Abstract
The AP-2 clathrin adaptor differs fundamentally from the related AP-1, AP-3, and AP-4 sorting complexes because membrane deposition does not depend directly on an Arf family GTPase. Instead phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) appears to act as the principal compartmental cue for AP-2 placement at the plasma membrane as well as for the docking of numerous other important clathrin coat components at the nascent bud site. This PtdIns(4,5)P2 dependence makes type I phosphatidylinositol 4-phosphate 5-kinases (PIPKIs) lynchpin enzymes in the assembly of clathrin-coated structures at the cell surface. PIPKIγ is the chief 5-kinase at nerve terminals, and here we show that the 26-amino acid, alternatively spliced C terminus of PIPKIγ661 is an intrinsically unstructured polypeptide that binds directly to the sandwich subdomain of the AP-2 β2 subunit appendage. An aromatic side chain-based, extended interaction motif that also includes the two bulky C-terminal residues of the short PIPKIγ635 variant is necessary for β2 appendage engagement. The clathrin heavy chain accesses the same contact surface on the AP-2 β2 appendage, but because of additional clathrin binding sites located within the unstructured hinge segment of the β2 subunit, clathrin binds the β2 chain with a higher apparent affinity than PIPKIγ661. A clathrin-regulated interaction with AP-2 could allow PIPKIγ661 to be strategically positioned for regional PtdIns(4,5)P2 generation during clathrin-coated vesicle assembly at the synapse.
The key regulatory activity of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2)2 during clathrin-mediated endocytosis is firmly established (1, 2). The heterotetrameric AP-2 adaptor complex and numerous clathrin-associated sorting proteins (CLASPs) display dedicated surfaces or domains that engage PtdIns(4,5)P2 with good selectively (3-5). PtdIns(4,5)P2, which is localized to the cell surface, thus biases the deposition and assembly of these coat components at the plasma membrane by synergizing with other low affinity interactions in a phenomenon termed coincidence detection (2, 4). Later acting endocytic regulatory proteins also bind to PtdIns(4,5)P2. The large GTPase dynamin contains a pleckstrin homology domain, which engages PtdIns(4,5)P2 and is required for vesicle scission (6). Similarly the clathrin uncoating cofactor, auxilin, has a PTEN homology domain that also binds to phosphoinositides and is necessary for targeting of this J-domain protein to clathrin-coated membranes (7). The lipid binding features of all these endocytic components is in full accord with PtdIns(4,5)P2 being necessary for both early and late stages of coated vesicle production (8).
PtdIns(4,5)P2 is a general, apparently ubiquitous marker of the plasma membrane, and the concept of functionally autonomous, stable PtdIns(4,5)P2-enriched microdomains within the cytosolic leaflet of the membrane has been challenged (9-11). This raises the question of whether the prevailing PtdIns(4,5)P2 concentration at the cell surface is simply permissive and sufficient for nucleation and sustained clathrin-coated vesicle assembly and budding or whether, in addition to basal PtdIns(4,5)P2 that might act as an initial compartmental cue, regional synthesis of this lipid is also necessary for clathrin coat assembly and progression. Supporting the first possibility is the general decrease in PtdIns(4,5)P2 levels in the brains of type Iγ phosphatidylinositol 4-phosphate 5-kinase (PIPKIγ) nullizygous mice that parallels major synaptic vesicle recycling aberrations in neurons of these animals, which die before (12, 13) or shortly after (14) birth. Also, activated P2Y purinergic receptors, which trigger phospholipase C-mediated cleavage of PtdIns(4,5)P2, diminish clathrin-mediated uptake of insulin (15), suggesting that signaling and endocytic processes can utilize a common phosphoinositide pool. PtdIns(4,5)P2 is rather uniformly dispersed over the plasma membrane of the budding yeast Saccharomyces cerevisiae (16, 17), and Mss4p, the only phosphatidylinositol 4-phosphate 5-kinase in this organism, is not localized to cortical, clathrin-containing endocytic structures (18). In mammalian cells, the subcellular positioning of PIPKI enzymes is, at least in part, dictated by substrate availability/concentration as switching the activation loop residues of a type II phosphatidylinositol 4-phosphate 5-kinase, which usually acts on phosphatidylinositol 5-phosphate, to that of a PIPKI induces the chimeric kinase to localize to the cell surface (19). In addition, ectopically expressed, tailored proteins that drive rapamycin-induced consumption of bulk PtdIns(4,5)P2 lead to a rapid and dramatic loss of the majority of surface-associated clathrin-coated structures and halt clathrin-dependent internalization (20-22). In fact, excess pleckstrin homology domain can block clathrin-mediated endocytosis in an in vitro reconstitution assay (8).
Yet the second idea of localized PtdIns(4,5)P2 synthesis is in accord with the subcellular localization of PIPKI isozymes depending upon more than just the location of phosphatidylinositol 4-phosphate (10) and with the PIPKI enzymes associating physically with the AP-2 adaptor complex (23-25) and with β-arrestin (26). That the interaction with AP-2 stimulates catalysis (24, 25) lends additional support for a feed-forward model for staged PtdIns(4,5)P2 generation at nascent clathrin assembly zones at the cell surface. The fact that ectopic expression of PIPKI enzymes in cultured cells increases both the number of surface clathrin-coated structures and the rate of internalization (27) also indicates that PtdIns(4,5)P2 on the cell surface can be limiting. Local production of PtdIns(4,5)P2 might counteract general competition of endocytic factors with other cell surface proteins for a limited phosphoinositide pool and thus may be important to sustain the rapid kinetics of clathrin-mediated endocytosis. This may be particularly relevant in vivo during signal transmission when PtdIns(4,5)P2 is consumed to generate diacylglycerol, inositol 1,4,5-trisphosphate, or phosphatidylinositol 3,4,5-trisphosphate (1, 2, 15).
Irrespective of precisely how PIPKI enzymes translocate to the plasma membrane, what is also clear is that temporal remodeling of PtdIns(4,5)P2 apparently accompanies coated vesicle biogenesis (18, 28). After targeted gene disruption of the phosphoinositide polyphosphatase synaptojanin 1, neurons exhibit excessive and prolonged clathrin coat associations with the membrane (29, 30). Somewhat analogously, S. cerevisiae synaptojanin-null mutants display mislocalized PtdIns(4,5)P2; the phospholipid now appears in endosomal structures (17, 18). These results show clearly that under normal conditions PtdIns(4,5)P2 within forming transport vesicles is dephosphorylated prior to, or rapidly following, scission from the cell surface. Recent time-resolved live cell imaging of the two splice isoforms of synaptojanin 1, termed SJ145 and SJ170 (31), reveals that although SJ145 masses at the bud site around the time of the fission event SJ170 populates the coat throughout the assembly process (28). Thus molecular mechanisms appear to exist to align cycles of PtdIns(4,5)P2 formation and hydrolysis with progression of the coated assemblage toward the final fission step. In this study, we confirm that PIPKIγ, a vital lipid kinase (12-14) and the major phosphatidylinositol 4-phosphate kinase at the synapse (32), binds to AP-2 chiefly through the functionally autonomous appendage domain of the large β2 chain. We show that this depends on an interaction surface positioned upon the sandwich subdomain of the β2 appendage, a site also engaged by clathrin and eps15. Binding of these proteins to the β2 appendage is mutually exclusive, leading to a model for spatial and temporal phosphoinositide remodeling managed by AP-2 appendages.
EXPERIMENTAL PROCEDURES
DNA Constructs—The recombinant proteins consisting of glutathione S-transferase (GST) fused to PIPKIγ (mouse residues 460-635, 460-661, 636-661, 630-661, and 624-661) were generated by PCR using a mouse PIPKIγ661 cDNA as a template followed by digestion and ligation into EcoRI/XhoI-cleaved pGEX-4T-1. Point mutations (I633A, Y634A, F635A, W642A, Y644A, Y649A, and S645E) were generated by QuikChange site-directed mutagenesis (Stratagene). The GST-PIPKIγ-(460-687) construct was generated by PCR using rat PIPKIγ687 cDNA kindly provided by Robin Irvine (University of Cambridge, Cambridge, UK) as a template followed by digestion and ligation into EcoRI/XhoI-cleaved pGEX-4T-1. A GST-human PIPKIγ668 C-terminal 28-amino acid fusion was generated by PCR utilizing overlapping synthetic oligonucleotides and cloned into pGEX-4T-1. The GST-conjugated AP-2 αC appendage (mouse residues 701-938) (33), AP-2 β2 appendage (rat residues 714-951) (34), eps15 (residues 622-736), and a cytosolic portion of the cation-independent mannose 6-phosphate receptor (CI-MPR YSKV, residues 2337-2372) (35) have been described previously. The FLAG-tagged AP-2 β2 subunit and μ2 subunit pFastBac Dual plasmid for baculovirus production was generated as described previously (35). The β2 trunk version was made from the full-length protein by deleting residues 593-951, placing the FLAG epitope after His592. PIPKIγ-(630-661) was cloned into EcoRI/BamHI-digested pGBKT7, whereas AP-2 β2 appendage (rat residues 700-937) was cloned into EcoRI/XhoI-cut pGADT7. Upon confirmation of the sequences, indicated mutations in the β2 appendage were created by QuikChange site-directed mutagenesis (Stratagene). The hexahistidine (His6)-tagged AP-2 β2 hinge and appendage (rat AP-2 β2 residues 592-951) was kindly provided by Tom Kirchhausen (Harvard University, Boston, MA). GST-conjugated epsin ENTH domain (rat epsin 1 residues 1-163) was described previously (100). All clones and mutations were verified by automated dideoxynucleotide sequencing.
Antibodies and Immunoblotting—Affinity-purified polyclonal antibody directed against epsin 1 (36) and HIP1 (37) were produced in our laboratory by standard procedures. Affinity-purified WI54 peptide antibodies directed against the 26-amino acid insert of PIPKIγ661 have been described previously (38). The anti-clathrin heavy chain monoclonal antibody (mAb) TD.1 was generously provided by Frances Brodsky (University of California, San Francisco, CA), and the light chain mAb Cl37.3 was provided by Reinhard Jahn. The anti-AP-1/2 β1/β2 subunit mAb 100/1 and rabbit polyclonal anti-eps15 serum were kind gifts from Ernst Ungewickell (Medizinische Hochschule, Hannover, Germany). The rabbit R11-29 polyclonal anti-AP-2 μ2 subunit serum was a generous gift from Juan Bonifacino (National Institutes of Health, Bethesda, MD), and the rabbit polyclonal anti-NECAP antibody was a kind gift from Peter McPherson (McGill University, Montreal, Quebec, Canada). The anti-PIPKIγ, anti-AP180, and anti-amphiphysin I/II mAbs were purchased from BD Transduction Laboratories. The anti-FLAG M1 and M2 and anti-talin mAbs were purchased from Sigma.
Samples were resolved by SDS-PAGE with an altered acrylamide-bisacrylamide (30:0.4) ratio stock solution. After electrophoresis, proteins were either stained with Coomassie Blue or transferred to nitrocellulose in ice-cold 15.6 mm Tris, 120 mm glycine. Blots were usually blocked overnight in 5% skim milk in 10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 0.1% Tween 20, and then portions were incubated with primary antibodies as indicated in individual figure legends. After incubation with horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G, immunoreactive bands were visualized with enhanced chemiluminescence.
Protein and Tissue Extract Preparation—GST and various GST fusion proteins were produced in Escherichia coli BL21 cells. Bacteria were induced by shifting log phase cultures (A600 ∼0.6) from 37 °C to room temperature and then adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 100 μm with constant shaking for 3 h or, in some instances, to 200 μm with constant shaking overnight (∼16 h). The bacteria were recovered by centrifugation at 15,000 × gmax at 4 °C for 15 min and used immediately or stored at -80 °C. Lysis was performed in 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 0.2% (w/v) Triton X-100, 10 mm β-mercaptoethanol with sonication on ice. Insoluble material was removed by centrifugation at 23,700 × gmax at 4 °C for 30 min, and then the GST fusions were collected on glutathione-Sepharose. After extensive washing in phosphate-buffered saline, GST fusions were eluted with 25 mm Tris-HCl, pH 8.0, 200 mm NaCl, 10 mm glutathione, 5 mm DTT on ice and dialyzed into phosphate-buffered saline, 1 mm DTT before use in binding assays. In some instances, purified fusion proteins were cleaved from the GST with thrombin (GE Healthcare) while still immobilized upon glutathione-Sepharose. Digestion was as recommended by the manufacturer followed by addition of the irreversible thrombin inhibitor d-Phe-Pro-Arg chloromethyl ketone (Calbiochem) to a final concentration of 25 μm. His6-β2 hinge + appendage was produced in E. coli BL21 (DE3) by induction of log phase cultures with 1 mm isopropyl 1-thio-β-d-galactopyranoside at room temperature for 3 h. Cleared lysates were incubated with Ni-NTA-agarose, and bound protein was eluted in 50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 200 mm imidazole.
