Abstract
Ikaros encodes a zinc finger protein that is involved in gene regulation and chromatin remodeling. The majority of Ikaros localizes at pericentromeric heterochromatin (PC-HC) where it regulates expression of target genes. Ikaros function is controlled by posttranslational modification. Phosphorylation of Ikaros by CK2 kinase determines its ability to bind DNA and exert cell cycle control as well as its subcellular localization. We report that Ikaros interacts with protein phosphatase 1 (PP1) via a conserved PP1 binding motif, RVXF, in the C-terminal end of the Ikaros protein. Point mutations of the RVXF motif abolish Ikaros-PP1 interaction and result in decreased DNA binding, an inability to localize to PC-HC, and rapid degradation of the Ikaros protein. The introduction of alanine mutations at CK2-phosphorylated residues increases the half-life of the PP1-nonbinding Ikaros mutant. This suggests that dephosphorylation of these sites by PP1 stabilizes the Ikaros protein and prevents its degradation. In the nucleus, Ikaros forms complexes with ubiquitin, providing evidence that Ikaros degradation involves the ubiquitin/proteasome pathway. In vivo, Ikaros can target PP1 to the nucleus, and a fraction of PP1 colocalizes with Ikaros at PC-HC. These data suggest a novel function for the Ikaros protein; that is, the targeting of PP1 to PC-HC and other chromatin structures. We propose a model whereby the function of Ikaros is controlled by the CK2 and PP1 pathways and that a balance between these two signal transduction pathways is essential for normal cellular function and for the prevention of malignant transformation.
The Ikaros gene encodes a C2H2 zinc finger protein that is essential for normal hematopoiesis (1). The absence of Ikaros expression leads to severely impaired lymphoid development as well as defects in myeloid and erythroid differentiation (2-5). Studies of mice with a disruption in the Ikaros gene demonstrate that Ikaros acts as a tumor suppressor (6). In humans, impaired Ikaros activity has been associated with the development of infant T-cell acute lymphoblastic leukemia (ALL) (7), adult B cell ALL (8), myelodysplastic syndrome (9), acute myeloid leukemia (10), and adult and juvenile chronic myelogenous leukemia (11). Recently, the deletion of Ikaros has been demonstrated in more than 30% of childhood leukemias, whereas a single copy or the complete absence of Ikaros was observed in more than 80% of patients with bcr-abl-positive acute lymphoblastic leukemia (12, 13). These studies established Ikaros as a key regulator of hematopoiesis and a major tumor suppressor whose loss of function is associated with the malignant transformation of hematopoietic cells.
The mechanism by which Ikaros exerts its tumor suppressor activity is less clear. Ikaros exhibits a punctate staining pattern in normal and leukemia cells (14). Co-staining for Ikaros and HP1 proteins showed that this punctate staining pattern is because of the localization of Ikaros to pericentromeric heterochromatin (PC-HC)3 (15). Ikaros binds in vitro and in vivo to the murine and human gamma satellite repeats that are located within the PC-HC, and Ikaros DNA binding ability is essential for its localization to PC-HC (16).
Ikaros binds to the regulatory elements of its target genes in a sequence-dependent manner. Ikaros binding leads to the activation or repression of target genes via chromatin remodeling (17, 18). Thus, optimal Ikaros DNA binding is essential both for its subcellular localization and for its regulation of target genes.
Ikaros associates with histone deacetylase (HDAC)-containing complexes (NuRD and Sin3) (19), although HDAC-independent binding to the transcriptional corepressor C-terminal binding protein has been reported as well (20). Ikaros also associates with Brg-1, a catalytic subunit of the SWI/SNF nucleosome remodeling complex that acts as an activator of gene expression (21, 22). The current hypothesis is that Ikaros binds the upstream region of target genes and aids in their recruitment to PC-HC, resulting in repression or activation of the gene (15, 23). Thus, Ikaros can act both as an activator and a repressor, depending on whether it associates with the NuRD, the C-terminal binding protein, or the SWI/SNF complex.
Ikaros activity is regulated at a posttranslational level by various mechanisms. The association of the full-length Ikaros protein with its dominant negative isoforms impairs its activity, whereas the formation of complexes with other active isoforms can modify its function (24, 25). Posttranslational modifications at specific residues regulate Ikaros activity. Sumoylation at two amino acids has been reported to regulate Ikaros repressor function (26). The cell cycle-specific phosphorylation of Ikaros at an evolutionarily conserved linker regulates its DNA binding ability and nuclear localization during mitosis (27). Phosphorylation of Ikaros by CK2 kinase at its C-terminal region regulates its ability to control G1/S cell cycle progression (28). Our previous work identified additional N-terminal phosphorylation sites that are phosphorylated either directly by CK2 kinase or by another kinase in the CK2 signal transduction pathway (29). Reversible phosphorylation of these amino acids regulates the subcellular localization of Ikaros to PC-HC as well as its ability to bind the upstream regulatory element of the Ikaros target gene, TdT (29). These data identified CK2 kinase as a major regulator of Ikaros function. Because the overexpression of CK2 kinase leads to T-cell leukemia, it has been hypothesized that the impairment of Ikaros function is one of the mechanisms by which CK2 kinase promotes malignant transformation.
In this report we demonstrate that Ikaros is dephosphorylated by protein phosphatase 1 (PP1) and identify the specific amino acids that are responsible for Ikaros-PP1 interactions. We show that the process of Ikaros dephosphorylation by PP1 is essential for normal Ikaros function and to prevent accelerated Ikaros degradation via the ubiquitin pathway. We also propose a novel function for Ikaros; that is, to target PP1 to PC-HC in hematopoietic cells.
EXPERIMENTAL PROCEDURES
Cells—The murine VL3-3M2 thymocyte leukemia cell line has been described previously (30). The human HEK 293T (293T) endothelial kidney cell line, the human T cell leukemia cell lines MOLT-4 and CCRF-CEM (CEM), and the human Burkitt's B cell lymphoma cell line, Ramos, were obtained from American Type Culture Collection (ATCC), Manassas, VA. The murine B cell lines BAL17 and HAFTL have been described previously (31, 32).
Antibodies—The antibodies used to detect the C terminus (Ikaros-CTS) of Ikaros (comprising amino acids (320-515 of the murine IK-VI isoform) have been described previously (33). The antibodies used to detect PP1 were purchased from Santa Cruz Biotechnology, Santa Cruz, CA (sc-7482). The antibodies against ubiquitin were from Assay Designs/Stressgen Bioreagents, Ann Arbor, MI (SPA-200). Antibodies to detect the hemagglutinin tag (12CA5) were purchased from Covance (Princeton, NJ).
In Vivo Labeling—For in vivo labeling, 293T cells were incubated with radioactive orthophosphate. Cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Cells were washed twice with phosphate-free RPMI 1640 medium and incubated for 4 h with 0.5 mCi/ml [32P]orthophosphate (PerkinElmer Life Sciences) in phosphate-free medium. Cells were collected by centrifugation, lysed on ice for 20 min in solubilizing buffer (50 mm Tris-HCL pH 7.2, 1% v/v Nonidet P-40, 150 mm NaCl, 5 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, and 5 μm leupeptin), and centrifuged at 15,000 rpm at 4 °C for 20 min. Lysates were incubated with anti-Ikaros-CTS antibodies for 1 h at 4 °C, and the resulting immune complexes were absorbed to protein G-Sepharose (Amersham Biosciences), washed 4 times with solubilizing buffer, separated by SDS-PAGE, transferred to a nylon membrane, and subjected to autoradiography.
Expression and Purification of Recombinant Ikaros Proteins—cDNA for the murine full-length Ikaros isoform, IK-VI, was amplified and cloned into a pGEX-2T GST-containing vector (Amersham Biosciences) that was cleaved with BamH1 and EcoR1 as described previously (34). IK-VI-GST-containing pGEX-2T plasmid was transformed and expressed in the SCS-1 strain of Escherichia coli (Stratagene, La Jolla, CA). E. coli were induced with 1 mm iospropylthiogalactopyranoside for 2 h in the presence of 100 μm ZnCl2. The GST-IK-VI fusion protein was purified from E. coli according to the standard procedures (Amersham Biosciences) except that all of the buffers contained 10 μm ZnCl2 as described previously (34). Fractions that contained purified GST-IK-VI protein were pooled and stored at -80 °C.
GST Pulldown Assay—The full-length Ikaros-GST fusion protein (10 μg) or GST protein, as a negative control, were incubated with glutathione-agarose beads and then mixed with cell lysate of VL3-3M2 cells (a total of 2 mg of protein in lysis buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 120 mm NaCl, protease inhibitors (phenylmethylsulfonyl fluoride, pepstatin and leupeptin)) for 2 h. After incubations, samples were washed with lysis buffer, and proteins were eluted with SDS sample buffer. Proteins were separated by SDS-PAGE, and the presence of PP1 was detected by Western blot with anti-PP1 antibody.
Plasmids and Transfection—Alanine substitution mutants for IK-VI were generated using the QuikChange method (Stratagene). The full-length Ikaros isoform IK-VI and its alanine mutants were cloned into the mammalian expression vector pcDNA3 (Invitrogen). For transfection, 1 μg of each construct was used to transfect 293T cells via the calcium phosphate method.
