Abstract
ArfGAP1 is a prototype of GTPase-activating proteins for ADP-ribosylation factors (ARFs) and has been proposed to be involved in retrograde transport from the Golgi apparatus to the endoplasmic reticulum (ER) by regulating the uncoating of coat protein I (COPI)-coated vesicles. Depletion of ArfGAP1 by RNA interference, however, causes neither a discernible phenotypic change in the COPI localization nor a change in the Golgi-to-ER retrograde transport. Therefore, we also examined ArfGAP2 and ArfGAP3, closely related homologues of ArfGAP1. Cells in which ArfGAP1, ArfGAP2, and ArfGAP3 are simultaneously knocked down show an increase in the GTP-bound ARF level. Furthermore, in these cells proteins resident in or cycling through the cis-Golgi, including ERGIC-53, β-COP, and GM130, accumulate in the ER-Golgi intermediate compartment, and Golgi-to-ER retrograde transport is blocked. The phenotypes observed in the triple ArfGAP knockdown cells are similar to those seen in β-COP-depleted cells. Both the triple ArfGAP- and β-COP-depleted cells accumulate characteristic vacuolar structures that are visible under electron microscope. Furthermore, COPI is concentrated at rims of the vacuolar structures in the ArfGAP-depleted cells. On the basis of these observations, we conclude that ArfGAP1, ArfGAP2, and ArfGAP3 have overlapping roles in regulating COPI function in Golgi-to-ER retrograde transport.
The ADP-ribosylation factors (ARFs)3 are a family of small GTPases. Once associated with organellar membranes in their GTP-bound form, these proteins trigger formation of coated carrier vesicles, e.g. coat protein I (COPI)-coated vesicles. ARFs cycle between a GDP-bound inactive state and a GTP-bound active state; in the latter form they recruit various effectors, including the COPI coat (1, 2). Exchange of bound GDP for GTP is catalyzed by guanine-nucleotide exchange factors, which constitute a large family of proteins that share a Sec7-like catalytic domain (3, 4). GTP hydrolysis in turn is stimulated by GTPase-activating proteins (GAPs), which constitute a large family that share a zinc finger-like catalytic domain (3, 5).
COPI-coated vesicles mediate retrograde transport from the cis-Golgi or endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) to the ER and probably intra-Golgi transport as well. In budding yeasts two ARF-GAPs, Gcs1 and Glo3, have been shown to play overlapping roles in COPI-mediated transport processes (6, 7). According to the prevailing view, ARF-GAPs (in particular, ArfGAP1, which is the founding member of mammalian ARF-GAPs and the counterpart of yeast Gcs1) (8) either induce dissociation of the coat from COPI-coated vesicles or antagonize formation of vesicles (for review, see Ref. 5). This view is based on several lines of evidence; first, blocking GTP hydrolysis on ARF1 by adding GTPγS or a GTPase-defective ARF1 mutant inhibits uncoating of COPI-coated vesicles in a cell-free reconstitution system (9), indirectly suggesting a role for ARF-GAP in vesicle uncoating; second, overexpression of the GTPase-defective ARF1 mutant stabilizes the COPI coat on Golgi membranes (10); third, overexpression of ArfGAP1 results in a phenotype similar to that induced by inhibiting ARF-guanine-nucleotide exchange factors; that is, cytosolic distribution of the COPI coat and disintegration of the Golgi apparatus (11); fourth, the addition of ArfGAP1 to an in vitro system inhibits formation of COPI-coated vesicles and induces uncoating of pre-existing vesicles (12); finally, ArfGAP1-mediated GTP hydrolysis is stimulated by the addition of the COPI coat in vitro (13, 14).