Cytosol was prepared from frozen rat brains (PelFreez) by sequential differential centrifugation after homogenization in 25 mm HEPES-KOH, pH 7.2, 250 mm sucrose, 2 mm EDTA, 2 mm EGTA supplemented with 1 mm phenylmethylsulfonyl fluoride and Complete (Roche Applied Science) protease inhibitor mixture. The 105,000 × gmax supernatant is defined as cytosol and was stored in small aliquots at -80 °C. Before use in binding assays, thawed samples of rat brain cytosol were adjusted to 25 mm HEPES-KOH, pH 7.2, 125 mm potassium acetate, 5 mm magnesium acetate, 2 mm EDTA, 2 mm EGTA, 1 mm DTT (assay buffer) by addition of a 10× stock and then centrifuged at 245,000 × gmax (TLA-100.4 rotor) at 4 °C for 20 min to remove insoluble particulate material. Lysates from infected Sf9 cells (35) were prepared by solubilization of pelleted cells in assay buffer with 0.4% Triton X-100 and supplemented with 1 mm phenylmethylsulfonyl fluoride and Complete (Roche Applied Science) protease inhibitor mixture. After incubation on ice for 15 min, the lysates were sonicated on ice three times for 15 s and centrifuged at 500 × gmax. The supernatants were carefully aspirated and stored in aliquots at -80 °C. Prior to use, thawed Sf9 cell lysates were centrifuged at 125,000 × gmax to remove aggregated material. Hemicomplexes were immunoprecipitated from 1 ml of clarified lysate using 35 μl of packed anti-FLAG M2-agarose (Sigma) equilibrated in assay buffer lacking DTT. After extensively washing the anti-FLAG-agarose, bound proteins were eluted with 200 μg/ml FLAG peptide.
Rat brain clathrin-coated vesicles were prepared exactly as described previously (37). Synaptosomes were prepared from rat brain homogenized in 5 mm HEPES-KOH, pH 7.4, 320 mm sucrose with a Potter-Elvehjem tissue disruptor on ice (39). The supernatant from a 2-min 3,000 × gmax spin at 4 °C was recentrifuged at 13,000 × gmax for 12 min, and the pellet was resuspended in homogenization buffer and again centrifuged at 13,000 × gmax for 12 min. The resuspended pellet was layered over a discontinuous gradient of 13%/9%/6% Ficoll 400 in homogenization buffer and centrifuged at 86,600 × gmax for 35 min at 4 °C. Cream/white-colored synaptosomes were harvested from the 6%/9% and 9%/13% interfaces and concentrated by centrifugation at 27,000 × gmax at 4 °C for 12.5 min after dilution with homogenization buffer. 10-fold dilution of synaptosomes into ice-cold 5 mm Tris-HCl, pH 8.0 followed by disruption in a Potter-Elvehjem homogenizer was used to generate synaptic plasma membranes. After adjusting the lysed membranes to 1.105 m sucrose, the sample was overlaid with 0.92 m and then 0.32 m sucrose and centrifuged at 60,000 × gave at 4 °C for 60 min. The turbid, white-colored synaptosome plasma membrane-enriched fraction at the 0.92 m/1.105 m interface was collected and concentrated by centrifugation after dilution in 5 mm Tris-HCl, pH 8.0.
Kinase Assays and TLC—Phosphoinositide synthesis on synaptic membranes was assayed in assay buffer in a final volume of 50 μl precisely as described previously (40). Briefly the final concentrations were 0.5 mg/ml for membranes, 5 mg/ml for gel-filtered cytosol, and 500 μm for [γ-32P]ATP (0.5-1 Ci/mmol). Reactions were terminated after 10 min at 37 °C by addition of chloroform:methanol:concentrated HCl (200:100: 0.75) followed by vigorous mixing. After carrier phosphoinositides (50 μg/tube) were added, a biphasic mixture was generated by addition of 0.6 m HCl. Samples were centrifuged at 200 × gave for 5 min, and the lower organic phase was removed and transferred to a new tube, washed twice with chloroform, methanol, 0.6 m HCl (3:48:47), and then dried under a stream of N2 gas at about 40 °C. Dried lipid films were resuspended in chloroform:methanol:H2O (75:25:1) and spotted onto oxalate-impregnated, heat-activated silica gel high performance TLC plates and resolved in chloroform:acetone:methanol:acetic acid:water (160:60:52:48:28). Autoradiographs were quantitated using ImageJ (41).
Limited Proteolysis—Purified, thrombin-cleaved PIPKIγ-(460-635) and PIPKIγ-(460-661) (20 μg) were incubated with 5-fold serial dilutions of trypsin (Worthington) in 50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm CaCl2, 1 mm DTT at 37 °C for 1 h. Proteolysis was stopped by addition of 95 °C 2× sample buffer, and 10% of each reaction was analyzed by SDS-PAGE.
Binding Assays—Typically 100-250 μg of GST or GST fusion proteins was first immobilized upon ∼25 μl of packed glutathione-Sepharose by incubation at 4 °C for 1 h with continuous mixing. The Sepharose beads containing the required immobilized proteins were then washed and resuspended to 50 μl in assay buffer. A 250-μl volume of clarified rat brain cytosol was usually added, and the tubes were incubated at 4 °C for 60 min with continuous mixing. For the peptide competition assay, thrombin-cleaved protein was added directly to the assay mixture in the presence of 25 μm d-Phe-Pro-Arg chloromethyl ketone. The glutathione-Sepharose beads were recovered by centrifugation at 10,000 × gmax for 1 min, and then an aliquot of each supernatant was removed and adjusted to 100 μl with SDS sample buffer. After washing the Sepharose pellets four times each with ∼1.5 ml of ice-cold phosphate-buffered saline by centrifugation, the supernatants were aspirated, and each pellet was resuspended in SDS sample buffer up to ∼80 μl. Unless otherwise noted, 10-μl aliquots, corresponding to ∼1.5% of each supernatant and 12.5% of each pellet, were analyzed.
For direct interaction assays between His6-β2 hinge + appendage and GST fusion proteins, typically 5 μg of His6-tagged protein was bound to 5 μl of packed Ni-NTA-agarose in 20 mm HEPES-HCl, pH 7.5, 20 mm imidazole, 120 mm potassium acetate, 0.1% (w/v) Triton X-100, 0.1 mg/ml bovine serum albumin (binding buffer) for 1 h at 4 °C with continuous mixing. After washing, immobilized proteins were incubated with 25 μg of GST or GST fusion proteins in a 300-μl volume for 1 h at 4 °C with continuous mixing. Proteins were recovered by centrifugation at 10,000 × gmax for 1 min. An aliquot of each supernatant was removed and adjusted to 100 μl with sample buffer. Pellets were washed in ∼1.5 ml of ice-cold binding buffer without bovine serum albumin by centrifugation, the supernatants were aspirated, and each pellet was resuspended in SDS sample buffer up to ∼40 μl. Unless otherwise noted 10-μl aliquots, corresponding to ∼2.5% of each supernatant and 25% of each pellet, were analyzed.
Circular Dichroism—Appropriate GST fusion proteins immobilized on glutathione-Sepharose were incubated with thrombin, and the soluble fraction was dialyzed into 25 mm potassium phosphate, pH 7.4, 1 mm DTT. CD spectra were measured on an AVIV Model 202 spectrometer. Five (PIPKIγ) or three (ENTH domain) reproducible spectra were obtained from samples of concentrations between 0.09 and 0.11 mg/ml at 25 °C in the near-UV wavelength region (190-280 nm). Spectra were averaged, smoothed (42), and base line-corrected by subtraction of similarly collected, averaged, and smoothed data for buffer alone.
Yeast Two-hybrid Assay—The yeast two-hybrid assay was done utilizing the Matchmaker GAL4 system (Clontech) according to the manufacturer's protocols. Appropriately transformed S. cerevisiae strain AH109 was selected first on synthetic defined minimal medium plates lacking Leu and Trp. Approximately 11-18 individual clones were selected and then streaked onto synthetic defined medium lacking Leu and Trp; on plates without His, Leu, and Trp; or onto plates lacking Ade, His, Leu, and Trp. Clones representative of the growth pattern for the interaction being tested were then resuspended, normalized by optical density, and spotted identically onto dropout plates, and yeast were grown at 30 °C for 4 days.
RESULTS
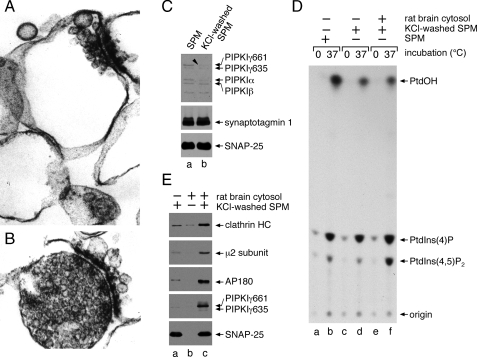
PIPKIγ661 Membrane Translocation and AP-2 Recruitment—Synaptic plasma membranes (Fig. 1A), purified from rat brain synaptosome preparations (Fig. 1B) by hypotonic lysis, contain trace levels of all three PIPKI isoforms, α, β, and γ (Fig. 1C). After extracting the membranes with 1 m KCl, predominantly the ∼90-kDa PIPKIγ isoforms and PIPKIα are removed, but the treatment has no effect on the membrane-associated proteins synaptotagmin 1 and SNAP-25 (Fig. 1C). Cell-free kinase assays show that the salt washing procedure diminishes the temperature-dependent synthesis of PtdIns(4,5)P2 to ∼67% (n = 3; Fig. 1D, compare lanes b and d) presumably because of the removal of PIPKI. By contrast, phosphatidylinositol 4-phosphate synthesis changes <10% on the extracted membranes. Using the salt-washed synaptic plasma membranes as a template for in vitro clathrin coat assembly assays, we find that the translocation of cytosolic clathrin, the AP-2 adaptor complex, and AP180 correlates with the accumulation of PIPKIγ661 on the synaptic membranes (Fig. 1E, lane c) and an increase in PtdIns(4,5)P2 synthesis (Fig. 1D, compare lanes d and f). The recruitment of PIPKIγ661 under these conditions suggests there may be linkage between PIPKIγ and AP-2/clathrin membrane translocation. In fact, there is evidence for PIPKI enzymes associating directly with AP-2 in three molecularly distinct manners. The long splice isoform of PIPKIγ contains a C-terminal 644YSPL sequence that can interact with the cargo-selective μ2 subunit of AP-2, akin to YXXØ-type (where Ø is a large hydrophobic amino acid) receptor sorting signals (23). The same general region of the 26-amino acid C-terminal insert of PIPKIγ661 can bind to the independently folded β2 appendage of AP-2 (25). In addition, the central kinase domain of all three PIPKI isoforms is proposed to associate with the μ2 subunit of AP-2 but in a manner that does not overlap with cargo binding; rather concomitant PIPKI and YXXØ sequence engagement by μ2 appears to stimulate the catalytic activity of the lipid kinase (24).
FIGURE 1.
