar]
- 2.Liu, S., Calderwood, D. A., and Ginsberg, M. H. (2000) J. Cell Sci. 113 3563-3571 [DOI] [PubMed] [Google Scholar]
- 3.Jiang, G., Giannone, G., Critchley, D. R., Fukumoto, E., and Sheetz, M. P. (2003) Nature 424 334-337 [DOI] [PubMed] [Google Scholar]
- 4.Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H., and Calderwood, D. A. (2003) Science 302 103-106 [DOI] [PubMed] [Google Scholar]
- 5.Nayal, A., Webb, D. J., and Horwitz, A. F. (2004) Curr. Opin. Cell Biol. 16 1-5 [DOI] [PubMed] [Google Scholar]
- 6.Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 [DOI] [PubMed] [Google Scholar]
- 7.Monkley, S. J., Zhou, X. H., Kinston, S. J., Giblett, S. M., Hemmings, L., Priddle, H., Brown, J. E., Pritchard, C. A., Critchley, D. R., and Fassler, R. (2000) Dev. Dyn. 219 560-574 [DOI] [PubMed] [Google Scholar]
- 8.Critchley, D. R. (2005) Biochem. Soc. Trans. 33 1308-1312 [DOI] [PubMed] [Google Scholar]
- 9.Petrich, B. G., Marchese, P., Ruggeri, Z. M., Spiess, S., Weichert, R. A., Ye, F., Tiedt, R., Skoda, R. C., Monkley, S. J., Critchley, D. R., and Ginsberg, M. H. (2007) J. Exp. Med. 204 3103-3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieswandt, B., Moser, M., Pleines, I., Varga-Szabo, D., Monkley, S., Critchley, D., and Fassler, R. (2007) J. Exp. Med. 204 3113-3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziegler, W. H., Liddington, R. C., and Critchley, D. R. (2006) Trends Cell Biol. 16 453-460 [DOI] [PubMed] [Google Scholar]
- 12.Chandrasekar, I., Stradal, T. E., Holt, M. R., Entschladen, F., Jockusch, B. M., and Ziegler, W. H. (2005) J. Cell Sci. 118 1461-1472 [DOI] [PubMed] [Google Scholar]
- 13.Cohen, D. M., Kutscher, B., Chen, H., Murphy, D. B., and Craig, S. W. (2006) J. Biol. Chem. 281 16006-16015 [DOI] [PubMed] [Google Scholar]
- 14.Lele, T. P., Pendse, J., Kumar, S., Salanga, M., Karavitis, J., and Ingber, D. E. (2006) J. Cell. Physiol. 207 187-194 [DOI] [PubMed] [Google Scholar]
- 15.Critchley, D. R. (2000) Curr. Opin. Cell Biol. 12 133-139 [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 [DOI] [PubMed] [Google Scholar]
- 17.Barsukov, I. L., Prescot, A., Bate, N., Patel, B., Floyd, D. N., Bhanji, N., Bagshaw, C. R., Letinic, K., Di Paolo, G., De Camilli, P., Roberts, G. C. K., and Critchley, D. R. (2003) J. Biol. Chem. 278 31202-31209 [DOI] [PubMed] [Google Scholar]
- 18.Gingras, A. R., Bate, N., Goult, B. T., Hazelwood, L., Canestrelli, I., Grossmann, J. G., Liu, H., Putz, N. S., Roberts, G. C., Volkmann, N., Hanein, D., Barsukov, I. L., and Critchley, D. R. (2007) EMBO J. 27 458-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tremuth, L., Kreis, S., Melchior, C., Hoebeke, J., Ronde, P., Plancon, S., Takeda, K., and Kieffer, N. (2004) J. Biol. Chem. 279 22258-22266 [DOI] [PubMed] [Google Scholar]
- 20.Gingras, A. R., Ziegler, W. H., Frank, R., Barsukov, I. L., Roberts, G. C., Critchley, D. R., and Emsley, J. (2005) J. Biol. Chem. 280 37217-37224 [DOI] [PubMed] [Google Scholar]
- 21.Lee, H. S., Bellin, R. M., Walker, D. L., Patel, B., Powers, P., Liu, H., Garcia-Alvarez, B., de Pereda, J. M., Liddington, R. C., Volkmann, N., Hanein, D., Critchley, D. R., and Robson, R. M. (2004) J. Mol. Biol. 343 771-784 [DOI] [PubMed] [Google Scholar]
- 22.Hemmings, L., Rees, D. J., Ohanian, V., Bolton, S. J., Gilmore, A. P., Patel, B., Priddle, H., Trevithick, J. E., Hynes, R. O., and Critchley, D. R. (1996) J. Cell Sci. 109 2715-2726 [DOI] [PubMed] [Google Scholar]
- 23.Brett, T. J., Legendre-Guillemin, V., McPherson, P. S., and Fremont, D. H. (2006) Nat. Struct. Mol. Biol. 13 121-130 [DOI] [PubMed] [Google Scholar]