Confocal Microscopy—293T cells were transfected with appropriate constructs and analyzed by confocal microscopy as described previously (16). Images were acquired at room temperature by a Leica TCS-SP MP Confocal and Multiphoton Microscope with a Leica DM-LFS body (upright fixed-stage microscope) using a 100× Leica HX PLAPO (Planapo) oil immersion lens with numerical aperture of 1.4 (Heidelberg, Germany).
Biochemical Experiments—Nuclear extractions, Western blots, and gel-shift experiments were performed as described previously (16, 35). The γ satellite A and γ satellite B gel shift probes have been described previously (16).
Degradation Assay—Pulse-chase analysis was performed in 293T cells as described previously (36, 37). The 293T cells were grown in 100-mm tissue culture dishes and transfected with full-length Ikaros constructs or the indicated mutants. Cells were grown in Dulbecco's modified Eagle's medium for 24 h, and then medium was replaced with cysteine- and methionine-free Dulbecco's modified Eagle's medium. After starvation for 1 h, cells were labeled with a [35S]methionine/[35S]cysteine mixture (PerkinElmer Life Sciences). Chase was performed in Dulbecco's modified Eagle's medium with 10% fetal bovine serum supplemented with 2 mm unlabeled methionine and cysteine. Cells were harvested at indicated time points and lysed in RIPA buffer with proteinase and phosphatase inhibitors. Ikaros proteins were immunoprecipitated using anti-Ikaros-CTS antibodies, separated by SDS-PAGE, and visualized by autoradiography.
In the degradation assay described in Fig. 4D, MOLT-4 cells were treated with cycloheximide (50 μg/ml) (Sigma-Aldrich catalogue #C1988) in the presence or absence of calyculin (10 nm) (Calbiochem catalogue #208851) and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB, 50 μm) (Calbiochem catalogue #287891) or 4,5,6,7-tetrabromo-1H-benzotriazole (TBB, 10 μm) (Sigma). Cells were harvested at the indicated times, nuclear extract was obtained, and proteins from 20 μg of nuclear extracts from indicated time points were separated by SDS-PAGE. The presence of Ikaros isoforms was detected by anti-Ikaros-CTS antibodies.
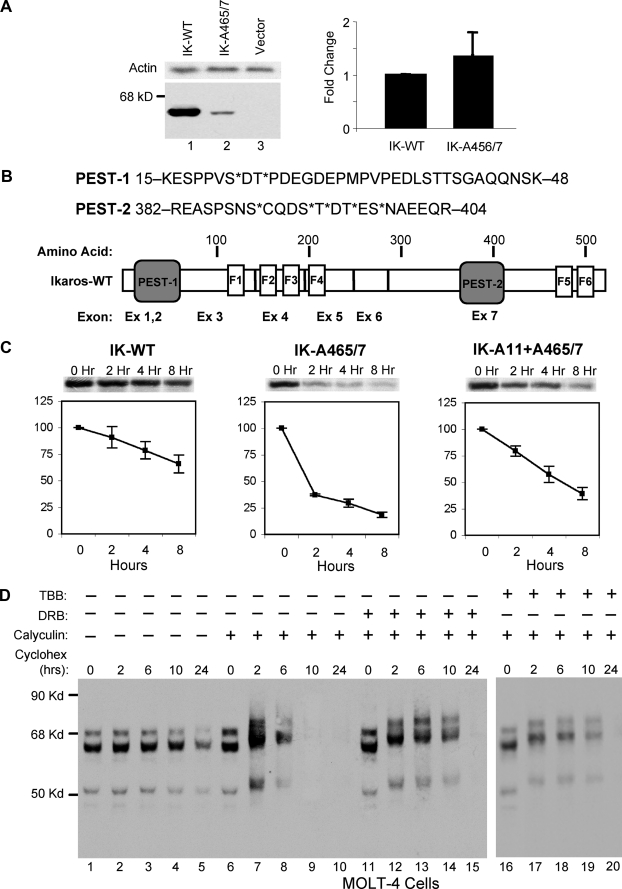
FIGURE 4.
Dephosphorylation of the Ikaros protein regulates its stability. A, the left panel shows Western blot analysis of 20 μg of whole cell lysate from 293T cells transfected with 5 μg of wild-type Ikaros (IK, lane 1) or 5 μg of Ikaros A465/7 mutant (lane 2) vector. Western blot with anti-β actin antibody was used as a loading control (top panel). The right panel shows a comparison of mRNA levels of wild-type (WT) Ikaros and the Ikaros A465/7 mutant in transduced cells as determined by real time PCR. The relative expression of mutant Ikaros as compared with wild-type Ikaros (assigned a value of 1) is shown. B, schematic representation of the two PEST sequences in the Ikaros protein (bottom panel). The sequence of both PEST sequences is shown (top panel). The amino acids that are known to be phosphorylated by CK2 kinase are marked by asterisk (*). C, pulse-chase analysis of degradation of wild-type Ikaros (left), the Ikaros-A465/7 mutant (middle), and the Ikaros A11+A465/7 mutant (right) in 293T cells, metabolically labeled with [35S]methionine/[35S]cysteine. Cells were harvested at indicated time points of chase with unlabeled methionine and cysteine. Ikaros proteins were immunoprecipitated with anti-Ikaros-CTS antibody and analyzed by autoradiography. Data shown are from one experiment performed in triplicate (mean ± S.D.) that is representative of five independent experiments. The insets above graphs show the levels of wild-type Ikaros and its mutants as assessed by immunoblotting with anti-Ikaros antibody. D, MOLT-4 cells were treated with cycloheximide to inhibit de novo protein synthesis, and Ikaros expression was determined by Western blot analysis of 20 μg of nuclear extract obtained from untreated MOLT-4 (lanes 1-5), MOLT-4 cells treated with the phosphatase inhibitor calyculin (lanes 6-10), or MOLT-4 cells treated with both calyculin and the CK2-kinase-selective inhibitors DRB (lanes 11-15) or TBB (lanes 16-20). Ikaros proteins were visualized by Western blot using anti-Ikaros-CTS antibody.
In Vitro Phosphatase Assay—Recombinant PP1 and PP2A were purchased from New England Biolabs, Ipswich, MA. Ikaros protein and its mutants were expressed in 293T cells. Cells were labeled in vivo with radioactive orthophosphate as described above and harvested in RIPA, and proteins were immunoprecipitated and washed three times in NTN buffer (100 mm NaCl, 20 mm Tris, pH 8.0, 0.5% NP-40), then once in phosphatase reaction buffer. Immunoprecipitated Ikaros proteins bound to protein A beads were incubated with phosphatase buffer (50 mm Tris-HCl, pH 7.0, 0.1% 2-mercaptoethanol, 0.3 mg/ml bovine serum albumin, 0.01% Brij 35, 5 mm caffeine, and 0.1 mm EGTA) in the presence or absence of PP1 or PP2A (∼150 pm) as indicated. The reaction was performed at 30 °C for 30 min and was terminated by the addition of SDS sample buffer. Proteins were separated by SDS-PAGE and visualized by autoradiography.
In Vivo Binding Assay—293T cells transfected with Ikaros or its mutant were lysed in RIPA lysis buffer. The Ikaros-PP1 interaction was tested by immunoprecipitation with anti-PP1 antibodies (Santa Cruz, sc 7482) followed by Western blot with anti-Ikaros CTS antibodies. The interaction between endogenous Ikaros and PP1 was tested in untransfected VL3-3M2, MOLT-4, CEM, HAFTL, and BAL17 cells. Cells were lysed in RIPA lysis buffer, the lysates were immunoprecipitated with anti-PP1 antibodies, and the presence of Ikaros was detected by Western blot using anti-Ikaros-CTS antibodies.
For experiments described in Fig. 1C, 293T cells expressing Ikaros were labeled in vivo with radioactive orthophosphates (PerkinElmer Life Science) for 4 h and lysed in RIPA lysis buffer, and lysates were immunoprecipitated with anti-Ikaros-CTS antibodies on protein A beads. Immunoprecipitates were washed three times with NTN buffer and once in phosphatase reaction buffer (described above). After washing, immunoprecipitates were incubated with phosphatase reaction buffer supplemented with phosphatase inhibitors as indicated: a phosphatase inhibitor mixture, PPi (Sigma-Aldrich) in lane 1;no phosphatase inhibitor (lane 2); or PP1-specific inhibitor (inhibitor 2, lane 3). The reactions shown in all lanes were supplemented with the CK2 kinase inhibitors DRB and heparin. Proteins were separated by SDS-PAGE and visualized by autoradiography.
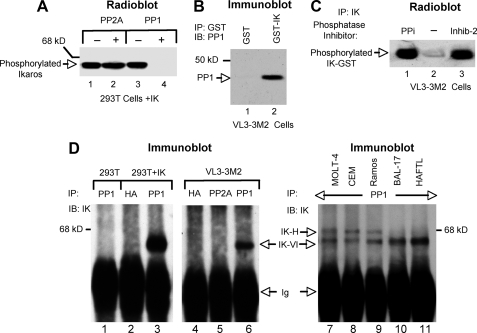
FIGURE 1.