PIPKIγ661 and AP-2 are coordinately recruited to the synaptic plasma membrane. A and B, aliquots of synaptic plasma membrane (A) or synaptosomes from which they were derived (B) were fixed with 2% glutaraldehyde and processed for electron microscopy. Thin section micrographs typical of the many fields examined are shown. C, samples of 50 μg of synaptic plasma membrane (SPM), suspended in assay buffer alone or supplemented with 1 m KCl, were sedimented after incubation on ice for 30 min. Aliquots of each resuspended membrane pellet were prepared for SDS-PAGE and immunoblotting. Portions of the blots were probed with an affinity-purified polyclonal anti-PIPKI antibody or mAbs directed against synaptotagmin 1 or SNAP-25. D, reactions containing 0.5 mg/ml untreated or salt-washed synaptic plasma membranes, 5 mg/ml cytosol, and 500 μm [γ-32P]ATP were prepared as indicated. After incubation at 37 °C for 10 min the lipids were extracted and analyzed by TLC and autoradiography. A representative experiment of three is shown, and the migration positions of authentic phospholipid standards are indicated. E, reactions containing 50 μg/ml salt-washed synaptic plasma membranes, 5 mg/ml rat brain cytosol, and an ATP-regenerating system were prepared as indicated. After incubation at 37 °C for 15 min, membranes were sedimented and prepared for SDS-PAGE and immunoblotting. Portions of the blots were probed with anti-clathrin subunit heavy chain (HC) mAb TD.1, rabbit R11-29 anti-μ2 serum, or an anti-AP180, anti-PIPKIγ, or anti-SNAP-25 mAb, and only relevant portions of the blots are shown. PtdIns(4)P, phosphatidylinositol 4-phosphate.
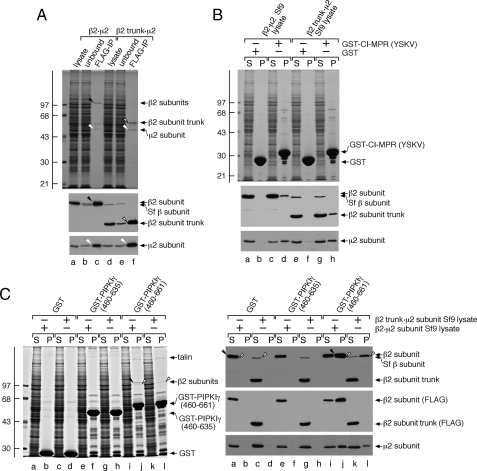
The PIPKIγ661 Binding Surface upon AP-2—To attempt to resolve whether the β2 or μ2 subunit of AP-2 represents a dominant interaction partner for the C-terminal 26-amino acid extension of PIPKIγ661, we used baculovirus-encoded AP-2 hemicomplexes (35). First we verified that the individual chains expressed in Sf9 cells associate into a macromolecular complex. Anti-FLAG immunoprecipitation shows that removal of the FLAG-tagged β2 subunit also depletes the μ2 subunit from the lysate (Fig. 2A, compare lanes a and b). The relative stoichiometry of the assembled β2·μ2 hemicomplex is seen on the Coomassie Blue-stained gel (lane c). Likewise a FLAG-tagged β2-subunit trunk, lacking the C-terminal 359 residues encoding the unstructured hinge as well as the appendage domain, immunoprecipitates along with the co-expressed μ2(lane f). This is expected as μ2 binds to the α-helical solenoid portion of the β2 subunit trunk (43). These experiments confirm the assembly of the β2·μ2 and β2 trunk·μ2 hemicomplexes. Notably the putative insect β subunit, recognized by the anti-β mAb, was not depleted from the lysates with the anti-FLAG mAb (lanes b and e).
FIGURE 2.
The C terminus of PIPKIγ661 but not PIPKIγ635 binds selectively to the AP-2 β2 appendage. A, equivalent volumes of Sf9 cell lysate overexpressing either the β2·μ2 or the β2 trunk·μ2 hemicomplex before (lanes a and d) or after (lanes b and e) immunoprecipitation with agarose-coupled anti-FLAG mAb M2 and aliquots from the washed and FLAG peptide-eluted immunoprecipitate (IP) pellet (lanes c and f) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-β1/β1-subunit mAb 100/1 or anti-μ2 serum, and only the relevant portions are shown. The migration position of the molecular mass standards is indicated on the left, and the location of the immunoprecipitated β2 subunit (black arrowhead), β2 trunk (open arrowhead), and μ2 subunit (white arrowheads) are shown. B, ∼250 μg of GST (lanes a, b, e, and f) or GST-CI-MPR (YSKV; lanes c, d, g, and h) immobilized on glutathione-Sepharose was incubated with Sf9 cell lysates overexpressing either the β2·μ2(lanes a-d) or β2 trunk·μ2(lanes e-h) hemicomplexes as indicated. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼12.5% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-β1/β1-subunit mAb 100/1 or anti-μ2 serum, and only the relevant portions are shown. C, ∼200 μg of GST (lanes a-d), GST-PIPKIγ-(460-635) (lanes e-h), or GST-PIPKIγ-(460-661) immobilized on glutathione beads was incubated with Sf9 cell lysates overexpressing either the β2·μ2(lanes a, b, e, f, i, and j) or β2 trunk·μ2(lanes c, d, g, h, k, and l) hemicomplexes. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼15% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-β1/β1-subunit mAb 100/1, anti-FLAG mAb M1, or anti-μ2 serum, and only the relevant portions are shown. The migration positions of the FLAG-tagged β2(arrowheads) and the presumptive Sf9 cell β subunit (open arrowheads) are indicated.
The capability of the two hemicomplexes to bind comparably to YXXØ-type sorting signals, which are recognized by the μ2 subunit (44, 45), can be seen with a portion of the CI-MPR cytoplasmic domain (residues 2337-2372) fused to GST (Fig. 2B) (35). Although there is no association of either the FLAG-β2 subunit or μ2 with GST (lanes b and f), some of both the β2 and μ2 proteins are recovered in the pellet fraction from GST-CI-MPR affinity isolations (lanes d and h). Still none of the presumptive Sf9 AP-2 (β subunit) binds to the GST-CI-MPR (lanes f and h), and the relative stoichiometry of the bound μ2 compared with the soluble fraction is greater than that of either the β2 full length or β2 trunk (lanes d and h). We interpret this to indicate that μ2 alone (uncomplexed) has the highest apparent affinity for YXXØ signals, although the strength of this interaction is still rather weak. Some of the binary β2·μ2 or β2 trunk·μ2 complexes also clearly associate with GST-CI-MPR, but the failure of heterotetrameric AP-2 to bind appreciably is fully consistent with the cytosolic pool of AP-2 assuming a basal, closed conformation that blocks the YXXØ binding site on μ2 (43).
Similar experiments using the C-terminal region of the PIPKIγ635 or PIPKIγ661 kinase variants (46) show that only the long splice isoform associates with the β2·μ2 hemicomplex (Fig. 2C, lane j). In fact, the presence of the full-length β2 subunit bound to GST-PIPKIγ-(460-661) can be seen by Coomassie Blue staining. By contrast, if incubated with either GST or GST-PIPKIγ-(460-635), there is complete recovery of the full length or the β2 trunk in the supernatant fraction (lanes a, c, e, and g). This shows that necessary AP-2 binding information is indeed located within the terminal 26 residues of PIPKIγ661. Nevertheless the β2 trunk·μ2 hemicomplex does not associate with the GST-PIPKIγ-(460-661) like the intact β2·μ2 hemicomplex (compare lanes l and j). There is a full-length β subunit recovered in the pulldown pellet after incubation with the β2 trunk lysate, but this chain does not contain a FLAG epitope (lane l), and the bound μ2 migrates slightly slower than the expressed mammalian μ2 subunit (lane l). We thus conclude that although the β2 trunk·μ2 fails to bind to the immobilized GST-PIPKIγ-(460-661) the endogenous Sf9 AP-2 heterotetramer specifically engages the 26-amino acid extension of PIPKIγ661 because no binding is seen with the PIPKIγ-(460-635) (lane h). This is in sharp contrast to the lack of association of the endogenous invertebrate AP-2 complex (Sf9 AP-2) with a GST-presented YXXØ sorting signal (Fig. 2B).
In other experiments to assure that the FLAG epitope-negative, ∼100-kDa β-immunoreactive band and the slower mobility μ2 subunit are indeed constituents of the Sf9 cell AP-2, we used affinity isolation with a C-terminal segment of ARH. ARH contains a single (D/E)nX1-2FXX(F/L)XXXR AP-2-binding motif that binds exclusively to the β2 subunit appendage (47-50). The GST-ARHM2 fusion contains a 20-amino acid tract that fully contains this interaction motif (34); the presumptive Sf9 cell β subunit is quantitatively removed from the supernatant by this motif and is recovered in the pellet fraction (data not shown). Furthermore, when bound to the immobilized ARH β-subunit interaction motif, the slower mobility μ2 form is detected (data not shown). These studies reveal that the β and μ2 bands observed indeed represent the endogenous invertebrate AP-2 present in the Sf9 cell lysates.
The 26-amino acid extension of PIPKIγ661 also binds physically to the FERM domain of the integrin-associated protein talin, allowing regulated PtdIns(4,5)P2 formation at focal adhesions (38, 51). Accordingly the ∼230-kDa insect talin in lysates of either β2·μ2 (Fig. 2C, lane j) or β2 trunk·μ2(lane l) binds to the GST-PIPKIγ-(460-661) but not GST-PIPKIγ-(460-635) (lanes f and h) as expected. Overall we conclude from this series of experiments that the β2 appendage of the AP-2 complex, and not the μ2 subunit, is the major site for PIPKIγ661 interaction under these conditions. This interpretation is in full accord with a recent independent study (25).
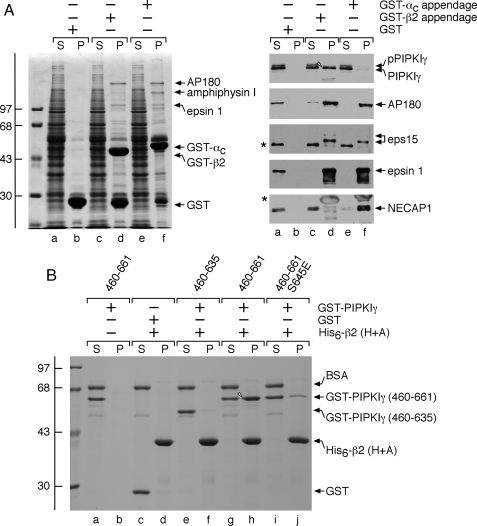
PIPKIγ661 Appendage Selectivity—The AP-2 complex has two independently folded appendages that project away from the central heterotetrameric core (see Fig. 10). Despite only ∼11% sequence identity, the α and β2 subunit appendages have an analogous overall fold (52) and share many binding partners (49, 50); therefore, the appendages could either be indiscriminately or selectively accessed by PIPKIγ661. In fact, PIPKIγ661 shows a striking selectivity for the β2 over the α appendage. Pulldown assays show that the GST-α and GST-β2 appendages bind to similar and different subsets of CLASPs and accessory factors (Fig. 3A, lanes d and f). Although both bind to AP180, epsin 1, and eps15 roughly equally, only GST-β2 binds to PIPKIγ661 (Fig. 3A, lane d, right). This is reminiscent of the strict preference of ARH and β-arrestin for the β2 appendage (47-50, 53). Significantly the α appendage also has distinctive interaction partners; the endocytic protein NECAP 1 binds only to GST-α, for example (Fig. 3A, lane f) (54). Cdk5 is known to phosphorylate PIPKIγ661 at Ser645, and dephosphorylation of this residue by calcineurin following calcium-induced synaptic vesicle exocytosis allows an interaction with AP-2 to promote compensatory endocytosis (25). Of the two bands detected by the anti-PIPKIγ mAb, only the lower band, which corresponds to the unphosphorylated protein, binds to β2, consistent with previous findings (25).
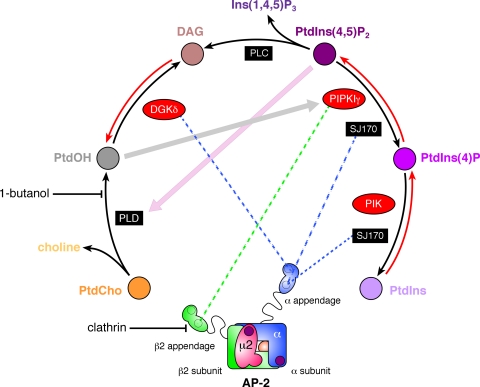
FIGURE 10.