However, additional evidence suggests roles of ArfGAP1 beyond that of a simple inactivator of ARFs (for review, see Ref. 5); first, GTP hydrolysis on ARF is required for proper sorting of cargo molecules into COPI-coated vesicles (15-17); second, ArfGAP1 promotes COPI-coated vesicle formation by coupling cargo sorting to vesicle formation (18-20); third, imaging studies have suggested that ArfGAP1 undergoes ARF1-dependent cycling between the cytosol and Golgi membranes independent of vesicle budding (21, 22); finally, Antonny and co-workers (23, 24) have proposed a model in which ArfGAP1 and Gcs1 sense the curvature of budding vesicles through a motif outside of their catalytic domain.
Despite the critical roles of ArfGAP1 in COPI-coated vesicle formation, most of the available data regarding their function have been obtained by in vitro experiments. We, therefore, attempted to determine the function of ArfGAP1 in the cell by exploiting RNA interference (RNAi). However, we could not detect any phenotypic change in ArfGAP1 knockdown cells. Because there are two poorly characterized mammalian ArfGAPs, ArfGAP2 and ArfGAP3 (25), both of which are more similar to Glo3 than Gcs1 (26-29), we then set out to determine the intracellular roles of these ArfGAPs. Here, we show that ArfGAP1, ArfGAP2, and ArfGAP3 play overlapping roles in COPI-mediated transport and in maintaining Golgi organization.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Plasmids—Antiserum to human ArfGAP1 was raised in rabbits against a synthetic peptide (amino acid residues 377-390) conjugated to keyhole limpet hemocyanin and affinity-purified using the immunized peptide immobilized on Sulfolink beads (Pierce). Antisera to human ArfGAP2 and ArfGAP3 were raised against their polypeptide regions (amino acid residues 329-430 and 329-426, respectively) fused to glutathione S-transferase and affinity-purified using the fusion proteins immobilized on Sulfolink beads. Monoclonal mouse anti-ERGIC-53 antibody (30) was originally provided by Hans-Peter Hauri (University of Basel) and later purchased from Alexis Biochemicals. Polyclonal rabbit antibodies to golgin-97 (31) and to GM130 (32) were kindly provided by Nobuhiro Nakamura (Kanazawa University, Japan) and Yoshio Misumi (Fukuoka University), respectively. Monoclonal mouse antibodies to golgin-245, GM130, and green fluorescent protein (GFP) were purchased from BD Biosciences. Monoclonal mouse anti-ARF1 antibody (3F1) was from Affinity Bioreagents. Polyclonal rabbit and monoclonal mouse anti-β-COP antibodies were from Affinity Bioreagents and Sigma-Aldrich, respectively. Polyclonal rabbit anti-syntaxin 5 antibody was from Synaptic Systems. AlexaFluor- and horseradish peroxidase-conjugated secondary antibodies were from Molecular Probes and Jackson Immuno-Research Laboratories, respectively.
An expression vector for N-terminal EGFP-tagged VSVG tsO45 (pCIpreEGFP-VSVG) was constructed by exchanging the mature VSVG tsO45 segment from pcDNA3-VSVG-EGFP (33) for the cation-independent mannose 6-phosphate receptor cDNA segment of pCIpreEGFP-CIMPRtail (34). Construction of an expression vector for C-terminal HA-tagged ARFRP1 was described previously (35). An expression vector for N-terminal EGFP+VSVG-tagged KDELR (pCIpreEGFP-VSVG-KDELR) was constructed by exchanging a cDNA segment of the mature human KDELR2 region (kindly provided by Victor Hsu, Harvard Medical School) (36, 37) for the region covering the transmembrane and cytoplasmic regions of VSVG in pCIpreEGFP-VSVG.
Cell Culture, RNAi Suppression, VSVG Transport Experiments, and Immunofluorescence Analysis—Culture of HeLa cells and transfection of expression plasmids were performed as described previously (35, 38). Knockdown of ArfGAP1, ArfGAP2, ArfGAP3, or β-COP was performed as previously described (39). Briefly, pools of siRNAs directed for the mRNA regions covering nucleotide residues 328-947, 400-1542, 397-1530, and 321-1040 (when the A residue of the initiation Met codon is assigned as residue 1) were prepared using a BLOCK-iT RNAi TOPO Transcription kit and a BLOCK-iT Dicer RNAi kit (Invitrogen). Cells were transfected with the siRNAs using Lipofectamine 2000 (Invitrogen) and incubated overnight. The transfected cells were then transferred to a culture dish containing coverslips, further incubated for up to 120 and 48 h in the case of ArfGAP knockdown and β-COP knockdown, respectively, and processed for immunofluorescence and immunoblot analyses and transport assays.