Ikaros interacts in vivo and is a substrate for PP1 phosphatase. A, Ikaros proteins phosphorylated in vivo were immunoprecipitated (IP) with anti-Ikaros-CTS antibodies and incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of PP2A or PP1 phosphatases. B, whole cell lysate from VL3-3M2 cells was mixed with glutathione-agarose beads preincubated with GST (lane 1) or GST-Ikaros (IK) fusion protein (lane 2). The bound proteins were eluted and separated by SDS-PAGE, and the presence of PP1 was detected by Western blot (IB). C, 293T cells expressing Ikaros were labeled in vivo with radioactive orthophosphates (PerkinElmer Life Science), and their whole cell lysates were immunoprecipitated with anti-Ikaros-CTS antibodies using protein A beads. Immunoprecipitates were incubated with phosphatase reaction buffer supplemented with phosphatase inhibitors as indicated: a phosphatase inhibitor mixture (PPi, lane 1), no phosphatase inhibitor (lane 2), or the PP1-specific inhibitor (Inhib-2) (lane 3). The reactions in all lanes were supplemented with the CK2 kinase inhibitors DRB and heparin. Proteins were separated by SDS-PAGE and visualized by autoradiography. D, whole cell lysates from untransfected 293T cells (lane 1), 293T cells transfected with Ikaros (lanes 2-3), VL3-3M2 cells (lanes 4-6), or other human (lanes 7-9) or murine (lanes 10-11) cell lines were immunoprecipitated with anti-PP1, anti-hemagglutinin (HA), or anti-PP2A antibodies as indicated. The presence of Ikaros in immunoprecipitates was detected by Western blot with anti-Ikaros-CTS antibody.
Analysis of Ikaros Ubiquitination—MOLT-4 or VL3-3M2 cells were treated with Cbz-LLL (MG132) proteasome inhibitor (50 μm) (Calbiochem, catalogue #474790) for 2 h or left untreated (as indicated). Cells were harvested in ice-cold phosphate-buffed saline and lysed in RIPA buffer along with protease and phosphatase inhibitors. Cell lysates were immunoprecipitated with anti-Ikaros-CTS antibody or with anti-ubiquitin antibody (Assay Designs/Stressgen Bioreagents, SPA-200) on protein A beads. Immunoprecipitates were eluted in SDS sample buffer and separated on SDS-PAGE. Ikaros-ubiquitin conjugates were detected by Western blot using anti-Ikaros-CTS or anti-ubiquitin antibodies as indicated.
RESULTS
PP1 Binds and Dephosphorylates Ikaros in Vitro and in Vivo—We tested the ability of different phosphatases to dephosphorylate Ikaros in vitro. Wild-type Ikaros was expressed in 293T cells, labeled in vivo by radioactive orthophosphate, and immunoprecipitated using anti-Ikaros-CTS antibody. The phosphatases PP1, PP2A, PP2B, and PP2C were used for in vitro reactions with immunoprecipitated Ikaros. Changes in Ikaros phosphorylation were visualized by autoradiography (Fig. 1A and data not shown). Results showed that after in vitro reaction with PP1, Ikaros lost its radioactive phosphate (Fig. 1A, lanes 3 versus 4), whereas PP2A (Fig. 1A, lanes 1 and 2) and PP2B and PP2C (data not shown) failed to dephosphorylate Ikaros. These data demonstrate that PP1 can dephosphorylate Ikaros.
The ability of Ikaros to associate with PP1 was tested using a GST pulldown assay. The nuclear extract from VL3-3M2 cells was mixed with glutathione-agarose beads that were preincubated with GST-Ikaros fusion protein (Fig. 1B, lane 2) or GST protein alone (Fig. 1B, lane 1). Proteins that were bound to the beads were eluted and tested for the presence of PP1 by Western blot (Fig. 1B). These results show that Ikaros interacts with PP1 in a GST pulldown assay.
Next, we tested whether Ikaros could be co-immunoprecipitated with PP1. First we examined the ability of Ikaros to be dephosphorylated by a co-immunoprecipitated protein and the ability of a PP1-specific inhibitor to abrogate this dephosphorylation. Ikaros-transfected 293T cells were labeled in vivo with radioactive orthophosphate, and whole cell lysates were immunoprecipitated with anti-Ikaros-CTS antibody. Immunoprecipitates were then incubated with phosphatase buffer. Incubations were performed in the presence of a phosphatase inhibitor mixture (PPi, Fig. 1C, lane 1), the absence of phosphatase inhibitor (Fig. 1C, lane 2), or in the presence of a PP1-specific inhibitor (Inhibit-2, Fig. 1C, lane 3). After incubation, samples were separated by SDS-PAGE and visualized by autoradiography. Results showed that the radioactive, in vivo phosphorylated Ikaros undergoes dephosphorylation when incubated with phosphatase buffer in the absence of phosphatase inhibitors (Fig. 1C lane 2), suggesting that a phosphatase was co-immunoprecipitated with Ikaros. The co-immunoprecipitated phosphatase activity is inhibited by the addition of a nonspecific phosphatase inhibitor mixture (PPi) or inhibitor-2, a PP1-specific inhibitor (Fig. 1C, lanes 1 and 3, respectively). These results show that Ikaros co-immunoprecipitates a phosphatase whose activity is inhibited by inhibitor-2, a PP1-specific inhibitor. These results suggest that Ikaros interacts in vivo with PP1.
To confirm this, we determined whether Ikaros could be co-immunoprecipitated with PP1-specific antibodies (Fig. 1D). Immunoprecipitation with anti-PP1 antibodies followed by Western blot with anti-IK antibodies showed that PP1 co-immunoprecipitated transfected Ikaros in 293T cells as well as endogenous Ikaros in VL3-3M2 cells (Fig. 1D, lanes 3 and 6, respectively). Immunoprecipitations of untransfected 293T cells with anti-PP1 antibodies (Fig. 1D, lane 1) of Ikaros-transfected 293T and VL3-3M2 cells with anti-hemagglutinin antibodies (Fig. 1D, lanes 2 and 4) and of VL3-3M2 cells with anti-PP2A antibodies (Fig. 1D, lane 5) were used as negative controls. The association of endogenous Ikaros with endogenous PP1 in vivo in other human and murine hematopoietic cell lines was confirmed by co-immunoprecipitation using anti-PP1 antibodies. The co-immunoprecipitated Ikaros isoforms were detected by Western blot with anti-IK antibodies (Fig. 1D, lanes 7-11). These experiments demonstrate that Ikaros associates with PP1 in hematopoietic cells in vivo.
PP1 Interacts with Ikaros via a Specific Recognition Motif—To identify the recognition site that is required for PP1-Ikaros interaction, we tested the ability of PP1 to dephosphorylate Ikaros deletion mutants (Fig. 2A). In vivo phosphorylated Ikaros deletion mutants were immunoprecipitated and then incubated with appropriate buffer in the absence (Fig. 2A, lanes 1, 3, 5, 7, and 9) or presence (Fig. 2A, lanes 2, 4, 6, 8, and 10) of PP1, and their phosphorylation status was visualized by radiography. Results showed that PP1 is able to dephosphorylate in vitro all Ikaros deletion mutants except the mutant that lacked the C-terminal part of the protein (N-458, Fig. 2A, lanes 9 and 10). These results suggest that residues that are essential for the dephosphorylation of Ikaros by PP1 are located at the C-terminal part of Ikaros (Fig. 2B).
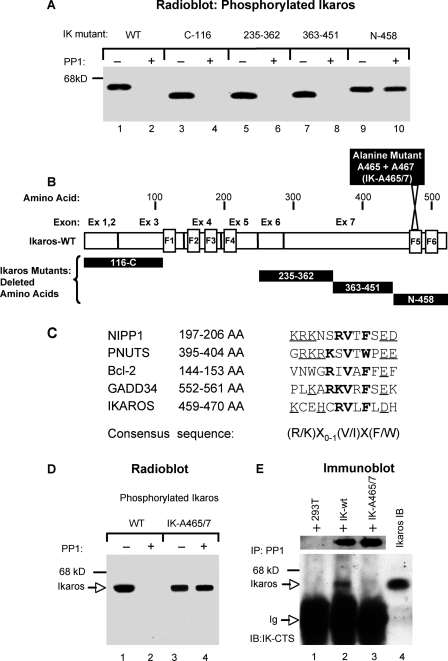
FIGURE 2.