Phosphoinositide metabolism at clathrin-coated buds. Schematic representation of key enzymes responsible for PtdIns(4,5)P2 synthesis and the mapped interactions with the AP-2 adaptor appendages. The location of the two separate PtdIns(4,5)P2 binding sites on the AP-2 heterotetramer is indicated with purple spots. DAG, diacylglycerol; PLD, phospholipase D; PLC, phospholipase C; PIK, phosphatidylinositol 4-kinase; PtdCho, phosphatidylcholine; Ins(1,4,5)P3, inositol 1,4,5-trisphosphate; PtdIns, phosphatidylinositol; PtdIns(4)P, phosphatidylinositol 4-phosphate.
FIGURE 3.
PIPKIγ661 discriminates between the AP-2α andβ2 appendages. A, ∼100 μg of GST (lanes a and b), GST-β2 appendage (lanes c and d), or GST-αC appendage (lanes e and f) immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-PIPKIγ mAb clone 12, anti-AP180 mAb clone 34, or affinity-purified anti-eps15, -epsin 1, or -NECAP 1 antibodies, and only the relevant portions are shown. The different migration of the non-phosphorylated (arrowhead) and phosphorylated (pPIPKIγ) forms of PIPKIγ is indicated. The asterisk demarcates a non-specific band detected by the anti-eps15 antibodies. B, ∼5 μg of His6-tagged β2 hinge + appendage (H+A; lanes c-j) immobilized on Ni-NTA-agarose was incubated with ∼25 μg of GST (lanes c and d), GST-PIPKIγ-(460-635) (lanes e and f), GST-PIPKIγ-(460-661) (lanes g and h), or GST-PIPKIγ-(460-661) with a phosphomimetic S645E mutation (lanes i and j) as indicated in the presence of carrier bovine serum albumin (BSA). GST-PIPKIγ-(460-661) was also incubated with Ni-NTA-agarose alone (lanes a and b). After centrifugation, aliquots of ∼2.5% of each supernatant (S) and ∼25% of each washed pellet (P) were resolved by SDS-PAGE and stained with Coomassie Blue.
PIPKIγ661 Directly Engages the β2 Appendage—In binary interaction assays, when immobilized His6-tagged AP-2 β2 hinge + appendage is incubated with soluble GST-PIPKIγ-(460-661), the two proteins interact, and PIPKIγ is recovered in the pellet with the β2 hinge + appendage (Fig. 3B, lane h). Conversely the C-terminal segment of the PIPKIγ short splice isoform (GST-PIPKIγ-(460-635)) does not bind the β2 hinge + appendage and remains in the supernatant (Fig. 3B, lane e) as does GST (lane c). This corroborates a direct interaction between PIPKIγ661 and the β2 appendage mediated by the 26-amino acid extension of the long splice isoform. Furthermore a phosphomimetic S645E mutant in a GST-PIPKIγ-(460-661) background has a sharply reduced ability to bind β2 (Fig. 3B, lane j) in the direct interaction assay.
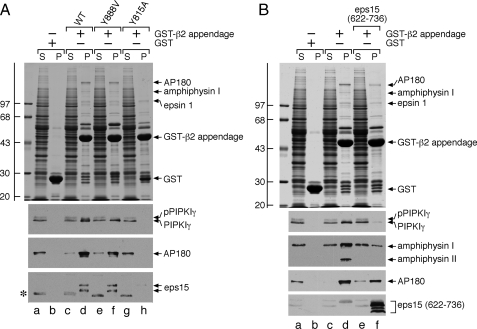
PIPKIγ661 Binds the β2 Appendage Sandwich Domain—Like the AP-2 α appendage, the β2 appendage plays an integral role in recruiting CLASPs and accessory factors to sites of endocytosis by acting as an organizational hub (49, 50, 52, 55). The β2 appendage contains two rigidly apposed functional surfaces: an N-terminal, β-sheet-containing sandwich subdomain and a C-terminal, α-helix- and β-sheet-containing platform subdomain (52). Mutation and co-crystallization studies reveal that the two sites bind to discrete sets of endocytic proteins (49, 50). To determine whether PIPKIγ661 interacts with either of these sites, we mutated select residues critical for binding at each surface. A Y888V mutation in the β2 platform subdomain has no effect on the binding of PIPKIγ or the established sandwich-binding partners AP180 and eps15 (Fig. 4A, lanes d and f). This alteration does reduce epsin 1 binding (49, 50). By contrast, a Y815A sandwich subdomain mutation completely eliminates the binding of PIPKIγ661, AP180, and eps15, whereas epsin 1 binding is essentially unaffected (Fig. 4A, lane h). These results indicate that PIPKIγ661 associates directly with the sandwich subdomain of the β2 appendage.
FIGURE 4.
PIPKIγ661 physically contacts the sandwich subdomain of the AP-2 β2 appendage. A, ∼100 μg of GST (lanes a and b), GST-β2 appendage (lanes c and d), or GST-β2 Y888V (lanes e and f) or Y815A (lanes g and h) mutant appendage immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-PIPKIγ mAb clone 12, anti-AP180 mAb clone 34, or affinity-purified anti-eps15 antibodies, and only the relevant portions are shown. The slowed migration of the phosphorylated from of PIPKIγ (pPIPKIγ) is indicated as is the location of the Coomassie Blue-stained epsin 1 band. The asterisk demarcates a non-specific band detected by the anti-eps15 antibodies. B, ∼100 μg of GST (lanes a and b) or GST-β2 appendage (lanes c-f) immobilized on glutathione-Sepharose was incubated with rat brain cytosol in the absence or presence of 2 μm eps15 (residues 622-736) competitor polypeptide (lanes e and f) as indicated. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-PIPKIγ mAb clone 12, anti-amphiphysin mAb clone 15, anti-AP180 mAb clone 34, or anti-eps15 antibodies, and only the relevant portions are shown. The slowed migration of the phosphorylated from of PIPKIγ (pPIPKIγ) is indicated, as is the location of the Coomassie Blue-stained epsin 1 band. WT, wild type.
Competition experiments in which the immobilized GST-β2 is incubated with cytosol and an eps15 polypeptide encompassing the region co-crystallized with β2 appendage at the sandwich site (50) confirm the contact site. Alone the GST-β2 recovers the typical cohort of soluble endocytic proteins (Fig. 4B, lane d). Addition of the eps15 polypeptide to the mixture substantially reduces PIPKIγ661 and amphiphysin I and II binding (Fig. 4B, lane f). AP180 binding is clearly reduced although not to the same extent as binding of the kinase (lane f).
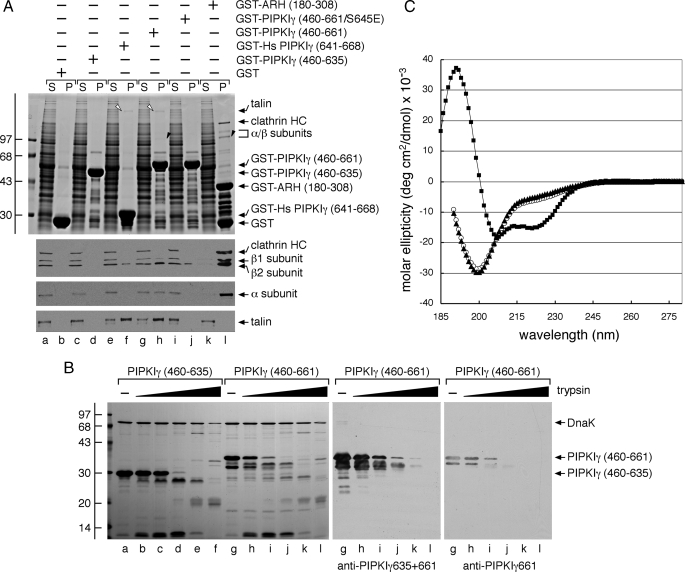
Delineation of the PIPKIγ661 AP-2 Interaction Motif—Because the PIPKIγ-(460-661) but not the PIPKIγ-(460-635) binds to the AP-2 β2 appendage, we evaluated whether just the terminal 28 amino acids of the human PIPKIγ668 variant (also termed PIP5K Iγ90 (51)) are sufficient for this interaction. AP-2 binds to only the 28-residue extension considerably more weakly than in the context of the 460-661 fragment in a GST pulldown assay (Fig. 5A, compare lanes f and h). Yet in the same assay, the extent of the association with talin is similar for both GST fusions (Fig. 5A). This could indicate that although talin binds only the 26/28-amino acid extension in PIPKIγ661/668, AP-2 may bind primarily to this same region but depend on a distinct secondary structural element or be stabilized by an adjacent region(s) within residues 460-635.
FIGURE 5.
The alternatively spliced C terminus of PIPKIγ alone does not contain all the AP-2 binding information. A, ∼200 μg of GST (lanes a and b), GST-PIPKIγ-(460-635) (lanes c and d), GST-Hs PIPKIγ-(641-668) (lanes e and f), GST-PIPKIγ-(460-661) (lanes g and h), GST-PIPKIγ-(460-661) containing S645E (lanes i and j), or GST-ARH-(180-308) (lanes k and l) immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1 and anti-β1/β2 subunit mAb 100/1, anti-AP-2 α subunit mAb C4, or anti-talin mAb 8d4, and only the relevant portions are shown. The position of talin (open arrowheads) and the AP-2 β2 and α subunits (black arrowheads) on the stained gel is shown. Note that PIPKIγ661 clearly binds to the β2 appendage of AP-2 more weakly than ARH, which has a KD for AP-2 of ∼2 μm (49, 50). B, aliquots (20 μg) of PIPKIγ-(460-635) (lanes a-f) or PIPKIγ-(460-661) (lanes g-l) were incubated alone or with 1:5 serial 5-fold dilutions of trypsin (0.6-375 ng) in 50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm CaCl2, 1 mm DTT at 37 °C for 1 h. Proteolysis was stopped by the addition of 95 °C SDS sample buffer, and aliquots of 10% of each reaction were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. The PIPKIγ-(460-661) portion of the blot was probed with anti-PIPKIγ mAb clone 12 that recognizes both isoforms or with an affinity-purified antibody that recognizes only the PIPKIγ661 isoform. C, circular dichroism spectra of PIPKIγ-(460-635) (open circles), PIPKIγ-(460-661) (triangles), and epsin 1 ENTH domain (squares) polypeptides were measured in 25 mm potassium phosphate + 1 mm DTT. Measurements were made from 280 to 185 nm at 1-nm increments, and the spectra were base line-corrected and represent the average of five (PIPKIγ) or three (ENTH) runs. Note the signature α-helical features of the globular ENTH domain compared with the unstructured PIPKIγ polypeptides. deg, degrees.