Transport of EGFP-VSVG from the ER to the cell surface was examined as described previously (35). HeLa cells transfected with pCIpreEGFP-VSVG were incubated at 40 °C overnight, then at 32 °C for up to 60 min. Retrograde transport of the EGFP-VSVG-KDELR chimera was assayed as follows; HeLa cells transfected with pCIpreEGFP-VSVG-KDELR were incubated at 40 °C overnight and at 20 °C for 3 h to accumulate the KDELR chimera in the Golgi. The cells were then incubated at 40 °C again for 2 h and processed for immunofluorescence analysis. Internalization of extracellularly applied transferrin was examined as described previously (38).
Determination of Intracellular Level of GTP-bound ARF—To determine the level of GTP-bound ARF, lysates of control cells or of siRNA-treated cells were subjected to pulldown assays using the GAT domain of GGA1, as described previously (40, 41). Briefly, lysates were pulled down with the glutathione S-transferase-GGA1(GAT) domain, pre-bound to glutathione-Sepharose 4B beads (GE Healthcare Biosciences), and bound materials were electrophoresed on a 12.5% SDS-polyacrylamide gel and subjected to immunoblot analysis with anti-ARF1 antibody (3F1).
Electron Microscopy—For conventional electron microscopy, control and siRNA-treated cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 m phosphate buffer. They were post-fixed with 1% OsO4, embedded in Epon812, and sectioned as previously described (42). Small vesicles/tubules near vacuoles were counted in 1.6 × 1.6-μm regions that were randomly selected within perinuclear region (40 areas from 20 cells). The count was normalized to the total area, excluding the vacuolar area, measured using the Meta-Morph image processing software (Universal Imaging Corporation; West Chester, PA), and expressed as the number of vesicles/tubules per 1 μm2. Immunoelectron microscopy on ultrathin cryosections was performed as described previously (35, 42).
RESULTS
Knockdown of ArfGAP1 Alone Has No Discernible Effect—In the prevailing model (5) ArfGAP1 is involved in cargo selection and formation of COPI-coated vesicles, as well as in their uncoating through inactivating ARFs. To explore the role of ArfGAP1 in COPI-mediated transport, we looked for phenotypic changes in cells depleted of ArfGAP1 by RNAi. By transfecting HeLa cells with a pool of siRNAs targeted against ArfGAP1, the ArfGAP1 protein was depleted by more than 95% (supplemental Fig. S1A, top panel), and a typical Golgi-like staining for ArfGAP1 was eliminated (supplemental Fig. S1B). We then compared subcellular localization of various proteins between cells depleted of ArfGAP1 (supplemental Fig. S2B) and control cells transfected with siRNAs for LacZ (supplemental Fig. S2A). However, we failed to detect a significant difference between the knockdown and control cells in the localization of proteins. The proteins examined included: β-COP (top panels), a COPI subunit; ERGIC-53 (middle panels), which is a major type I membrane protein containing a C-terminal di-lysine motif and which cycles between the cis-Golgi and ER (43); GM130 (bottom panels), a cis-Golgi matrix protein (44). We also examined the effects of ArfGAP1 knockdown on retrograde and anterograde transport of model cargo proteins. However, ArfGAP1 knockdown affected neither retrieval of a VSVG-KDELR construct from the cis-Golgi to the ER nor anterograde transport of a GFP-VSVG construct from the ER to the plasma membrane through the Golgi (data not shown; see below).