Interaction of Ikaros and PP1 requires a specific motif. A, wild-type (WT) Ikaros or its corresponding mutants were transfected into 293T cells and labeled in vivo with radioactive [32P]orthophosphate. The whole cell lysates were immunoprecipitated with anti-Ikaros-CTS antibodies. Immunoprecipitates were incubated with PP1 reaction buffer in the absence or presence of recombinant PP1 as indicated. The samples were resolved by SDS-PAGE and visualized by autoradiography. IK, Ikaros. B, an Ikaros protein map showing deleted segments of the corresponding Ikaros deletion mutants used in experiments above as well the position of the Ikaros point mutant (IK-A465/7) used in other experiments. C, a comparison of PP1-interacting motifs among known PP1 interactors and the Ikaros protein. The amino acids that constitute the core element are presented in bold letters, whereas the contributing amino acids (lysine/arginine proximal to the interacting motif and aspartate/glutamate distal to the interacting motif) are underlined. The consensus sequence of the PP1-interacting motif is at the bottom. D, 293T cells were transfected with wild-type Ikaros or an Ikaros gene that is mutated at the conserved PP1 binding motif (IK-A465/7) as indicated and were in vivo labeled with radioactive [32P]orthophosphate. Ikaros proteins were immunoprecipitated with anti-Ikaros CTS antibody and incubated in the presence or absence of recombinant PP1 as indicated. After the dephosphorylation reaction, samples were resolved on SDS-PAGE and visualized by autoradiography. E, whole cell lysate from untransfected 293T cells (lane 1) or transfected with wild-type Ikaros (lane 2) or with the IK-A465/7 mutant (lane 3) were immunoprecipitated (IP) with anti-PP1 antibody. Before immunoprecipitation, the whole cell lysates of 293T cells transfected with wild-type Ikaros or the IK-A465/7 mutant were normalized for the presence of Ikaros protein as evidenced by Western blot with anti-Ikaros CTS antibody (top panel). The presence of Ikaros in immunoprecipitates was visualized by Western blot with anti-Ikaros antibodies. The nuclear extract from 293T cells transfected with wild-type Ikaros was used in Western blot as a marker for the presence of Ikaros (lane 4).
It has been demonstrated that PP1 interacts with its substrate via a specific recognition motif, (R/K)X0-1(V/I)X(F/W) (38-40). A search for potential residues involved in PP1-Ikaros interaction at the C-terminal end of the Ikaros protein revealed the presence of an evolutionarily conserved PP1 recognition motif at amino acids 459-470. A sequence comparison of this motif in the Ikaros protein to the PP1 recognition site in proteins known to interact with PP1 is provided in Fig. 2C. The Ikaros PP1 recognition motif is located within one of the C-terminal zinc fingers involved in protein-protein interactions with other Ikaros family members (Fig. 2B). Because valine and phenylalanine have previously been shown to be essential for interaction with PP1 (38), we determined whether mutation of these two amino acids would affect the ability of PP1 to dephosphorylate Ikaros. Results show that PP1 dephosphorylates wild-type Ikaros (Fig. 2D, lanes 1 and 2) but is unable to dephosphorylate an Ikaros mutant where valine and phenylalanine at positions 465 and 467, respectively (IK-A465/7), are mutated into alanine (Fig. 2D, lanes 3 and 4). These data show that the presence of the PP1 recognition motif is essential for dephosphorylation of Ikaros by PP1.
Next, we determined by immunoprecipitation whether the presence of the PP1 recognition motif is essential for the direct interaction of Ikaros with PP1 (Fig. 2E). Results showed that the wild-type Ikaros expressed in 293T cells was precipitated with anti-PP1 antibodies (Fig. 2E, lane 2), whereas the same antibodies failed to immunoprecipitate Ikaros protein with a mutated PP1 recognition motif, IK-465/7 (Fig. 2E, lane 3). Together these results demonstrate that Ikaros directly interacts with PP1 via the PP1-recognition motif located at amino acids 459-470 and that this interaction is essential for the dephosphorylation of Ikaros by PP1.
Dephosphorylation of Ikaros by PP1 Regulates Its DNA Binding Ability and Pericentromeric Localization—The phosphorylation of Ikaros by CK2 kinase has previously been shown to regulate Ikaros DNA binding affinity toward promoters of its target genes as well as toward repetitive sequences located at PC-HC (29). The binding of Ikaros to γ satellite A and γ satellite B probes has also been shown to determine its subcellular localization (16). We tested whether the loss of interaction with PP1 could affect Ikaros binding affinity toward these probes that are derived from PC-HC sequences (Fig. 3, A and B). Electromobility shift assays showed that the wild-type Ikaros expressed in 293T cells binds strongly to the γ satellite A probe, whereas its mutant, that is unable to interact with PP1 (IK-A465/7), does not bind to γ satellite A (Fig. 3A, lanes 2 and 3). When the IK-A465/7 mutant was incubated with calf intestinal alkaline phosphatase during the DNA binding assay, its DNA binding was restored to the level of wild-type Ikaros, whereas incubation with PP1 failed to restore its DNA binding ability (Fig. 3A, lanes 5 and 6). These results suggest that the lack of dephosphorylation by PP1 leads to hyperphosphorylation of Ikaros and that this increased phosphorylation is responsible for its loss of DNA binding activity.
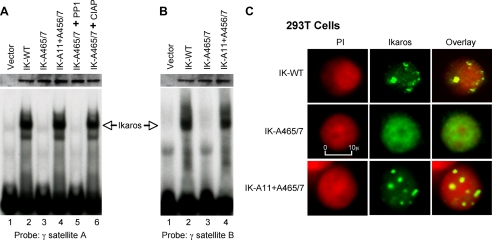
FIGURE 3.
The ability of Ikaros to interact with PP1 regulates its DNA binding and subcellular localization. A, nuclear extracts were prepared from 293T cells expressing wild-type or mutant Ikaros (IK) proteins. Equal amounts of Ikaros proteins were used in gel shift reactions as confirmed by Western blot (upper panel). Where indicated, recombinant PP1 (lane 5) or calf intestinal alkaline phosphatase (CIAP, lane 6) were added to the DNA binding reaction. Gel shifts were performed using radiolabeled probes derived from the murine γ satellite A repeat (A) or the murine γ satellite B repeat (B). C, confocal microscopy was used to examine the subnuclear localization of wild-type (WT) and mutant Ikaros proteins. 293T cells that do not express endogenous Ikaros were transduced with wild-type Ikaros (top panel), the Ikaros mutant that is unable to interact with PP1-IK-A465/7 (middle panel), or with a combined Ikaros mutant that is unable to interact with PP1 and is also resistant to phosphorylation by CK2 kinase (IK-A11+A465/7). DNA was visualized by staining with propidium iodine (PI, red), and Ikaros proteins were visualized by anti-Ikaros-CTS antibodies (green). The right panel shows the combined images, with yellow indicating co-localization of Ikaros and DNA.
Because CK2 kinase is the major kinase that regulates Ikaros DNA binding ability, we tested whether an Ikaros mutant with alanine substitutions (mimic constitutive dephosphorylation) at 11 CK2 phosphorylation sites within the Ikaros protein would restore DNA binding to the Ikaros mutant, that is unable to interact with PP1. Results showed that an Ikaros mutant containing alanine mutations at the 11 CK2 target sites along with the PP1 recognition motif (IK-A11+A465/7) is able to bind the γ satellite A probe (Fig. 3A, lane 4). Thus, the introduction of 11 alanine mutations at CK2 phosphorylation sites restored the DNA binding ability of an Ikaros mutant that is unable to interact with PP1. This suggests that PP1 binds Ikaros via the PP1 recognition motif and dephosphorylates CK2-phosphorylated amino acids, thereby increasing the ability of Ikaros to bind DNA. Similarly, the Ikaros mutant IK-A465/7 was unable to bind the γ satellite B probe, whereas the introduction of alanine mutations of CK2-phosphorylated amino acids restored the ability of Ikaros to bind the same probe (Fig. 3B, lanes 1-4).
Ikaros DNA binding toward pericentromeric repeats has been shown to regulate its subcellular localization to PC-HC (16). Increased phosphorylation of Ikaros by CK2 kinase leads to the loss of Ikaros pericentromeric localization (29). We tested whether dephosphorylation of Ikaros by PP1 is necessary for localization of Ikaros to PC-HC. Ikaros exhibits a punctate staining pattern in the nucleus that has been shown to be due to its to PC-HC localization (14, 15). We demonstrate that wild type Ikaros also produces a punctate staining pattern in 293T cells, indicative of its PC-HC localization (Fig. 3C, top panel). In contrast, the Ikaros mutant that is unable to interact with PP1 exhibited a diffuse nuclear staining pattern, suggesting the loss of PC-HC localization (Fig. 3C, middle panel). The presence of alanine mutations at CK2-phosphorylated residues restored PC-HC localization of Ikaros mutant IK-A465/7, as evidenced by a punctate staining pattern (Fig. 3C, bottom panel).
These data show that the dephosphorylation of Ikaros by PP1 is essential for optimal DNA binding ability as well as for subcellular localization at PC-HC. These results suggest that amino acids phosphorylated by CK2 kinase are among the targets for dephosphorylation by PP1 and that constitutive dephosphorylation of these amino acids restores Ikaros DNA binding ability as well as its subcellular localization even in the absence of interaction with PP1.
Lack of Dephosphorylation Leads to Increased Degradation of Ikaros Due to Phosphorylation by CK2 Kinase at PEST Regions—When 293T cells that do not express endogenous Ikaros were transfected with the vectors expressing wild-type Ikaros, the expression level of Ikaros protein was high. When an Ikaros mutant that was unable to interact with PP1 phosphatase (IK-A465/7) was expressed in the same cells, its protein level was more than 5-fold decreased compared with the wild-type Ikaros (Fig. 4A, lanes 1 and 2). Real time PCR of the same transfected cells showed that the reduction in expression of the IK-A465/7 mutant was not due to a decrease in Ikaros mRNA levels (Fig. 4A). These data suggest that the decreased protein level observed for the IK-A465/7 mutant is due to an increase in its degradation. Another, although less likely possibility, is that the reduction in expression of the IK-A465/7 mutant is due to its lower translational efficiency.