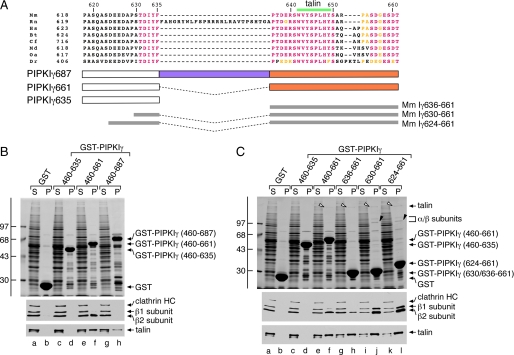
Using controlled tryptic proteolysis of bacterially expressed 460-635 and 460-661 polypeptides derived from PIPKIγ, a hierarchical sequence of degradation fragments is observed (Fig. 5B). Immunoblots with either a mAb that detects both PIPKIγ635 and -661 or affinity-purified antibodies that detect the 26-amino acid extension of PIPKIγ661 reveal that both epitopes are efficiently removed by low concentrations of trypsin. This is in contrast to the stably folded bacterial chaperone DnaK, co-purified with the PIPKIγ peptides, that is unaffected by lower trypsin concentrations (Fig. 5B). Neither antibody reacts with the more stable PIPKIγ fragments. Additionally, circular dichroism analysis of the 460-635 and 460-661 protein segments indicate that both are essentially unstructured (Fig. 5C). Indeed the 460-661 polypeptide is resistant to denaturation at 100 °C as are the C-terminal portions of the natively unstructured endocytic proteins epsin 1 and AP180 (56). We believe the intrinsic disorder of the C-terminal region of PIPKIγ makes it unlikely that the difference in AP-2 binding between the GST-PIPKIγ-(460-661) and GST-Hs PIPK1γ-(641-668) (equivalent to the mouse residues 636-661) is due to secondary structural elements. Instead residues at the junction of the 635 and 661 splice isoforms may contribute to AP-2 β2 appendage binding. To examine this further, we used a third rodent PIPKIγ splice variant, PIPKIγ687 (also termed PIPkin IγC (57) or PIP-kin Iγ93(58)), which contains a distinct 26-amino acid insert positioned between the C terminus of PIPKIγ635 and the N terminus of PIPKIγ661 26-amino acid insert (Fig. 6A). Interestingly separating the C-terminal end of the 635 splice isoform from the 661 insert in this configuration diminishes AP-2 binding to roughly that seen with the GST-PIPKIγ-(460-635) alone (Fig. 6B). This indicates that residues proximal to the 26-amino acid insert may contribute to the engagement of the β2 appendage of the AP-2 adaptor. Indeed a pair of constructs that include the last six (GST-PIPKIγ-(630-661)) or 12 (GST-PIPKIγ-(624-661)) residues of PIPKIγ635 have dramatically increased AP-2 binding capability compared with the mouse PIPKIγ661 C terminus alone fused to GST (GST-PIPKIγ-(636-661)) (Fig. 6C). There is little change in the interaction with talin for these fusion proteins, however. These results confirm then that the extreme C-terminal region of the PIPKIγ635 splice isoform contributes to binding to the AP-2 β2 appendage but alone is insufficient for associating with AP-2.
FIGURE 6.
Differing partner binding properties of the three PIPKIγ splice variants. A, primary sequence alignment of the C-terminal segment of PIPKIγ isoforms from various species: murine (Mm; NCBI accession number NP_032870.1), rat (Rn; NP_001009967.2), human (Hs; NP_036530.1), bovine (Bt; XP_585653.4), feline (Cf; XP_542172.2), opossum (Md; XP_001363745.1), platypus (Oa; XP_001511349.1), and zebrafish (Dr; XP_683392.3). Identical residues are colored pink, and conservatively substituted residues are yellow. The location of the talin-binding motif is indicated above, and the GST fusion proteins spanning the junction between the PIPKIγ635 and -661 isoforms tested is indicated below. B, ∼200 μg of GST (lanes a and b), GST-PIPKIγ-(460-635) (lanes c and d), GST-PIPKIγ-(460-661) (lanes e and f), or GST-PIPKIγ-(460-687) (lanes g and h) immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of 1.5% of each supernatant (S) and 10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1 and anti-β1/β2 subunit mAb 100/1 or with anti-talin mAb 8d4, and only the relevant portions are shown. C, ∼200 μg of GST (lanes a and b), GST-PIPKIγ-(460-635) (lanes c and d), GST-PIPKIγ-(460-661) (lanes e and f), GST-PIPKIγ-(636-661) (lanes g and h), GST-PIPKIγ-(630-661) (lanes i and j), or GST-PIPKIγ-(624-661) (lanes k and l) immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of 1.5% of each supernatant(S) and 10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1 and anti-β1/β2 subunit mAb 100/1 or anti-talin mAb 8d4, and only the relevant portions are shown. The position of talin (open arrowheads) and the AP-2 β2 and αC subunits (black arrowheads) on the stained gel is shown.
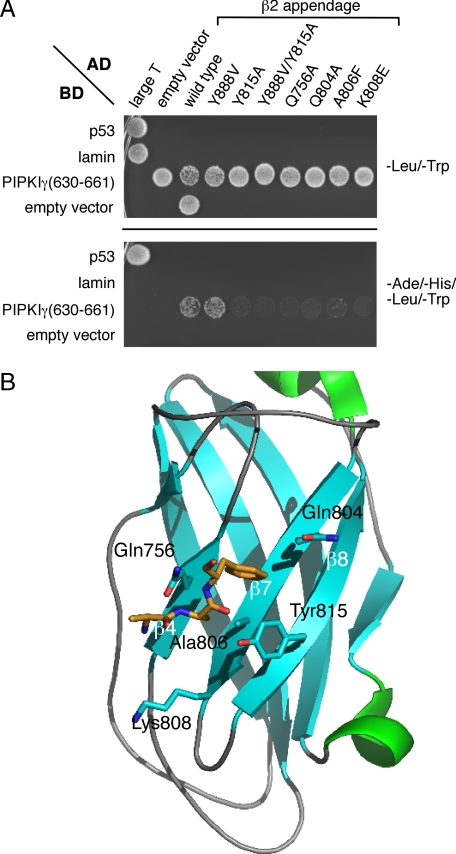
The β2 Sandwich Subdomain Contact Surface—A yeast two-hybrid interaction screen with the PIPKIγ-(630-661) peptide fused to the DNA binding domain of Gal4p corroborates that this sequence engages the sandwich subdomain of the β2 appendage. Transformed AH109 yeast grow on quadruple dropout selection plates only when expressing both the PIPKIγ-(630-661) and β2 appendage but not either protein alone (Fig. 7A). Site-directed mutagenesis of selected residues on the β2 appendage shows that a Y888V mutation, which disrupts the platform interaction site (52), has no effect on PIPKIγ interaction. By contrast the Y815A substitution as well as alteration of several other side chains that contribute to the sandwich site interaction surface (Gln756, Gln804, Ala806, and Lys808; Fig. 7B) (49, 50) strongly impedes the binding to the C-terminal PIPKIγ-(630-661) polypeptide (Fig. 7A).
FIGURE 7.
AP-2-PIPKIγ interaction in a yeast two-hybrid assay. A, S. cerevisiae strain AH109 transformed with the indicated Gal4 pGBKT7 binding domain (BD) and pGADT7 activation domain (AD) plasmid combinations were spotted onto synthetic defined minimal medium plates lacking either Leu and Trp or Ade, His, Leu, and Trp and grown at 30 °C. B, ribbon representation (Protein Data Bank code 2G30) of the AP-2 β2 appendage sandwich subdomain indicating the location of important side chains (blue, nitrogen; red, oxygen) involved in accommodating the PIPKIγ661 C-terminal interaction motif. Shown in stick representation (gold) is the location of co-crystallizing AAF peptide that demarcates a portion of the binding surface upon the β2 appendage sandwich subdomain.
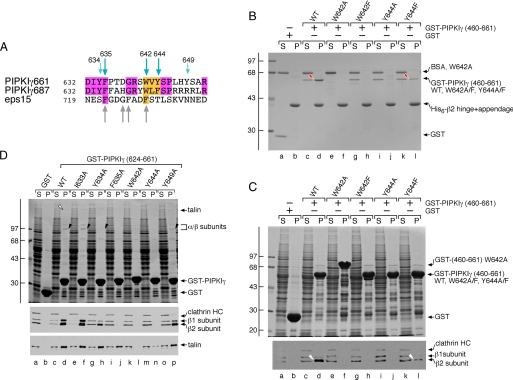
Pinpointing Key Anchor Residues in PIPKIγ Necessary for β2 Appendage Engagement—Having delineated the AP-2 β2 interaction motif in PIPKIγ661 to a tract of ∼20 residues, we next tested the importance of selected hydrophobic side chains within this region. This is because for all characterized adaptors and accessory factors that associate with the α and β2 appendages, aromatic residue-containing motifs are critically important binding determinants, such as the DP(F/W) motif in eps15 and epsin (3); the WXX(F/W)X(D/E)n motif in SJ170 (59), NECAP (54), and stonin 2 (60, 61); and the (D/E)nX1-2FXX(F/L)XXXR motif in ARH and β-arrestin (49, 50). There are five phylogenetically conserved aromatic residues in the mapped PIPKIγ AP-2 binding region (Figs. 6A and 8A). Two, Trp642 and Tyr644, are vicinal to Ser645, which inhibits AP-2 binding when phosphorylated (Fig. 3) (25). Both a W642A mutation and a more conservative phenylalanine substitution (W642F) eliminate β2 binding (Fig. 8, B and C, lanes f and h) in either binary GST pulldown assays (Fig. 8B) or with rat brain cytosol (Fig. 8C). Likewise a Y644A change eliminates β2 appendage binding (Fig. 8, B and C, lane j), but the conservative Y644F mutation reduces, but does not abolish, β2 engagement (Fig. 8, B and C, lane l). This finding is significant because alignment of the sequences that follow the C-terminal IYF of the PIPKIγ635 isoform in either the PIPKIγ661 or PIPKIγ687 variants shows some obvious similarities (Fig. 8A). The longest PIPKIγ687 insert has a Trp positioned analogously to Trp642 in PIPKIγ661 and a Phe at the position equivalent to Tyr644. In fact, five of the 11 N-terminal residues are identical between the two alternatively spliced proteins, and two are conservatively substituted (Fig. 8A). Nevertheless the PIPKIγ687 protein does not bind to AP-2 efficiently (Fig. 8B) presumably in part because of the natural Phe-for-Tyr substitution.
FIGURE 8.
Delineation of key anchor residues that mediate PIPKIγ661 binding to the β2 appendage. A, primary sequence alignment of the two PIPKIγ 26-amino acid inserts and comparison with a tract from eps15 that also binds to the AP-2 β2 appendage sandwich subdomain. B, ∼40 μg of His6-tagged β2 hinge + appendage immobilized on Ni-NTA-agarose was incubated with ∼10 μg of GST (lanes a and b), GST-PIPKIγ-(460-661) wild type (WT; lanes c and d), or GST-PIPKIγ-(460-661) containing W642A (lanes e and f), W642F (lanes g and h), Y644A (lanes i and j), or Y644F (lanes k and l) mutations in the presence of carrier bovine serum albumin (BSA). After centrifugation, aliquots of ∼4% of each supernatant (S) and ∼25% of each washed pellet (P) were resolved by SDS-PAGE and stained with Coomassie Blue. Bound protein (red arrowhead) is shown. C, ∼250 μg of GST (lanes a and b), GST-PIPKIγ-(460-661) wild type (WT; lanes c and d), or GST-PIPKIγ-(460-661) containing W642A (lanes e and f), W642F (lanes g and h), Y644A (lanes i and j), or Y644F (lanes k and l) mutations immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of ∼3.5% of each supernatant (S) and ∼10% of each pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. A portion of the blot was probed with anti-β1/β2-subunit mAb 100/1 and anti-clathrin heavy chain (HC) mAb TD.1. Bound protein (white arrowhead) is shown. D, ∼100 μg of GST (lanes a and b), GST-PIPKIγ-(624-661) wild type (WT; lanes c and d), or GST-PIPKIγ-(624-661) containing I633A (lanes e and f), Y634A (lanes g and h), F635A (lanes i and j), W642A (lanes k and l), Y644A (lanes m and n), or Y649A (lanes o and p) mutations immobilized on glutathione-Sepharose was incubated with rat brain cytosol as indicated. After centrifugation, aliquots of ∼1.5 supernatant (S) and ∼10% of each pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blot were probed with anti-β1/β2-subunit mAb 100/1 and anti-clathrin heavy chain (HC) mAb TD.1 or anti-talin mAb 8d4. The position of talin (open arrowhead) and the AP-2 α and β2 subunits (black arrowhead) on the stained gel are shown.