- 24.Goksoy, E., Ma, Y. Q., Wang, X., Kong, X., Perera, D., Plow, E. F., and Qin, J. (2008) Mol. Cell 31 124-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martel, V., Racaud-Sultan, C., Dupe, S., Marie, C., Paulhe, F., Galmiche, A., Block, M. R., and Albiges-Rizo, C. (2001) J. Biol. Chem. 276 21217-21227 [DOI] [PubMed] [Google Scholar]
- 26.Legate, K. R., Montanez, E., Kudlacek, O., and Fassler, R. (2006) Nat. Rev. Mol. Cell Biol. 7 20-31 [DOI] [PubMed] [Google Scholar]
- 27.Moes, M., Rodius, S., Coleman, S. J., Monkley, S. J., Goormaghtigh, E., Tremuth, L., Kox, C., van der Holst, P. P., Critchley, D. R., and Kieffer, N. (2007) J. Biol. Chem. 282 17280-17288 [DOI] [PubMed] [Google Scholar]
- 28.Franco, S. J., Senetar, M. A., Simonson, W. T., Huttenlocher, A., and McCann, R. O. (2006) Cell Motil. Cytoskeleton 63 563-581 [DOI] [PubMed] [Google Scholar]
- 29.van der Ven, P. F., Wiesner, S., Salmikangas, P., Auerbach, D., Himmel, M., Kempa, S., Hayess, K., Pacholsky, D., Taivainen, A., Schroder, R., Carpen, O., and Furst, D. O. (2000) J. Cell Biol. 151 235-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, W., Baribault, H., and Adamson, E. D. (1998) Development 125 327-337 [DOI] [PubMed] [Google Scholar]
- 31.Rabut, G., and Ellenberger, J., in Goldman, R. D., and Spector, D. L. (Eds.) (2005) Live Cell Imaging: A Laboratory Manual, pp. 101-126, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 32.Tanentzapf, G., and Brown, N. H. (2006) Nat. Cell Biol. 8 601-606 [DOI] [PubMed] [Google Scholar]
- 33.Patel, B., Gingras, A. R., Bobkov, A. A., Fujimoto, L. M., Zhang, M., Liddington, R. C., Mazzeo, D., Emsley, J., Roberts, G. C., Barsukov, I. L., and Critchley, D. R. (2006) J. Biol. Chem. 281 7458-7467 [DOI] [PubMed] [Google Scholar]
- 34.McCann, R. O., and Craig, S. W. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5679-5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senetar, M. A., Foster, S. J., and McCann, R. O. (2004) Biochemistry 43 15418-15428 [DOI] [PubMed] [Google Scholar]
- 36.Bakolitsa, C., Cohen, D. M., Bankston, L. A., Bobkov, A. A., Cadwell, G. W., Jennings, L., Critchley, D. R., Craig, S. W., and Liddington, R. C. (2004) Nature 430 583-586 [DOI] [PubMed] [Google Scholar]
- 37.Bois, P. R., O'Hara, B. P., Nietlispach, D., Kirkpatrick, J., and Izard, T. (2006) J. Biol. Chem. 281 7228-7236 [DOI] [PubMed] [Google Scholar]
- 38.Bulinski, J. C., Odde, D. J., Howell, B. J., Salmon, T. D., and Waterman-Storer, C. M. (2001) J. Cell Sci. 114 3885-3897 [DOI] [PubMed] [Google Scholar]
- 39.Beaudouin, J., Mora-Bermudez, F., Klee, T., Daigle, N., and Ellenberg, J. (2006) Biophys. J. 90 1878-1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, S. J., and McCann, R. O. (2007) Biochemistry 46 10886-10898 [DOI] [PubMed] [Google Scholar]
- 41.Rodius, S., Chaloin, O., Moes, M., Schaffner-Reckinger, E., Landrieu, I., Lippens, G., Lin, M., Zhang, J., and Kieffer, N. (2008) J. Biol. Chem. 283 24212-24223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bass, M. D., Smith, B. J., Prigent, S. A., and Critchley, D. R. (1999) Biochem. J. 341 257-263 [PMC free article] [PubMed] [Google Scholar]
- 43.Bass, M. D., Patel, B., Barsukov, I. G., Fillingham, I. J., Mason, R., Smith, B. J., Bagshaw, C. R., and Critchley, D. R. (2002) Biochem. J. 362 761-768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphries, J. D., Wang, P., Streuli, C., Geiger, B., Humphries, M. J., and Ballestrem, C. (2007) J. Cell Biol. 179 1043-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, H., Choudhury, D. M., and Craig, S. W. (2006) J. Biol. Chem. 281 40389-40398 [DOI] [PubMed] [Google Scholar]