We examined whether the phosphatase-resistant Ikaros mutant undergoes increased degradation. We noted that previously identified Ikaros phosphorylation sites are located within regions that are rich in proline, glutamine, and serine. Within these regions we have identified, using the PEST-finder software, two typical PEST sequences (PEST-1 and PEST-2, Fig. 4B) that scored 17 and 16, respectively; scores above zero suggest a possible PEST region, whereas scores above five indicate the strong probability that a particular sequence represents a PEST region. The PEST motifs have been demonstrated to function as targets for phosphorylation-mediated protein degradation (41, 42). The phosphorylated residues within both PEST sequences have been shown to be substrates for CK2 kinase (28, 29).
To determine whether CK2 kinase-mediated phosphorylation at PEST sequences causes increased degradation of Ikaros protein, we compared the half-life of wild-type Ikaros to those of IK-A465/7 and the IK-A465/7 that has alanine mutations at all 11 amino acids known to be phosphorylated by CK2 kinase in vivo (IK-A11+A465/7) using a pulse-chase degradation assay. Results showed that the loss of interaction with PP1 results in a severely shortened Ikaros half-life (Fig. 4C, IK-WT versus IK-A465/7). The presence of alanine mutations at CK2 phosphorylation sites stabilized the IK-A465/7 mutant protein and extended its half-life (from 1 h for IK-A465/7A to 4 h for IK-A11+A465/7). The partial rather than full restoration of half-life observed in the IK-A11+A465/7 mutant suggests that phosphorylation of additional amino acids contributes to Ikaros degradation.
To confirm that CK2 kinase and PP1 phosphatase activities regulate Ikaros degradation, we studied the expression of Ikaros in the human leukemia cell line, MOLT-4 (Fig. 4D). MOLT-4 cells were treated with cycloheximide to inhibit de novo protein synthesis, and Ikaros expression was studied by Western blot in the absence (Fig. 4D, lanes 1-5) and in presence of the phosphatase inhibitor, calyculin (Fig. 4D, lanes 6-10) or in the presence of both calyculin and DRB (Fig. 4D, lanes 11-15) or TBB (Fig. 4D, lanes 16-20), two kinase inhibitors that are highly selective for inhibition of CK2. Data showed that treatment with calyculin results in a short Ikaros half-life as compared with untreated cells. Treatment with the CK2 inhibitors DRB or TBB stabilizes Ikaros protein and extends its half-life, although it does not completely restore Ikaros half-life to that observed in untreated cells. Calyculin was used in this assay because of its potency and ability to penetrate cells. Although calyculin inhibits both PP1 and PP2A in vivo, PP2A was unable to dephosphorylate Ikaros in vitro (Fig. 1A). Therefore, the impact of calyculin on Ikaros half-life should reflect the impact of PP1-mediated effects only. The concentrations of DRB and TBB that we used (50 and 10 μm, respectively) result in a partial inhibition of CK2. This explains why cells treated with both calyculin and DRB or TBB had an Ikaros half-life that was slightly less than that observed in untreated cells. Higher concentrations of DRB or TBB or the addition of other CK2 inhibitors resulted in growth arrest with a mild apoptotic effect on MOLT-4 cells, which preclude their use in this assay.
Together, the data presented in Fig. 4, C and D, suggest that the phosphorylation of Ikaros by CK2 kinase at PEST regions promotes its degradation, whereas dephosphorylation by PP1 stabilizes Ikaros protein and extends its half-life. Results in Fig. 4C also suggest that a large percentage of Ikaros protein undergoes phosphorylation and dephosphorylation in vivo, as the loss of interaction with PP1 decreases protein levels by more than 5-fold.
Degradation of Ikaros Is Mediated by the Ubiquitin/Proteasome Pathway—The evidence that Ikaros degradation is regulated by its phosphorylation at PEST sequences suggested that degradation might involve the ubiquitin/proteasome pathway. Protein degradation via the ubiquitin/proteasome pathway involves the addition of multiple ubiquitin residues to the substrate preceding its degradation. If Ikaros is degraded via the ubiquitin/proteasome pathway, it should form a complex with ubiquitin residues in vivo. We tested whether Ikaros/ubiquitin conjugates can be detected in vivo and whether the presence of the proteasome inhibitor Cbz-LLL (MG132) (43, 44) would allow us to detect higher molecular weight Ikaros/ubiquitin conjugates.
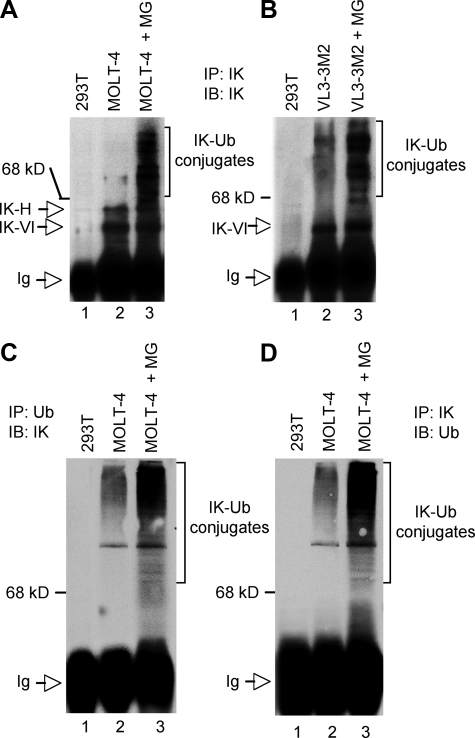
Human MOLT-4 T lymphocyte leukemia cells and murine VL3-3M2 thymocyte leukemia cells were treated with MG132 or left untreated. The whole cell lysate from MG132-treated or untreated cells was used to immunoprecipitate endogenous Ikaros protein, and immunoprecipitates were analyzed by Western blot using anti-Ikaros antibodies (Fig. 5, A and B). Results showed that in the absence of MG132, immunoprecipitates contained mostly large Ikaros isoforms, IK-VI and IK-H, with low levels of higher molecular weight forms of Ikaros (Fig. 5, A, lane 2, and B, lane 2). Immunoprecipitates of MG132-treated MOLT-4 and VL3-3M2 cells demonstrated high levels of the high molecular weight complexes that are recognized by anti-Ikaros antibodies (Fig. 5, A, lane 3, and B, lane 3). The observed Ikaros-containing high molecular weight complexes exhibited a ladder appearance that is typical for multi-ubiquitinated protein conjugates. Immunoprecipitations with anti-Ikaros antibodies from cell lysates of 293T cells that do not express Ikaros were used as a negative control (Fig. 5, A, lane 1, and B, lane 1).
FIGURE 5.
Ikaros degradation is mediated by the ubiquitin/proteasome pathway. A-D, whole cell lysates were prepared from 293T cells, as a negative control, or as indicated from untreated MOLT-4 or VL3-3M2 cells or MOLT-4 or VL3-M2 cells that had been treated with the MG132 proteasome inhibitor (MG). Whole cell lysates were immunoprecipitated (IP) with the indicated antibodies, and immunoprecipitates were analyzed by 9% SDS-PAGE followed by immunoblot (IB) with the indicated antibodies. The high molecular weight complexes containing Ikaros (IK)-ubiquitin (Ub) conjugates are marked. The Ikaros detected in MOLT-4 and VL3-3M2 cells in all panels is endogenous Ikaros.
To test whether high molecular weight forms of Ikaros correspond to multi-ubiquitinated Ikaros, the whole cell lysates from untreated or MG132-treated MOLT-4 cells were immunoprecipitated with anti-ubiquitin antibodies, and immunoprecipitates were subjected to Western blot with anti-Ikaros antibodies. Results showed that the high molecular weight complexes immunoprecipitated by anti-ubiquitin antibodies are recognized by anti-Ikaros antibodies and, thus, represent Ikaros/ubiquitin conjugates (Fig. 5C, lanes 2 and 3). As expected, treatment with MG132 increased the levels of detected Ikaros/ubiquitin conjugates (Fig. 5C, lane 3), which provided further evidence that this complex is degraded by the proteasome.
To provide additional evidence that Ikaros is ubiquitinated in vivo, the whole cell lysates of untreated and MG132-treated MOLT-4 cells were immunoprecipitated with anti-Ikaros antibodies, and the presence of Ikaros/ubiquitin conjugates was tested by Western blot using anti-ubiquitin antibodies. Results showed the presence of high molecular weight Ikaros ladder forms that are representative of Ikaros/ubiquitin conjugates (Fig. 5D).
Taken together, the results presented in Fig. 4 suggest that the phosphorylation of Ikaros by CK2 kinase induces its degradation, whereas dephosphorylation by PP1 interferes with this process. The data presented in Fig. 5 show that Ikaros is ubiquitinated in vivo, which suggests that the mechanism of Ikaros degradation involves the ubiquitin/proteasome pathway.
Ikaros Co-localizes with PP1 at PC-HC—In cells, PP1 associates with more than 70 different proteins (45). These proteins are direct substrates for PP1, modulate PP1 activity, or target PP1 to its substrates. The presence of a well preserved PP1 binding motif at the C-terminal end of Ikaros as well as the ability of Ikaros to co-immunoprecipitate with PP1 (demonstrated in Fig. 1) raised the question of whether the Ikaros-PP1 complex is of sufficient stability to allow Ikaros to function in targeting PP1 to other substrates.