The pair of aromatics at the end of the PIPKIγ635 sequence are also important; a double Tyr634 and Phe635 to Ala substitution (YF → AA) in the context of the GST-PIPKIγ-(460-661) fusion protein diminishes AP-2 binding to the level seen with just the 26-amino acid extension of the PIPKIγ661 isoform (data not shown). Separate Y634A and F635A mutations in the GST-PIPKIγ-(624-661) fusion backbone reveal that Phe635 is critical whereas Tyr634 is somewhat less so as there is still some AP-2 bound to the fusion protein with a Y364A mutation (Fig. 8D). By contrast, converting Ile633 to Ala does not diminish the binding of AP-2 when compared with the wild-type protein. As seen with the GST-PIPKIγ-(460-661) fusions, Trp642 and Tyr644 are vital residues for β subunit engagement. The Y649A switch, located 13 residues after the intersection between the 635 and 661 splice forms, shows that this amino acid also contributes weakly to AP-2 binding. The lack of a bulky aromatic residue equivalent to Tyr649 in the PIPKIγ687 longest splice isoform as well as other substitutions may also contribute to the poor interaction with AP-2. This set of mutations has a different effect on talin binding. Only the W642A and Y644A mutants exhibit greatly diminished binding to cytosolic talin (Fig. 8D), which is in accord with the mapped talin binding region (see Fig. 6A) (38, 51, 62) and structural studies of the talin FERM F3 subdomain engaged with PIPKIγ peptide (63, 64). Altogether these data indicate that aromatic residues contribute to β2 sandwich subdomain binding possibly through inserting into complementary hydrophobic surfaces in the appendage. The results also confirm the important role of Tyr644 in AP-2 binding (23) but argue strongly against the kinase-AP-2 interaction being YXXØ-mediated because those interactions are critically dependent on the proximal tyrosine that cannot be replaced functionally by phenylalanine (65, 66).
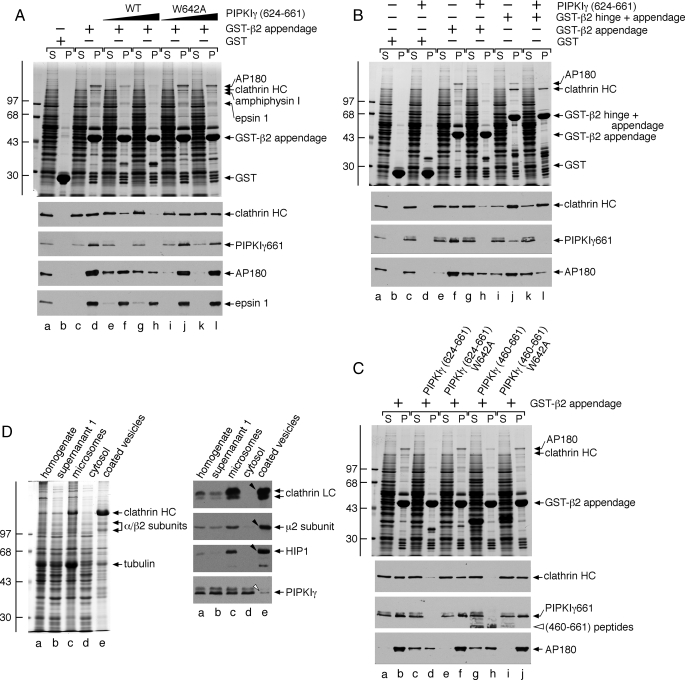
Clathrin Is Displaced from the AP-2 β2 Appendage by PIPKIγ661—An additional endocytic coat protein that binds to the sandwich subdomain of the AP-2 β2 appendage is the clathrin heavy chain (49, 52, 55, 67). Rather than utilizing a short unstructured interaction motif to contact the β2 appendage, the clathrin heavy chain interaction surface is located within the distal leg (67) comprised of repeating antiparallel α-helices in a solenoid-type fold (68, 69). Although clathrin is a functional trimer, we nevertheless examined whether PIPKIγ661 and clathrin compete for a common binding surface on the β2 appendage. Supplementing whole cytosol with a PIPKIγ-(624-661) polypeptide interferes with clathrin binding to immobilized GST-β2 appendage in a dose-dependent manner (Fig. 9A). The peptide competitor also abolishes endogenous PIPKIγ661 and AP180 binding but leaves epsin 1 still attached to the β2 appendage, further arguing that it binds to the sandwich subdomain. By contrast, adding a PIPKIγ-(624-661) peptide containing a W642A substitution has a weak, if any, effect on clathrin, PIPKIγ661, or AP180 binding (Fig. 9A).
FIGURE 9.
Mutually exclusive engagement of the β2 appendage sandwich by either PIPKIγ661 or clathrin. A, ∼100 μg of GST (lanes a and b) or GST-β2 appendage (lanes c-l) immobilized on glutathione-Sepharose was incubated with rat brain cytosol alone (lanes a-d) or cytosol supplemented with 46 μm wild type (WT; lanes e and f) or W642A (lanes i and j) PIPKIγ-(624-661) peptide or 139 μm wild type (lanes g and h) or W642A (lanes k and l) PIPKIγ-(624-661) peptide. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1, anti-PIPKIγ mAb clone 12, anti-AP180 mAb clone 34, or affinity-purified anti-epsin 1 antibodies. Notice that although epsin 1 binding to the β2 appendage is affected by the addition of the PIPKIγ peptide, particularly at the highest concentration, the sandwich-binding partners are clearly much more sensitive to the competitor. B, ∼100 μg of GST (lanes a-d), GST-β2 appendage (lanes e-h), or GST-β2 hinge + appendage (lanes i-l) immobilized on glutathione-Sepharose was incubated with rat brain cytosol alone (lanes a, b, e, f, i, and j) or rat brain cytosol supplemented with 113 μm PIPKIγ-(624-661) polypeptide (lanes c, d, g, h, k, and l). After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1, anti-PIPKIγ mAb clone 12, or anti-AP180 mAb clone 34. C, ∼100 μg of GST-β2 appendage immobilized on glutathione-Sepharose was incubated with rat brain cytosol alone (lanes a and b) or cytosol supplemented with 113 μm wild type (lanes c and d) or W642A (lanes e and f) PIPKIγ-(624-661) polypeptide or 113 μm wild type (lanes g and h) or W642A (lanes i and j) PIPKIγ-(460-661) polypeptide. After centrifugation, aliquots of ∼1.5% of each supernatant (S) and ∼10% of each washed pellet (P) were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin heavy chain (HC) mAb TD.1, anti-PIPKIγ mAb clone 12, or anti-AP180 mAb clone 34. D, fractions (20 μg) from a preparation of rat brain clathrin-coated vesicles were resolved by SDS-PAGE and either stained with Coomassie Blue or transferred to nitrocellulose. Portions of the blots were probed with anti-clathrin light chain (LC) mAb Cl57.3, anti-μ2 subunit serum, affinity-purified anti-HIP1 antibodies, or anti-PIPKIγ mAb clone 12. Only the relevant portions are shown. Note the strong enrichment of clathrin, AP-2, and HIP1 (arrowheads) but exclusion of PIPKIγ (open arrowhead) in the coated vesicle fraction.
If clathrin engagement by the larger β2 subunit hinge + appendage domains is followed under the same conditions, the triskelia bind much more avidly to the immobilized GST-β2 fragment (Fig. 9B, compare lanes f and j). The PIPKIγ-(624-661) peptide now does compete with clathrin only weakly (Fig. 9B, compare lanes j and l). Yet the added excess peptide does effectively compete PIPKIγ and AP180 off the β2 hinge + appendage, however (Fig. 9B, compare lanes j and l). This selectively refractory behavior is because the extra β2 hinge region contains additional clathrin binding determinants that contact the N-terminal β-propeller of the clathrin heavy chain (52, 70, 71). The additional points of contact increase the apparent affinity of clathrin for the β2 chain versus PIPKIγ661 or eps15 (49, 72). Thus, the AP-2 β2 subunit binds more tightly to multivalent clathrin trimers than to monovalent PIPKIγ661.
Like the PIPKIγ-(624-661) segment, the whole C-terminal PIPKIγ-(460-661) polypeptide also interferes with clathrin binding in the presence of cytosol, whereas the W642A mutant does not (Fig. 9C). A corollary of these results is that clathrin engagement could displace prebound PIPKIγ661 from AP-2 as the lattice assembly process progresses toward the deeply invaginated state. Circumstantial evidence for this contention comes from the marked de-enrichment of PIPKIγ661 in preparations of rat brain clathrin-coated vesicles (Fig. 9D) (32). These results imply that the temporal residence of PIPKIγ661 at the bud site may be determined by the extent of clathrin lattice polymerization.
DISCUSSION
Phosphoinositides are biologically useful signaling molecules in part because they fail to transfer rapidly between different membrane compartments in the absence of vesicular transport. These phospholipids can rapidly move laterally within the plane of the membrane. Bound to PtdIns(4,5)P2, the diffusion coefficient of the phospholipase C δ1 pleckstrin homology domain (10, 73) approximates that of lipid alone within the plasma membrane bilayer (11, 74); this could impact the action of PtdIns(4,5)P2 at discrete membrane patches on the cell surface. In addition, the inositol lipid head group can be rapidly remodeled upon delivery to a new organelle or even while within a forming coated bud or membrane-bounded transport vesicle. This plasticity of the head group, coupled with vectorial membrane flow, allows for differentially phosphorylated phosphatidylinositol phosphates to play critical regulatory roles along the secretory and endosomal pathways. Here we demonstrate that the PIPKIγ661 isoform binds physically to the sandwich subdomain of the AP-2 β2 subunit appendage. This interaction allows for the 5-kinase to be strategically positioned during clathrin coat assembly and to augment PtdIns(4,5)P2 production focally. A precedent for this type of localized regulation already exists; the talin FERM domain engages the longer splice isoforms of PIPKIγ (using a sequence that overlaps the β2 appendage interaction motif), and this association coordinates PtdIns(4,5)P2 production at focal adhesion sites (38, 51).
In eukaryotes, alternative splicing can increase genomic complexity and diversity. The organization of the C-terminal splice sites allows the synthesis of three distinct PIPKIγ variants: one (PIPKIγ687) can bind selectively to talin via the C-terminal extension, one (PIPKIγ661) can bind to both talin and AP-2, and one (PIPKIγ635) can bind to neither protein. This flexibility could impart importantly different biological functions to each isoform (75). An additional level of regulation can fine tune this variation as phosphorylation of Ser645 impedes AP-2 (Fig. 3 and Ref. 25) and talin (76) binding, whereas Tyr644 (Tyr649 in the human isoform) phosphorylation enhances the association with talin (62, 64, 76). Intriguingly Tyr634 is also subject to phosphorylation in response to growth factors (77). In the brain, PIPKIγ661 is the major isoform at the presynaptic plasma membrane (14, 32) suggesting that the endocytic function of the kinase is paramount in this tissue.
We mapped the AP-2 interaction motif in PIPKIγ to a more expansive sequence than those typically recognized by the α appendage (DP(F/W), FXDXF, and WXX(F/W)X(D/E)n) or even the platform site on the β2 appendage ((D/E)nX1-2FXX(F/L)XXXR). The sequence differs too from the 721SFGDGFADF region of murine eps15 that also binds to the β2 sandwich subdomain (50). A co-crystal with the β2 appendage shows that the eps15 sequence tract forms a tight turn that depends critically on the central Gly for proper positioning of the three Phe side chains (50). Because of the lack of a Gly within the β2 binding region of PIPKIγ661 and the altered spacing of the aromatic residues (Fig. 8A), it seems unlikely that the kinase and eps15 engage the β2 sandwich site in precisely the same manner. Yet the strict selectivity of PIPKIγ661 for the sandwich subdomain makes the binding to AP-2 subject to interference by clathrin, which can occupy the same interaction site (49, 55). More important is that in the context of the assembled polyhedral lattice the clathrin heavy chains project a profusion of terminal domains down into the interior of the assemblage (49, 68). In our view, this high density of clathrin coupled with the capability of the β2 chain of AP-2 to engage the clathrin heavy chain through at least two binding sites will strongly favor AP-2 associations with clathrin in regions of extensively assembled coat. In fact, in biochemical assays eps15, which also binds to the AP-2 β2 appendage sandwich (49, 50) but not to clathrin, is displaced from AP-2 by polymerizing clathrin (72). Accordingly eps15 is restricted to the outer edge of clathrin lattices on the cell surface (49, 78). We propose that this hierarchical binding phenomenon offers a mechanism for temporal and spatial control of PtdIns(4,5)P2 formation as a function of the extent of polymerized clathrin lattice.