Simultaneous Depletion of ArfGAP1, ArfGAP2, and ArfGAP3 Disorganizes the Golgi Structure—We then examined a possibility that there might be another ARFGAP(s) that also participates in the COPI-mediated process. In mammals there are less well characterized paralogs (ArfGAP2 and ArfGAP3) of ArfGAP1. At the sequence level, ArfGAP1 is similar to yeast Gcs1, whereas ArfGAP2 and ArfGAP3 are more similar to Glo3 (26-29).
We raised and affinity-purified polyclonal antibodies to ArfGAP2 and ArfGAP3 and used them to compare protein localization with that of various marker proteins. As shown in supplemental Fig. S1A, the antibodies each recognize a single band in immunoblot analysis; band intensities were specifically decreased by the appropriate siRNA treatment. When HeLa cells were doubly stained for any one of the ArfGAPs and GM130, the staining for ArfGAPs overlapped almost completely with the GM130 staining in the perinuclear Golgi region (supplemental Fig. S3, A-C). In contrast, the ArfGAP staining was juxtaposed to, but not significantly overlapping with staining for golgin-245 (supplemental Fig. S3, D-F), a protein associated with the trans-Golgi (45). To unequivocally show cis-Golgi localization of these ArfGAPs, HeLa cells were treated with nocodazole to fragment the Golgi structure; the fragmented Golgi structures are composed of mini-stacks and are suitable for examining cis and trans polarity (46). In the fragmented Golgi structures, the staining for any ArfGAP overlapped almost completely with that for GM130 (supplemental Fig. S3, G-I) but juxtaposed to that for golgin-245 (supplemental Fig. S3, J-L), indicating that these three ArfGAPs are associated predominantly with the cis-Golgi.
We then examined whether depletion of ArfGAP2 or ArfGAP3 affected the localization of Golgi proteins and transport between the ER and Golgi. Despite the successful depletion of ArfGAP2 or ArfGAP3 by RNAi (supplemental Fig. S1), however, we failed to detect any difference in either the subcellular localization of Golgi proteins examined, anterograde transport of GFP-VSVG from the ER to the Golgi, or retrograde transport of VSVG-KDELR from the cis-Golgi to the ER (data not shown) between control cells and cells knocked down of ArfGAP2 or ArfGAP3.
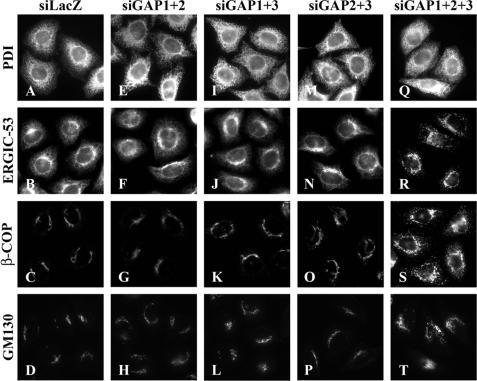
We next examined effects of depletion of any pairwise combination of ArfGAPs but failed to detect any significant difference in the subcellular distribution of the Golgi proteins, including ERGIC-53, β-COP, and GM130, between control cells and double-depleted cells (ArfGAP1 + 2, ArfGAP1 + 3, or ArfGAP2 + 3) (Fig. 1).
FIGURE 1.
Disorganization of the Golgi apparatus in cells simultaneously depleted of ArfGAP1, ArfGAP2, and ArfGAP3. HeLa cells were treated for 120 h with siRNAs for LacZ (A-D) or for ArfGAP1+ArfGAP2 (E-H), ArfGAP1+ArfGAP3 (I-L), ArfGAP2+ArfGAP3 (M-P), or ArfGAP1+ArfGAP2+ArfGAP3 (Q-T) and stained for protein disulfide isomerase (A, E, I, M, and Q), ERGIC-53 (B, F, J, N, and R), β-COP (C, G, K, O, and S), or GM130 (D, H, L, P, and T).