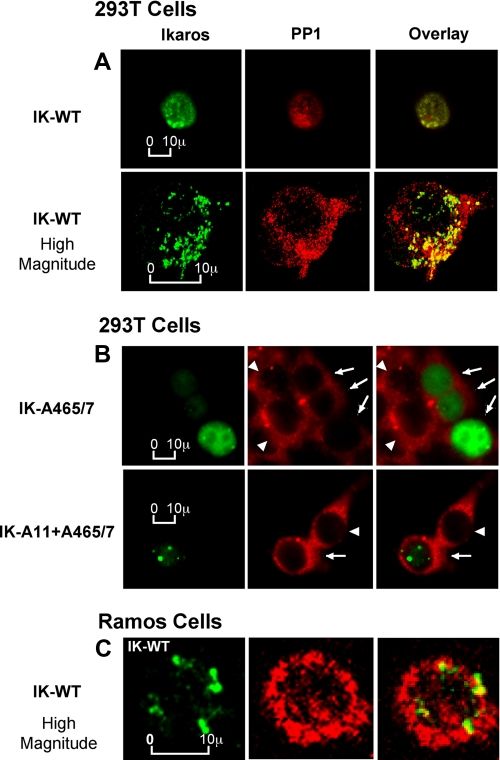
We used 293T cells that do not express endogenous Ikaros to study whether the expression of Ikaros affects the subcellular localization of PP1. When wild-type Ikaros is transfected into 293T cells, it displays a nuclear punctate staining pattern similar to its typical localization in hematopoietic cells, as observed with immunofluorescent microscopy (16) (Fig. 6A, left panels, green staining). In Ikaros-transfected 293T cells, PP1 localizes to the nucleus (Fig. 6A, middle panels, red staining). A fraction of PP1 protein co-localize with Ikaros, as evidenced in the overlay image of Ikaros and PP1 (Fig. 6A, right panels, yellow staining). The co-localization of a fraction of Ikaros with a fraction of the PP1 proteins in the nucleus was confirmed by using high magnitude confocal microscopy (Fig. 6A, bottom panels). These data suggest that a part of cellular PP1 forms a complex with Ikaros in the nucleus in vivo. These results correspond with the co-immunoprecipitation experiments showing that PP1 and Ikaros proteins associate in vivo (Fig. 1).
FIGURE 6.
Ikaros targets PP1 into the nucleus and to pericentromeric heterochromatin. A, 293T cells were transfected with wild-type (WT) Ikaros. Localization of transfected Ikaros proteins and endogenous PP1 was examined by immunofluorescence microscopy (top panel) and high magnitude confocal microscopy (bottom panel). Ikaros proteins were detected with anti-Ikaros-CTS antibody (green), whereas endogenous PP1 was detected with anti-PP1 antibody (red). The right panel shows combined images, with yellow indicating co-localization of Ikaros and PP1 proteins. B, 293T cells were transfected with the Ikaros-A465/7 mutant (top panel) or the Ikaros-A11+A465/7 mutant (bottom panel). Confocal microscopy was used to examine localization of transfected Ikaros (IK) proteins and endogenous PP1. Ikaros proteins were detected with anti-Ikaros-CTS antibody (green), whereas endogenous PP1 was detected with anti-PP1 antibody (red). The right panel shows combined images, with yellow indicating co-localization of Ikaros and PP1 proteins. Arrows indicate transfected cells, and arrowheads indicate nontransfected cells within the field in the middle and right panels. C, high magnitude confocal microscopy was used to examine the localization of endogenous Ikaros and endogenous PP1 protein in the Ramos human lymphoma cell line. Endogenous Ikaros proteins were detected with anti-Ikaros-CTS antibody (green), whereas endogenous PP1 was detected with anti-PP1 antibody (red). The right panel shows combined images with yellow indicating co-localization of Ikaros and PP1 proteins.
Next, we determined whether the loss of the ability of Ikaros to bind PP1 affects the subcellular localization of PP1. Immunofluorescence microscopy showed that in 293T cells transfected with the Ikaros PP1 binding domain mutant (IK-A465/67, Fig. 6B, top panels, transfected cells are indicated by arrows) PP1 exhibits a cytoplasmic localization with the absence of the nuclear localization observed in cells transfected with wild-type Ikaros (Fig. 6A, top panels). The Ikaros mutant protein exhibited a diffuse nuclear staining with the loss of punctate PC-HC localization (Fig. 6B, top panel, green staining) as shown previously in Fig. 3C. The subcellular localization of PP1 in 293T cells that are transfected with the Ikaros mutant (indicated by arrows) appears to be similar to that observed in 293T cells that are not transfected (indicated by arrowheads). This suggests that in these cells, the interaction of PP1 with Ikaros is necessary for the nuclear localization of PP1.
To confirm that the changes in PP1 localization are due to the lack of its interaction with Ikaros and not due to poor Ikaros expression and/or DNA binding ability, 293T cells were transfected with Ikaros containing alanine mutations at the 11 CK2 phosphorylation sites and in the PP1 binding motif (IK-A11+A465-7). Confocal microscopy showed that in transfected cells (indicated by arrows), the Ikaros mutant exhibits a punctate, pericentromeric nuclear localization (Fig. 6B, bottom panel, green staining), whereas PP1 localization remains cytoplasmic (Fig. 6B, bottom panel, red staining) similar to its localization in non-transfected 293T cells (indicated by arrowheads). These data further suggest that in 293T cells Ikaros can regulate localization of PP1 within the nucleus and to PC-HC.
Ikaros is not physiologically expressed in 293T cells. In hematopoietic cells the majority of Ikaros is localized in PC-HC. We determined whether Ikaros and PP1 co-localize in hematopoietic cells using confocal microscopy. In the human B lymphoma cell line Ramos, Ikaros localizes in PC-HC (Fig. 6C, green staining), whereas PP1 is abundant in the nucleus (Fig. 6C, red staining). Overlay staining demonstrated that a fraction of PP1 co-localizes with Ikaros at PC-HC (Fig. 6C, yellow staining). These data together with data in Fig. 6A suggest that a fraction of PP1 can form a stable complex with Ikaros and that a fraction of PP1 localizes to PC-HC in the nucleus.
DISCUSSION
The function of Ikaros is regulated by its post-translational modifications. Ikaros is phosphorylated at multiple sites. Previous reports showed that phosphorylation by CK2 kinase controls the ability of Ikaros to bind DNA and regulate cell cycle progression as well as its subcellular localization (27-29). Phosphorylation is a reversible process, and optimal Ikaros function requires a balance between phosphorylated and dephosphorylated protein forms. We have demonstrated that Ikaros is a substrate for PP1 phosphatase. The PP1 phosphatase is a highly conserved threonine-serine phosphatase that is involved in multiple cellular functions including cell cycle progression, apoptosis, and energy metabolism (45, 46). In the cell PP1 forms multimeric holoenzymes with interactor/regulatory proteins. The best characterized regulatory binding site for PP1 is the RVXF motif, which is part of the consensus sequence (R/K)X0-1(V/I){P}(W/F), where X is any amino acid, and {P} can be any residue except proline (38, 40). Most PP1 interactors share this motif, which likely serves as an anchor for the binding of regulatory proteins to PP1 and, thus, promotes the formation of a multimeric holoenzyme. We have identified the well conserved RVXF motif at the C-terminal end of the Ikaros protein and demonstrate that the presence of this sequence is required for Ikaros-PP1 interaction. Although only a fraction of Ikaros is in a complex with PP1, the ability to detect these complexes with standard co-immunoprecipitation techniques suggests that these complexes are more stable than a typical phosphatase-substrate interaction.
Our previous studies identified the functional significance of Ikaros phosphorylation at specific amino acids but left two important questions unanswered; 1) what percentage of Ikaros protein undergoes phosphorylation/dephosphorylation in the cell, and 2) how does an alteration in the balance of phosphorylation/dephosphorylation affect Ikaros cellular functions? The presence of a specific sequence that is essential for the Ikaros-PP1 interaction allowed us to study both the significance of Ikaros phosphorylation/dephosphorylation status and the importance of Ikaros-PP1 interactions for Ikaros function. Here we use an Ikaros mutant that is unable to bind PP1 and, thus, is resistant to dephosphorylation (IK-A465/7) together with a combined Ikaros mutant that in addition to being unable to interact with PP1 is resistant to phosphorylation by CK2 kinase to determine several functions that are controlled by phosphorylation/dephosphorylation (summarized in Fig. 7).
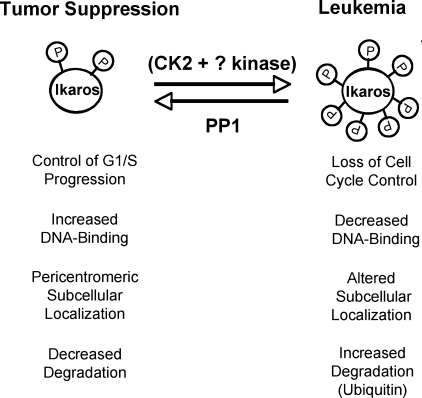
FIGURE 7.