Coincidence detection is a common theme in the coordinated placement of clathrin coat components necessary to drive bud progression (2). We therefore cannot rule out that PIPKIγ661 utilizes this by establishing secondary associations with the μ2 subunit of AP-2 either through the YXXØ interaction surface (23) or by binding of the catalytic core region to a distinct portion of μ2 (24). Conceivably these additional contacts could be precisely regulated by phosphorylation as exposure of the μ2 subunit to accommodate YXXØ sorting signals requires phosphorylation of Thr156 (79). Perhaps this allows PIPKIγ661 binding to be variably regulated at different points in the coat assembly cycle. However, because PIPKIγ661 is not enriched in clathrin-coated vesicles purified from rat brain (Fig. 9 and (32)) and immunoelectron microscopy reveals PIPKIγ on synaptic plasma membrane adjacent to, but not within, clathrin-coated buds (32), we favor the β2 appendage as the site most able to manage this spatial positioning of the 5-kinase during clathrin-mediated endocytosis at the nerve terminal. Intriguingly PIPKIγ661 binds to the AP-2 β2 appendage as well as the β1 subunit of the AP-1 heterotetramer (25). In this regard, it is interesting that AP-1 is implicated in return of certain integral membrane proteins from the recycling endosome compartment and that PtdIns(4,5)P2 is the necessary phosphoinositide (80). Similar regulation by the clathrin heavy chain and/or possibly eps15 could occur as eps15 is located on endosomes and the trans-Golgi network (81), and the key residues that constitute the interaction surface on the β2 appendage sandwich subdomain are all conserved in the AP-1 β1 chain (55).
The structurally related AP-2 α and β2 appendages contact a rich constellation of binding partners, some of which are common to both but nevertheless engage structurally non-equivalent surfaces on each appendage (49, 50). A major, currently unresolved question is why certain proteins have evolved specific combinations of dissimilar AP-2 (and clathrin) interaction motifs as opposed to utilizing standardized, generic connection elements? Also given the relatively low affinity of these interactions in general, how does a timed succession of protein occupancies lead to progressive coat assembly without misdirected protein placement or catastrophic entanglement? One explanation is that binding events that occur at different locations upon the appendages utilizing particular peptide motifs generate different functional consequences, enabling the two proximate domains to act in concert to coordinate coat assembly. The coordination of phosphatidylinositol phosphate metabolism at coated buds seems to be emblematic of this type of regulation. The 5-kinase activity of PIPKIs is stimulated severalfold by phosphatidic acid (PtdOH) (46, 82). Clathrin-mediated endocytosis ceases rapidly (<5 min) upon administration of 1-2% 1-butanol to cultured cells, and new lattice assembly is prevented (83), illustrating the likely involvement of PtdOH in coat assembly and progression. We have shown previously that, in vitro, PtdOH can promote limited translocation of AP-2 to biological membranes in the absence of PtdIns(4,5)P2 synthesis (40). In fact, a positive feedback loop operates as the product of PIPKI activity, PtdIns(4,5)P2, is a required activator of phospholipase D (Fig. 10) (40, 84). Primary alcohols interfere with phospholipase D-catalyzed hydrolysis of phosphatidylcholine because a phosphatidylalcohol and not PtdOH is produced (85). An alternative pathway for the generation of PtdOH is through the phosphorylation of diacylglycerol by diacylglycerol kinase (DGK; Fig. 10) (86). Intriguingly the pleckstrin homology domain-containing type II DGK, DGKδ2, partly colocalizes with clathrin-coated structures and binds directly to AP-2, in this case to the platform subdomain of the α appendage (87). Although DGKδ2 is widely expressed, mRNA transcripts for the enzyme are not particularly abundant in brain (88). Therefore, at the synapse, this enzyme may not be recruited to presynaptic coated buds concomitantly with PIPKIγ661. Perhaps in different cell types, PtdIns(4,5)P2 and PtdOH production at clathrin-coated regions is managed by different enzymes associating with distinct sites on the AP-2 appendages.
Like PIPKIγ, the phosphoinositide polyphosphatase synaptojanin 1 is subject to alternative splicing (31). Somewhat unexpectedly, the ubiquitously expressed large splice isoform, SJ170, is present within clathrin-coated structures at the cell surface throughout the assembly cycle (28). SJ170 also associates physically with AP-2 (59, 89), and similarly to PIPKIγ661, we mapped the principal interaction motif to a WXX(F/W)X(D/E)n motif within the C-terminal extension of SJ170 (59). This type of interaction motif selectively engages the sandwich subdomain of the AP-2 α appendage (54, 60, 61). Thus, PIPKIγ661, DGKδ2, and SJ170 can all associate directly with AP-2 albeit through different contact sites on the appendages (Fig. 10). By binding to different surfaces of the α or β2 appendage, the various enzymes that modulate phosphatidylinositol phosphate metabolism at the growing bud can be subject to different regulatory inputs. Therefore, the α and β2 appendages appear to operate as organizational scaffolds in part to optimize the regional lipid environment necessary for clathrin coat formation. Indeed this seems to be a general requirement for efficient receptor internalization because β-arrestin 2, a CLASP that promotes the uptake of stimulated G protein-coupled receptors (90, 91), also associates directly with DGK (92). β-Arrestin 2 also binds physically to PIPKI in a PtdIns(4,5)P2-sensitive fashion (26). The ability of other CLASPs to engage PIPKIs is in accord with much data showing that, under certain experimental circumstances, internalization of discrete transmembrane cargo molecules continues in an AP-2 gene-silenced background (93-99).
Acknowledgments
We thank Luisa Giudici and Robin Irvine for the PIPkin1γ93 cDNA and Mike Cascio for expert guidance with the CD spectroscopy. We are grateful to Juan Bonifacino, Frances Brodsky, Reinhard Jahn, Peter McPherson, and Ernst Ungewickell for generously providing antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK53249 (to L. M. T.), T32 DK061296 (to J. R. T.), and R01 CA104708 (to R. A. A.). This work was also supported in part by American Heart Association Established Investigator Award 0540007N.
Footnotes
The abbreviations used are: PtdIns(4,5)P2, phosphatidylinositol 4,5-bisphosphate; ARH, autosomal recessive hypercholesterolemia protein; CI-MPR, cation-independent mannose 6-phosphate receptor; CLASP, clathrin-associated sorting protein; DGK, diacylglycerol kinase; FERM, band 4.1, ezrin, radixin, moesin; GST, glutathione S-transferase; mAb, monoclonal antibody; PIPKI, type I phosphatidylinositol 4-phosphate 5-kinase; PtdOH, phosphatidic acid; ENTH, epsin N-terminal homology; DTT, dithiothreitol; Ni-NTA, nickel-nitrilotriacetic acid.
References
- 1.Krauss, M., and Haucke, V. (2007) EMBO Rep. 8 241-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Paolo, G., and De Camilli, P. (2006) Nature 443 651-657 [DOI] [PubMed] [Google Scholar]
- 3.Maldonado-Baez, L., and Wendland, B. (2006) Trends Cell Biol. 16 505-513 [DOI] [PubMed] [Google Scholar]
- 4.Schmid, E. M., and McMahon, H. T. (2007) Nature 448 883-888 [DOI] [PubMed] [Google Scholar]
- 5.Ungewickell, E. J., and Hinrichsen, L. (2007) Curr. Opin. Cell Biol. 19 417-425 [DOI] [PubMed] [Google Scholar]
- 6.Vallis, Y., Wigge, P., Marks, B., Evans, P. R., and McMahon, H. T. (1999) Curr. Biol. 9 257-260 [DOI] [PubMed] [Google Scholar]
- 7.Lee, D. W., Wu, X., Eisenberg, E., and Greene, L. E. (2006) J. Cell Sci. 119 3502-3512 [DOI] [PubMed] [Google Scholar]
- 8.Jost, M., Simpson, F., Kavran, J. M., Lemmon, M. A., and Schmid, S. L. (1998) Curr. Biol. 8 1399-1402 [DOI] [PubMed] [Google Scholar]
- 9.van Rheenen, J., Mulugeta Achame, E., Janssen, H., Calafat, J., and Jalink, K. (2005) EMBO J. 24 1664-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brough, D., Bhatti, F., and Irvine, R. F. (2005) J. Cell Sci. 118 3019-3025 [DOI] [PubMed] [Google Scholar]
- 11.Yaradanakul, A., and Hilgemann, D. W. (2007) J. Membr. Biol. 220 53-67 [DOI] [PubMed] [Google Scholar]
- 12.Narkis, G., Ofir, R., Landau, D., Manor, E., Volokita, M., Hershkowitz, R., Elbedour, K., and Birk, O. S. (2007) Am. J. Hum. Genet. 81 530-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, Y., Lian, L., Golden, J. A., Morrisey, E. E., and Abrams, C. S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11748-11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Paolo, G., Moskowitz, H. S., Gipson, K., Wenk, M. R., Voronov, S., Obayashi, M., Flavell, R., Fitzsimonds, R. M., Ryan, T. A., and De Camilli, P. (2004) Nature 431 415-422 [DOI] [PubMed] [Google Scholar]
- 15.Carvou, N., Norden, A. G., Unwin, R. J., and Cockcroft, S. (2006) Cell. Signal. 19 42-51 [DOI] [PubMed] [Google Scholar]
- 16.Yu, J. W., Mendrola, J. M., Audhya, A., Singh, S., Keleti, D., DeWald, D. B., Murray, D., Emr, S. D., and Lemmon, M. A. (2004) Mol. Cell 13 677-688 [DOI] [PubMed] [Google Scholar]
- 17.Stefan, C. J., Audhya, A., and Emr, S. D. (2002) Mol. Biol. Cell 13 542-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, Y., Carroll, S., Kaksonen, M., Toshima, J. Y., and Drubin, D. G. (2007) J. Cell Biol. 177 355-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunz, J., Wilson, M. P., Kisseleva, M., Hurley, J. H., Majerus, P. W., and Anderson, R. A. (2000) Mol. Cell 5 1-11 [DOI] [PubMed] [Google Scholar]
- 20.Varnai, P., Thyagarajan, B., Rohacs, T., and Balla, T. (2006) J. Cell Biol. 175 377-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoncu, R., Perera, R. M., Sebastian, R., Nakatsu, F., Chen, H., Balla, T., Ayala, G., Toomre, D., and De Camilli, P. V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3793-3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abe, N., Inoue, T., Galvez, T., Klein, L., and Meyer, T. (2008) J. Cell Sci. 121 1488-1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bairstow, S. F., Ling, K., Su, X., Firestone, A. J., Carbonara, C., and Anderson, R. A. (2006) J. Biol. Chem. 281 20632-20642 [DOI] [PubMed] [Google Scholar]
- 24.Krauss, M., Kukhtina, V., Pechstein, A., and Haucke, V. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11934-11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano-Kobayashi, A., Yamazaki, M., Unoki, T., Hongu, T., Murata, C., Taguchi, R., Katada, T., Frohman, M. A., Yokozeki, T., and Kanaho, Y. (2007) EMBO J. 26 1105-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, C. D., Kovacs, J. J., Nobles, K. N., Whalen, E. J., and Lefkowitz, R. J. (2008) J. Biol. Chem. 283 21093-21101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padron, D., Wang, Y. J., Yamamoto, M., Yin, H., and Roth, M. G. (2003) J. Cell Biol. 162 693-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perera, R. M., Zoncu, R., Lucast, L., De Camilli, P., and Toomre, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19332-19337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cremona, O., Di Paolo, G., Wenk, M. R., Luthi, A., Kim, W. T., Takei, K., Daniell, L., Nemoto, Y., Shears, S. B., Flavell, R. A., McCormick, D. A., and De Camilli, P. (1999) Cell 99 179-188 [DOI] [PubMed] [Google Scholar]