However, in a subpopulation (∼30%) of cells in which ArfGAP1, ArfGAP2, and ArfGAP3 were simultaneously knocked down, we found significant changes in the distribution of the Golgi proteins. ERGIC-53 disappeared from reticular ER-like structures and instead became predominant in punctate ERGIC-like structures (Fig. 1R). In contrast, the reticular staining for protein disulfide isomerase, an ER marker, was unchanged in the triple knockdown cells (Fig. 1Q), indicative of the integrity of the ER. These observations suggest that retrieval of ERGIC-53 from the ERGIC/cis-Golgi to the ER is blocked in cells simultaneously depleted of ArfGAP1, ArfGAP2, and ArfGAP3. Furthermore, β-COP and GM130 were redistributed from perinuclear Golgi-like structures to punctate ERGIC-like structures in a subpopulation of the triple knockdown cells (Fig. 1, S and T).
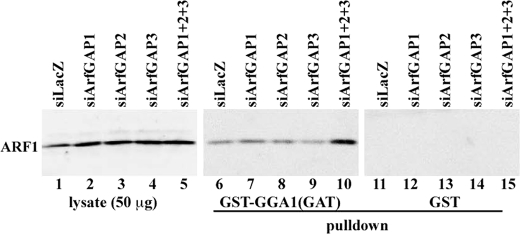
To support the hypothesis that these phenotypic changes are indeed induced by ArfGAP depletion, we then compared levels of GTP-bound active ARF in the control and knockdown cells. To this end we employed a pulldown assay using the GAT domain of GGA1, which specifically interacts with GTP-bound but not GDP-bound ARF (38, 47) under the assumption that GTP hydrolysis on ARF would be slowed (i.e. that the level of GTP-bound ARF would be increased) if the ArfGAP levels were decreased. As shown in Fig. 2, the level of GTP-bound ARF was not significantly changed in cells depleted of single ArfGAP (lanes 7-9) as compared with the control cells (lane 6). In striking contrast, the level of GTP-bound ARF was robustly increased in the triple ArfGAP knockdown cells (lane 10). Thus, the phenotypic changes observed in the triple knockdown cells are correlated with an increase in the level of GTP-bound ARF, i.e. with a decrease in the ArfGAP levels.
FIGURE 2.
Increase in the GTP-bound ARF level in cells simultaneously knocked down of ArfGAP1, ArfGAP2, and ArfGAP3. Lysates were prepared from HeLa cells treated for 120 h with siRNAs for LacZ (lanes 1, 6, and 11) or for ArfGAP1 (lanes 2, 7, and 12), ArfGAP2 (lanes 3, 8, and 13), ArfGAP3 (lanes 4, 9, and 14), or ArfGAP1+ArfGAP2+ArfGAP3 (lanes 5, 10, and 15) and subjected to pulldown with glutathione S-transferase (GST)-GGA1(GAT) (lanes 6-10) or GST (lanes 11-15). The data are representatives of five independent experiments for the control and triple ArfGAP knockdown cells and two independent experiments for the single ArfGAP knockdown cells.