Proposed regulation of Ikaros activity by CK2 and PP1 signal transduction pathways. The proposed model summarizes the opposing effects of the CK2 kinase and the PP1 pathways on Ikaros function. Our hypothesis is that an optimal balance of these two pathways insures normal cellular proliferation and enables Ikaros to function as a tumor suppressor, whereas increased CK2 kinase activity (as observed in many forms of leukemia) leads to the loss of Ikaros function, increased Ikaros degradation, and malignant transformation.
Regulation of DNA Binding Ability and Subcellular Localization—A mutation in Ikaros that prevents its interaction with PP1 results in the loss of DNA binding and the loss of pericentromeric localization. In combined Ikaros mutants that lack the ability to bind PP1 and which also have mutations that prevent phosphorylation by CK2 kinase both functions are restored. This suggests that (a) dephosphorylation of Ikaros is essential for its DNA binding activity and subcellular localization, (b) PP1 is the major phosphatase responsible for maintaining these functions, and (c) a large fraction of Ikaros protein undergoes phosphorylation by CK2 kinase and dephosphorylation by PP1. If the latter was not true we would expect the PP1 non-binding IK-A465/7 mutant to show dual localization; Ikaros protein-phosphorylated by CK-2 (unable to bind DNA) would exhibit diffuse nuclear staining, whereas Ikaros protein that had not interacted with CK2 (unphosphorylated and able to bind DNA) would be localized to PC-HC. Instead, the PP1 non-binding mutant displays diffuse nuclear staining without obvious localization to PC-HC. This correlates with a severely decreased DNA binding affinity observed in electromobility shift assay (Fig. 3, A and B). This indicates that most of the Ikaros protein in the cell is phosphorylated by CK2, and in the absence of PP1 activity, Ikaros remains phosphorylated and unable to bind DNA. Because the PP1 nonbinding IK-A465/7 mutants could not be detected in PC-HC, the probable explanation for these results is that the majority of cellular Ikaros during cell cycle undergoes phosphorylation by CK2 and loses its PC-HC localization due to an inability to be dephosphorylated by PP1. These data underscore the importance of the continuous process of reversible phosphorylation/dephosphorylation of Ikaros by CK2 and PP1 for its subcellular localization and its function in regulating the expression of genes by mediating their localization to PC-HC.
Regulation of Ikaros Stability—Our data suggest that dephosphorylation by PP1 stabilizes the Ikaros protein, whereas phosphorylation by CK2 kinase has the opposite effect. An Ikaros mutant that is deficient in PP1 binding ability has a much shorter half-life compared with wild-type Ikaros, suggesting that dephosphorylation by PP1 stabilizes Ikaros protein. The introduction of alanine mutations at CK2-phosphorylated residues increases the stability of Ikaros, suggesting that phosphorylation by CK2 promotes Ikaros degradation. Many residues phosphorylated by CK2 are located within two strong PEST sequences in the Ikaros protein, which provides further support for the hypothesis that CK2-mediated phosphorylation promotes Ikaros degradation.
The use of proteasome inhibitors provided evidence that Ikaros is ubiquitinated. Treatment with proteasome inhibitors was required to detect Ikaros-ubiquitin complexes, which suggests that ubiquitinated Ikaros is a good substrate for proteasome-mediated degradation. These data provided evidence that the mechanism for Ikaros degradation is likely to include the ubiquitin pathway. Phosphorylation in a PEST domain followed by ubiquitin-mediated degradation is a known mechanism by which protein activity is regulated and has been reported for other proteins, like Bcl6, that are known to regulate cellular proliferation (47). Our data provide evidence that the phosphorylation of Ikaros plays a novel role that is significant for Ikaros function, that of regulating protein stability. In addition, the studies described here suggest that increased degradation of the Ikaros tumor suppressor downstream of CK-2-mediated phosphorylation provides an additional mechanism for the pro-oncogenic activity of CK2 kinase.
Does Ikaros Act as a Regulatory Subunit of PP1 in Vivo?—PP1 forms complexes with many different regulatory subunits. These subunits target PP1 to particular substrates and regulate the activity and specificity of PP1. In cells, PP1 exhibits both cytoplasmic and nuclear localization (48), and it has been detected in chromosome-associated protein complexes (49, 50), suggesting that PP1 is involved in transcriptional control (50). It has been demonstrated that the NIPP1 and PNUTS proteins can target PP1 to the nucleus (51). Our data suggest that expression of Ikaros regulates subcellular localization of PP1 in 293T cells and that Ikaros and a fraction of PP1 co-localize at PC-HC in human lymphoma cells. It should be taken into account that Ikaros is not physiologically expressed in 293T cells, and its overexpression might lead to the abnormal distribution of PP1 in cells and that only fractions of each protein were co-localized in hematopoietic cells. The presented data led us to hypothesize that the Ikaros protein functions as a regulatory subunit of the PP1 holoenzyme. Ikaros associates with histone deacetylase 1 and with the SWI/SNF complex, whereas PP1 has been shown to be a component of class I histone deacetylase (52) as well as the SWI/SNF complex (53, 54). We hypothesize that Ikaros can function to target PP1 to its substrates, to PC-HC, or to other chromatin-remodeling complexes. This hypothesis will be tested in future studies.
In summary, our data suggest that two signaling pathways, one pro-oncogenic, involving CK2 kinase, and the other tumor suppressive, involving PP1 phosphatase, converge on the Ikaros protein, which is a substrate for both enzymes. CK2 kinase and PP1 have opposite effects on Ikaros function (summarized in Fig. 7). Because Ikaros regulates cellular proliferation and the loss of Ikaros function is an important step in the malignant transformation of hematopoietic cells, a balance between the two signal transduction pathways would be essential for normal cellular function and for the prevention of malignant transformation. Because Ikaros regulates normal hematopoiesis and the immune response, we hypothesize that CK2 kinase and PP1 influence these processes by regulating Ikaros activity. Future studies that test the model shown in Fig. 7 will aid in understanding the function of Ikaros in hematopoiesis and in the immune response.
Acknowledgments
We thank Vladimir Spiegelman and Bhatia Neehar for valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 HL07899 and K12 CA 087718 (to M. P.), 5K01 DK066163 (to K. J. P.), and K22 CA 111392 (to S. D.). This project was supported in part by the University of Wisconsin Institute for Clinical and Translational Research, funded through National Institutes of Health Clinical and Translational Science Award 1UL1RR025011. This work was also supported by a Midwest Athletes Against Childhood Cancer Award, St. Baldrick's Foundation Career Development grant, and by the University of Wisconsin Medical Education and Research Committee New Investigator Program (to S. D.).
Footnotes
The abbreviations used are: PC-HC, pericentromeric heterochromatin; PP1, protein phosphatase 1; DRB, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole; TBB, 4,5,6,7-tetrabromo-1H-benzotriazole; GST, glutathione S-transferase; RIPA, radioimmune precipitation assay buffer.