- 30.Harris, T. W., Hartwieg, E., Horvitz, H. R., and Jorgensen, E. M. (2000) J. Cell Biol. 150 589-600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramjaun, A. R., and McPherson, P. S. (1996) J. Biol. Chem. 271 24856-24861 [DOI] [PubMed] [Google Scholar]
- 32.Wenk, M. R., Pellegrini, L., Klenchin, V. A., Di Paolo, G., Chang, S., Daniell, L., Arioka, M., Martin, T. F., and De Camilli, P. (2001) Neuron 32 79-88 [DOI] [PubMed] [Google Scholar]
- 33.Traub, L. M., Downs, M. A., Westrich, J. L., and Fremont, D. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8907-8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra, S. K., Keyel, P. A., Edeling, M. A., Owen, D. J., and Traub, L. M. (2005) J. Biol. Chem. 280 19270-19280 [DOI] [PubMed] [Google Scholar]
- 35.Doray, B., Lee, I., Knisely, J., Bu, G., and Kornfeld, S. (2007) Mol. Biol. Cell 18 1887-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drake, M. T., Downs, M. A., and Traub, L. M. (2000) J. Biol. Chem. 275 6479-6489 [DOI] [PubMed] [Google Scholar]
- 37.Mishra, S. K., Agostinelli, N. R., Brett, T. J., Mizukami, I., Ross, T. S., and Traub, L. M. (2001) J. Biol. Chem. 276 46230-46236 [DOI] [PubMed] [Google Scholar]
- 38.Ling, K., Doughman, R. L., Firestone, A. J., Bunce, M. W., and Anderson, R. A. (2002) Nature 420 89-93 [DOI] [PubMed] [Google Scholar]
- 39.Fischer von Mollard, G., Sudhof, T. C., and Jahn, R. (1991) Nature 349 79-81 [DOI] [PubMed] [Google Scholar]
- 40.Arneson, L. S., Kunz, J., Anderson, R. A., and Traub, L. M. (1999) J. Biol. Chem. 274 17794-17805 [DOI] [PubMed] [Google Scholar]
- 41.Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004) Biophotonics Int. 11 36-42 [Google Scholar]
- 42.Savitzky, A., and Golay, M. J. E. (1964) Anal. Chem. 36 1627-1639 [Google Scholar]
- 43.Collins, B. M., McCoy, A. J., Kent, H. M., Evans, P. R., and Owen, D. J. (2002) Cell 109 523-535 [DOI] [PubMed] [Google Scholar]
- 44.Ohno, H., Stewart, J., Fournier, M.-C., Bosshart, H., Rhee, I., Miyatake, S., Saito, T., Galluser, A., Kirchhausen, T., and Bonifacino, J. S. (1995) Science 269 1872-1875 [DOI] [PubMed] [Google Scholar]
- 45.Owen, D. J., and Evans, P. R. (1998) Science 282 1327-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishihara, H., Shibasaki, Y., Kizuki, N., Wada, T., Yazaki, Y., Asano, T., and Oka, Y. (1998) J. Biol. Chem. 273 8741-8748 [DOI] [PubMed] [Google Scholar]
- 47.He, G., Gupta, S., Yi, M., Michaely, P., Hobbs, H. H., and Cohen, J. C. (2002) J. Biol. Chem. 277 44044-44049 [DOI] [PubMed] [Google Scholar]
- 48.Mishra, S. K., Watkins, S. C., and Traub, L. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16099-16104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edeling, M. A., Mishra, S. K., Keyel, P. A., Steinhauser, A. L., Collins, B. M., Roth, R., Heuser, J. E., Owen, D. J., and Traub, L. M. (2006) Dev. Cell 10 329-342 [DOI] [PubMed] [Google Scholar]
- 50.Schmid, E. M., Ford, M. G., Burtey, A., Praefcke, G. J., Peak Chew, S. Y., Mills, I. G., Benmerah, A., and McMahon, H. T. (2006) PLoS Biol. 4 e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Paolo, G., Pellegrini, L., Letinic, K., Cestra, G., Zoncu, R., Voronov, S., Chang, S., Guo, J., Wenk, M. R., and De Camilli, P. (2002) Nature 420 85-89 [DOI] [PubMed] [Google Scholar]
- 52.Owen, D. J., Vallis, Y., Pearse, B. M., McMahon, H. T., and Evans, P. R. (2000) EMBO J. 19 4216-4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laporte, S. A., Miller, W. E., Kim, K. M., and Caron, M. G. (2002) J. Biol. Chem. 277 9247-9254 [DOI] [PubMed] [Google Scholar]
- 54.Ritter, B., Denisov, A. Y., Philie, J., Deprez, C., Tung, E. C., Gehring, K., and McPherson, P. S. (2004) EMBO J. 23 3701-3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keyel, P. A., Thieman, J. R., Roth, R., Erkan, E., Everett, E. T., Watkins, S. C., and Traub, L. M. (2008) Mol. Biol. Cell 19 5309-5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalthoff, C., Alves, J., Urbanke, C., Knorr, R., and Ungewickell, E. J. (2002) J. Biol. Chem. 277 8209-8216 [DOI] [PubMed] [Google Scholar]
- 57.Giudici, M. L., Emson, P. C., and Irvine, R. F. (2004) Biochem. J. 379 489-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giudici, M. L., Lee, K., Lim, R., and Irvine, R. F. (2006) FEBS Lett. 580 6933-6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jha, A., Agostinelli, N. R., Mishra, S. K., Keyel, P. A., Hawryluk, M. J., and Traub, L. M. (2004) J. Biol. Chem. 279 2281-2290 [DOI] [PubMed] [Google Scholar]
- 60.Walther, K., Diril, M. K., Jung, N., and Haucke, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 964-969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra, S. K., Hawryluk, M. J., Brett, T. J., Keyel, P. A., Dupin, A. L., Jha, A., Heuser, J. E., Fremont, D. H., and Traub, L. M. (2004) J. Biol. Chem. 279 46191-46203 [DOI] [PubMed] [Google Scholar]
- 62.Ling, K., Doughman, R. L., Iyer, V. V., Firestone, A. J., Bairstow, S. F., Mosher, D. F., Schaller, M. D., and Anderson, R. A. (2003) J. Cell Biol. 163 1339-1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Pereda, J. M., Wegener, K. L., Santelli, E., Bate, N., Ginsberg, M. H., Critchley, D. R., Campbell, I. D., and Liddington, R. C. (2005) J. Biol. Chem. 280 8381-8386 [DOI] [PubMed] [Google Scholar]
- 64.Kong, X., Wang, X., Misra, S., and Qin, J. (2006) J. Mol. Biol. 359 47-54 [DOI] [PubMed] [Google Scholar]
- 65.Collawn, J. F., Stangel, M., Kuhn, L. A., Esekogwu, V., Jing, S. Q., Trowbridge, I. S., and Tainer, J. A. (1990) Cell 63 1061-1072 [DOI] [PubMed] [Google Scholar]
- 66.Jadot, M., Canfield, W. M., Gregory, W., and Kornfeld, S. (1992) J. Biol. Chem. 267 11069-11077 [PubMed] [Google Scholar]
- 67.Knuehl, C., Chen, C. Y., Manalo, V., Hwang, P. K., Ota, N., and Brodsky, F. M. (2006) Traffic 7 1688-1700 [DOI] [PubMed] [Google Scholar]
- 68.ter Haar, E., Musacchio, A., Harrison, S. C., and Kirchhausen, T. (1998) Cell 95 563-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ybe, J. A., Brodsky, F. M., Hofmann, K., Lin, K., Liu, S. H., Chen, L., Earnest, T. N., Fletterick, R. J., and Hwang, P. K. (1999) Nature 399 371-375 [DOI] [PubMed] [Google Scholar]
- 70.Gallusser, A., and Kirchhausen, T. (1993) EMBO J. 12 5237-5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.ter Haar, E., Harrison, S. C., and Kirchhausen, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1096-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cupers, P., Jadhav, A. P., and Kirchhausen, T. (1998) J. Biol. Chem. 273 1847-1850 [DOI] [PubMed] [Google Scholar]
- 73.Hammond, G. R. V., Sim, Y., Lagnado, L., and Irvine, R. F. (2009) J. Cell Biol. 184 297-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Golebiewska, U., Nyako, M., Woturski, W., Zaitseva, I., and McLaughlin, S. (2008) Mol. Biol. Cell 19 1663-1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao, Y. S., Yamaga, M., Zhu, X., Wei, Y. J., Sun, H. Q., Wang, J., Yun, M., Wang, Y., Di Paolo, G., Bennett, M., Mellman, I., Abrams, C. S., De Camilli, P., Lu, C. Y., and Yin, H. L. (2009) J. Cell Biol. 184 281-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee, S. Y., Voronov, S., Letinic, K., Nairn, A. C., Di Paolo, G., and De Camilli, P. (2005) J. Cell Biol. 168 789-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun, Y., Ling, K., Wagoner, M. P., and Anderson, R. A. (2007) J. Cell Biol. 178 297-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tebar, F., Sorkina, T., Sorkin, A., Ericsson, M., and Kirchhausen, T. (1996) J. Biol. Chem. 271 28727-28730 [DOI] [PubMed] [Google Scholar]
- 79.Olusanya, O., Andrews, P. D., Swedlow, J. R., and Smythe, E. (2001) Curr. Biol. 11 896-900 [DOI] [PubMed] [Google Scholar]
- 80.Crottet, P., Meyer, D. M., Rohrer, J., and Spiess, M. (2002) Mol. Biol. Cell 13 3672-3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chi, S., Cao, H., Chen, J., and McNiven, M. (2008) Mol. Biol. Cell 19 3564-3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenkins, G. H., Fisette, P. L., and Anderson, R. A. (1994) J. Biol. Chem. 269 11547-11554 [PubMed] [Google Scholar]
- 83.Boucrot, E., Saffarian, S., Massol, R., Kirchhausen, T., and Ehrlich, M. (2006) Exp. Cell Res. 312 4036-4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morris, A. J. (2007) Biochem. Soc. Symp. 74 247-257 [DOI] [PubMed] [Google Scholar]
- 85.Jenkins, G. M., and Frohman, M. A. (2005) CMLS Cell. Mol. Life Sci. 62 2305-2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Merida, I., Avila-Flores, A., and Merino, E. (2008) Biochem. J. 409 1-18 [DOI] [PubMed] [Google Scholar]
- 87.Kawasaki, T., Kobayashi, T., Ueyama, T., Shirai, Y., and Saito, N. (2007) Biochem. J. 409 471-479 [DOI] [PubMed] [Google Scholar]
- 88.Sakane, F., Imai, S., Yamada, K., Murakami, T., Tsushima, S., and Kanoh, H. (2002) J. Biol. Chem. 277 43519-43526 [DOI] [PubMed] [Google Scholar]
- 89.Haffner, C., Paolo, G. D., Rosenthal, J. A., and de Camilli, P. (2000) Curr. Biol. 10 471-474 [DOI] [PubMed] [Google Scholar]
- 90.Lefkowitz, R. J., and Shenoy, S. K. (2005) Science 308 512-517 [DOI] [PubMed] [Google Scholar]
- 91.Marchese, A., Paing, M. M., Temple, B. R., and Trejo, J. (2008) Annu. Rev. Pharmacol. Toxicol. 48 601-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nelson, C. D., Perry, S. J., Regier, D. S., Prescott, S. M., Topham, M. K., and Lefkowitz, R. J. (2007) Science 315 663-666 [DOI] [PubMed] [Google Scholar]
- 93.Motley, A., Bright, N. A., Seaman, M. N., and Robinson, M. S. (2003) J. Cell Biol. 162 909-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hinrichsen, L., Harborth, J., Andrees, L., Weber, K., and Ungewickell, E. J. (2003) J. Biol. Chem. 278 45160-45170 [DOI] [PubMed] [Google Scholar]
- 95.Barriere, H., Nemes, C., Lechardeur, D., Khan-Mohammad, M., Fruh, K., and Lukacs, G. L. (2006) Traffic 7 282-297 [DOI] [PubMed] [Google Scholar]
- 96.Keyel, P. A., Mishra, S. K., Roth, R., Heuser, J. E., Watkins, S. C., and Traub, L. M. (2006) Mol. Biol. Cell 17 4300-4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maurer, M. E., and Cooper, J. A. (2006) J. Cell Sci. 119 4235-4246 [DOI] [PubMed] [Google Scholar]
- 98.Eden, E. R., Sun, X. M., Patel, D. D., and Soutar, A. K. (2007) Hum. Mol. Genet. 16 2751-2759 [DOI] [PubMed] [Google Scholar]
- 99.Huang, F., Khvorova, A., Marshall, W., and Sorkin, A. (2004) J. Biol. Chem. 279 16657-16661 [DOI] [PubMed] [Google Scholar]
- 100.Hawryluk, M. J., Keyel, P. A., Mishra, S. K., Watkins, S. C., Heuser, J. E., and Traub, L. M. (2006) Traffic 7 262-281 [DOI] [PubMed] [Google Scholar]