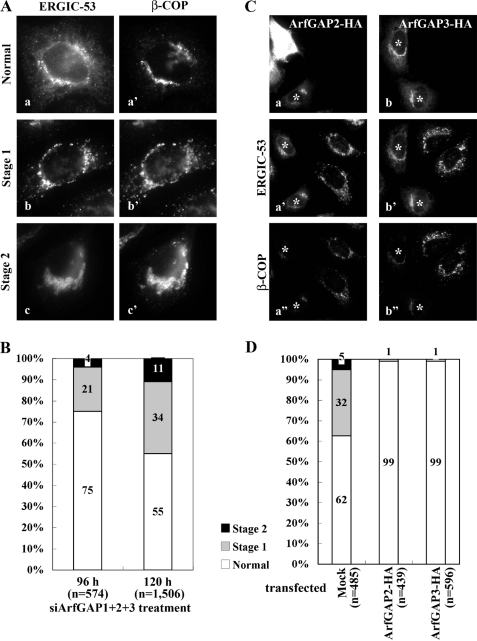
In the course of these experiments, we noticed that the triple knockdown cells showed two distinct phenotypes in terms of subcellular distribution of ERGIC-53 and β-COP. As shown in Fig. 3A, a fraction of these cells showed a punctate ERGIC-like distribution of ERGIC-53 and β-COP (stage 1), whereas a minor fraction showed perinuclear aggregates of ERGIC-53 and β-COP (stage 2). Extending the period of siRNA treatment (96 to 120 h) resulted in a significant increase in the fraction of both the stage 1 (21-34%) and stage 2 (4-11%) cells (Fig. 3B), suggesting that the stage 2 phenotype is the more severe of the two. However, further siRNA treatment resulted in a decrease in the stage 1 and stage 2 populations (data not shown), probably because of the death of cells completely depleted of all the three ArfGAPs, as reported previously (27), and because of selective growth of cells incompletely depleted of any of the three ArfGAPs. For the following experiments we, therefore, used cells treated for 120 h with siRNAs for ArfGAP1, ArfGAP2, and ArfGAP3.
FIGURE 3.
Two distinct phenotypic stages in cells triple-depleted of ArfGAP1, ArfGAP2, and ArfGAP3. A, HeLa cells treated for 120 h with siRNAs for ArfGAP1+ArfGAP2+ArfGAP3 were double-stained for ERGIC-53 (a-c) and β-COP (a′-c′). Cells with normal distribution of ERGIC-53 and β-COP (a and a′) or with typical stage 1 (b and b′) or stage 2 (c and c′) distributions are shown. B, HeLa cells treated with siRNAs for ArfGAP1+ArfGAP2+ArfGAP3 for 96 or 120 h were classified as having normal, stage 1, and stage 2 distributions of ERGIC-53, and the number of cells with each distribution was counted. The number (n) is the sum of counted cells in three independent experiments. C, HeLa cells treated with siRNAs for ArfGAP1+ArfGAP2+ArfGAP3 for 96 h were transfected with an expression vector for C-terminal-HA-tagged ArfGAP2 (a-a″) or ArfGAP3 (b-b″) and incubated for 24 h, then triply stained for HA (a and b), ERGIC-53 (a′ and b′) and β-COP (a′ and b′). Cells overexpressing ArfGAP2-HA or ArfGAP3-HA are indicated by asterisks. D, the number of cells with normal, stage 1, and stage 2 distribution of ERGIC-53 in mock-transfected cells or cells overexpressing either ArfGAP2-HA or ArfGAP3-HA. The number (n) is the sum of counted cells in three independent experiments.
The stage 1 and 2 phenotypes observed in the triple ArfGAP knockdown cells were restored by overexpressing HA-tagged ArfGAP2 or ArfGAP3 (Fig. 3, C and D). These results indicate that the phenotypes result from simultaneous depletion of these three ArfGAPs but not from off-target effects. The fact that the overexpression of any one of the three ArfGAPs is able to restore the triple knockdown phenotypes is consistent with the lack of significant effects observed in the single or double knockdown cells (Fig. 1 and supplemental Fig. S2). However, we could not examine the effects of ArfGAP1 overexpression because overexpression of ArfGAP1 on its own induces disintegration of the Golgi structure as reported previously (11). The differential effects of overexpressing ArfGAP1 and of overexpressing ArfGAP2 and ArfGAP3 on the Golgi organization may be because of their structural difference outside of the catalytic domain; namely, the former, but not the latter two, has a membrane curvature-sensing motif (23, 24).