References
- 1.Nichogiannopoulou, A., Trevisan, M., Neben, S., Friedrich, C., and Georgopoulos, K. (1999) J. Exp. Med. 190 1201-1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu, L., Nichogiannopoulou, A., Shortman, K., and Georgopoulos, K. (1997) Immunity 7 483-492 [DOI] [PubMed] [Google Scholar]
- 3.Avitahl, N., Winandy, S., Friedrich, C., Jones, B., Ge, Y., and Georgopoulos, K. (1999) Immunity 10 333-343 [DOI] [PubMed] [Google Scholar]
- 4.Winandy, S., Wu, L., Wang, J. H., and Georgopoulos, K. (1999) J. Exp. Med. 190 1039-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumortier, A., Kirstetter, P., Kastner, P., and Chan, S. (2003) Blood 101 2219-2226 [DOI] [PubMed] [Google Scholar]
- 6.Winandy, S., Wu, P., and Georgopoulos, K. (1995) Cell 83 289-299 [DOI] [PubMed] [Google Scholar]
- 7.Sun, L., Heerema, N., Crotty, L., Wu, X., Navara, C., Vassilev, A., Sensel, M., Reaman, G. H., and Uckun, F. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 680-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakase, K., Ishimaru, F., Avitahl, N., Dansako, H., Matsuo, K., Fujii, K., Sezaki, N., Nakayama, H., Yano, T., Fukuda, S., Imajoh, K., Takeuchi, M., Miyata, A., Hara, M., Yasukawa, M., Takahashi, I., Taguchi, H., Matsue, K., Nakao, S., Niho, Y., Takenaka, K., Shinagawa, K., Ikeda, K., Niiya, K., and Harada, M. (2000) Cancer Res. 60 4062-4065 [PubMed] [Google Scholar]
- 9.Crescenzi, B., La Starza, R., Romoli, S., Beacci, D., Matteucci, C., Barba, G., Aventin, A., Marynen, P., Ciolli, S., Nozzoli, C., Martelli, M. F., and Mecucci, C. (2004) Haematologica 89 281-285 [PubMed] [Google Scholar]
- 10.Yagi, T., Hibi, S., Takanashi, M., Kano, G., Tabata, Y., Imamura, T., Inaba, T., Morimoto, A., Todo, S., and Imashuku, S. (2002) Blood 99 1350-1355 [DOI] [PubMed] [Google Scholar]
- 11.Nakayama, H., Ishimaru, F., Avitahl, N., Sezaki, N., Fujii, N., Nakase, K., Ninomiya, Y., Harashima, A., Minowada, J., Tsuchiyama, J., Imajoh, K., Tsubota, T., Fukuda, S., Sezaki, T., Kojima, K., Hara, M., Takimoto, H., Yorimitsu, S., Takahashi, I., Miyata, A., Taniguchi, S., Tokunaga, Y., Gondo, H., Niho, Y., Nakao, S., Kgo, T., Dohy, H., Kamada, N., and Harada, M. (1999) Cancer Res. 59 3931-3934 [PubMed] [Google Scholar]
- 12.Mullighan, C. G., Goorha, S., Radtke, I., Miller, C. B., Coustan-Smith, E., Dalton, J. D., Girtman, K., Mathew, S., Ma, J., Pounds, S. B., Su, X., Pui, C. H., Relling, M. V., Evans, W. E., Shurtleff, S. A., and Downing, J. R. (2007) Nature 446 758-764 [DOI] [PubMed] [Google Scholar]
- 13.Mullighan, C. G., Miller, C. B., Radtke, I., Phillips, L. A., Dalton, J., Ma, J., White, D., Hughes, T. P., Le Beau, M. M., Pui, C. H., Relling, M. V., Shurtleff, S. A., and Downing, J. R. (2008) Nature 453 110-114 [DOI] [PubMed] [Google Scholar]
- 14.Klug, C. A., Morrison, S. J., Masek, M., Hahm, K., Smale, S. T., and Weissman, I. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 657-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown, K. E., Guest, S. S., Smale, S. T., Hahm, K., Merkenschlager, M., and Fisher, A. G. (1997) Cell 91 845-854 [DOI] [PubMed] [Google Scholar]
- 16.Cobb, B. S., Morales-Alcelay, S., Kleiger, G., Brown, K. E., Fisher, A. G., and Smale, S. T. (2000) Genes Dev. 14 2146-2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koipally, J., Heller, E. J., Seavitt, J. R., and Georgopoulos, K. (2002) J. Biol. Chem. 277 13007-13015 [DOI] [PubMed] [Google Scholar]
- 18.Thompson, E. C., Cobb, B. S., Sabbattini, P., Meixlsperger, S., Parelho, V., Liberg, D., Taylor, B., Dillon, N., Georgopoulos, K., Jumaa, H., Smale, S. T., Fisher, A. G., and Merkenschlager, M. (2007) Immunity 26 335-344 [DOI] [PubMed] [Google Scholar]
- 19.Koipally, J., Renold, A., Kim, J., and Georgopoulos, K. (1999) EMBO J. 18 3090-3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koipally, J., and Georgopoulos, K. (2000) J. Biol. Chem. 275 19594-19602 [DOI] [PubMed] [Google Scholar]
- 21.O'Neill, D. W., Schoetz, S. S., Lopez, R. A., Castle, M., Rabinowitz, L., Shor, E., Krawchuk, D., Goll, M. G., Renz, M., Seelig, H. P., Han, S., Seong, R. H., Park, S. D., Agalioti, T., Munshi, N., Thanos, D., Erdjument-Bromage, H., Tempst, P., and Bank, A. (2000) Mol. Cell. Biol. 20 7572-7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, J., Sif, S., Jones, B., Jackson, A., Koipally, J., Heller, E., Winandy, S., Viel, A., Sawyer, A., Ikeda, T., Kingston, R., and Georgopoulos, K. (1999) Immunity 10 345-355 [DOI] [PubMed] [Google Scholar]
- 23.Liberg, D., Smale, S. T., and Merkenschlager, M. (2003) Trends Immunol. 24 567-570 [DOI] [PubMed] [Google Scholar]
- 24.Molnár, A., and Georgopoulos, K. (1994) Mol. Cell. Biol. 14 8292-8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronni, T., Payne, K. J., Ho, S., Bradley, M. N., Dorsam, G., and Dovat, S. (2007) J. Biol. Chem. 282 2538-2547 [DOI] [PubMed] [Google Scholar]
- 26.Gomez-del Arco, P., Koipally, J., and Georgopoulos, K. (2005) Mol. Cell. Biol. 25 2688-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dovat, S., Ronni, T., Russell, D., Ferrini, R., Cobb, B. S., and Smale, S. T. (2002) Genes Dev. 16 2985-2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-del Arco, P., Maki, K., and Georgopoulos, K. (2004) Mol. Cell. Biol. 24 2797-2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurel, Z., Ronni, T., Ho, S., Kuchar, J., Payne, K. J., Turk, C. W., and Dovat, S. (2008) J. Biol. Chem. 283 8291-8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves, T., Katis, P., Madden, Z., Manickam, K., Ramsden, D., Wu, G., and Guidos, C. J. (1995) J. Immunol. 154 5011-5022 [PubMed] [Google Scholar]
- 31.Alessandrini, A., Pierce, J. H., Baltimore, D., and Desiderio, S. V. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 1799-1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuguchi, J., Tsang, W., Morrison, S. L., Beaven, M. A., and Paul, W. E. (1986) J. Immunol. 137 2162-2167 [PubMed] [Google Scholar]
- 33.Hahm, K., Cobb, B. S., McCarty, A. S., Brown, K. E., Klug, C. A., Lee, R., Akashi, K., Weissman, I. L., Fisher, A. G., and Smale, S. T. (1998) Genes Dev. 12 782-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahm, K., Ernst, P., Lo, K., Kim, G. S., Turck, C., and Smale, S. T. (1994) Mol. Cell. Biol. 14 7111-7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trinh, L. A., Ferrini, R., Cobb, B. S., Weinmann, A. S., Hahm, K., Ernst, P., Garraway, I. P., Merkenschlager, M., and Smale, S. T. (2001) Genes Dev. 15 1817-1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, Y., Suresh Kumar, K. G., Tang, W., Spiegelman, V. S., and Fuchs, S. Y. (2004) Mol. Cell Biol. 24 4038-4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia, N., Thiyagarajan, S., Elcheva, I., Saleem, M., Dlugosz, A., Mukhtar, H., and Spiegelman, V. S. (2006) J. Biol. Chem. 281 19320-19326 [DOI] [PubMed] [Google Scholar]
- 38.Wakula, P., Beullens, M., Ceulemans, H., Stalmans, W., and Bollen, M. (2003) J. Biol. Chem. 278 18817-18823 [DOI] [PubMed] [Google Scholar]
- 39.Cohen, P. T. (2002) J. Cell Sci. 115 241-256 [DOI] [PubMed] [Google Scholar]
- 40.Zhao, S., and Lee, E. Y. (1997) J. Biol. Chem. 272 28368-28372 [DOI] [PubMed] [Google Scholar]
- 41.Rogers, S., Wells, R., and Rechsteiner, M. (1986) Science 234 364-368 [DOI] [PubMed] [Google Scholar]
- 42.Rechsteiner, M., and Rogers, S. W. (1996) Trends Biochem. Sci. 21 267-271 [PubMed] [Google Scholar]
- 43.Kim, T. K., and Maniatis, T. (1996) Science 273 1717-1719 [DOI] [PubMed] [Google Scholar]
- 44.Rock, K. L., Gramm, C., Rothstein, L., Clark, K., Stein, R., Dick, L., Hwang, D., and Goldberg, A. L. (1994) Cell 78 761-771 [DOI] [PubMed] [Google Scholar]
- 45.Ceulemans, H., and Bollen, M. (2004) Physiol. Rev. 84 1-39 [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., Liu, B., Li, N., Li, H., Qiu, J., Zhang, Y., and Cao, X. (2008) J. Biol. Chem. 283 12076-12084 [DOI] [PubMed] [Google Scholar]
- 47.Niu, H., Ye, B. H., and Dalla-Favera, R. (1998) Genes Dev. 12 1953-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreassen, P. R., Lacroix, F. B., Villa-Moruzzi, E., and Margolis, R. L. (1998) J. Cell Biol. 141 1207-1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudenko, A., Bennett, D., and Alphey, L. (2003) EMBO Rep. 4 59-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett, D. (2005) Biochem. Soc. Trans. 33 1444-1446 [DOI] [PubMed] [Google Scholar]
- 51.Lesage, B., Beullens, M., Nuytten, M., Van Eynde, A., Keppens, S., Himpens, B., and Bollen, M. (2004) J. Biol. Chem. 279 55978-55984 [DOI] [PubMed] [Google Scholar]
- 52.Canettieri, G., Morantte, I., Guzman, E., Asahara, H., Herzig, S., Anderson, S. D., Yates, J. R., III, and Montminy, M. (2003) Nat. Struct. Biol. 10 175-181 [DOI] [PubMed] [Google Scholar]
- 53.Adler, H. T., Chinery, R., Wu, D. Y., Kussick, S. J., Payne, J. M., Fornace, A. J., Jr., and Tkachuk, D. C. (1999) Mol. Cell. Biol. 19 7050-7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, D. Y., Tkachuck, D. C., Roberson, R. S., and Schubach, W. H. (2002) J. Biol. Chem. 277 27706-27715 [DOI] [PubMed] [Google Scholar]