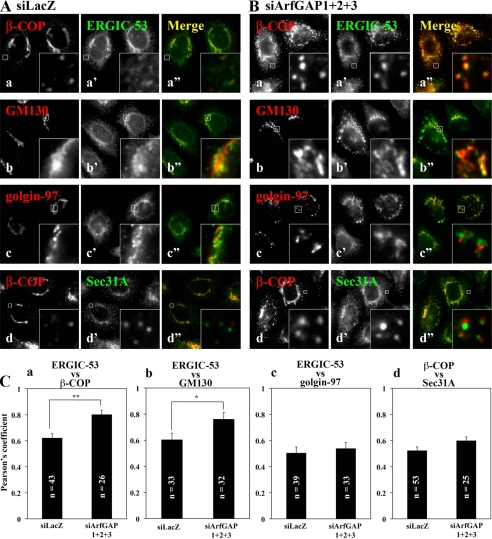
When we compared the subcellular distributions of ERGIC-53 and β-COP, we observed an almost complete overlap between the two proteins on punctate ERGIC-like structures in the stage 1 population of the triple knockdown cells (Fig. 4B, a-a″), in contrast to a rare overlap on punctate structures in the control cells (Fig. 4A, a-a″; also see Fig. 4C). In the triple ArfGAP knockdown cells, staining for ERGIC-53 showed significant, albeit not complete, overlap with that of GM130 (Fig. 4B, b-b″; also see Fig. 4C). The staining for golgin-97, a trans-Golgi marker, was also fragmented but not significantly overlapping with the punctate ERGIC-53 staining in the triple knockdown cells (Fig. 4B, c-c″), suggesting that, by the simultaneous depletion of the three ArfGAPs, the entire Golgi structure is fragmented, but its cis-trans polarity is maintained.
FIGURE 4.
Redistribution of Golgi proteins in cells triple-depleted of ArfGAPs. HeLa cells were treated for 120 h with siRNAs for LacZ (A) or ArfGAP1+ArfGAP2+ArfGAP3 (B) and processed for double staining for β-COP (a and d), GM130 (b), or golgin-97 (c), and either ERGIC-53 (a′-c′) or Sec31A (d′). Merged images are shown in a″-d″. Boxed regions are enlarged and shown in the corresponding insets. C, colocalization of the indicated proteins in cells treated with siRNAs for LacZ and ArfGAP1+ArfGAP2+ArfGAP3 was estimated using an IPLab 4.0 software (Solution Systems) and expressed as a Pearson coefficient. **, p < 0.0001; *, p < 0.001.
The punctate staining for Sec31A, which is a component of the COPII coat and which is associated exclusively with ER exit sites, was not altered in the triple knockdown cells (compare Fig. 4, Ad′ and Bd′), indicating that ER exit sites were not affected by the triple knockdown. When we compared the localization of β-COP with that of Sec31A we found that the punctate structures positive for these two proteins did not overlap significantly but were often juxtaposed with each other in the triple knockdown cells. It is likely that in the triple ArfGAP-depleted cells COPI-coated intermediates were arrested in the vicinity of the ER exit sites for block in the uncoating process. These observations together are consistent with the assumption that these ArfGAPs are all responsible for inactivation of ARFs and, thereby, for uncoating of the COPI coat. Thus, preventing the COPI uncoating through interfering with ARF inactivation by depletion of ArfGAPs might result in accumulation of COPI-coated intermediates.
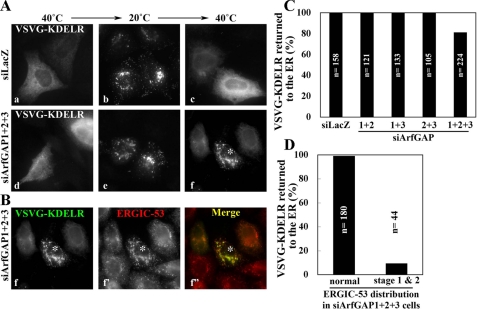
Simultaneous Depletion of ArfGAP1, ArfGAP2, and ArfGAP3 Blocks Retrograde Transport from the Golgi to the ER—Given the punctate ERGIC-like distribution of ERGIC-53 in cells simultaneously depleted of ArfGAP1, ArfGAP2, and ArfGAP3, in contrast to its ER- and Golgi-like distribution in control cells, we speculated that retrograde transport from the Golgi to the ER is blocked in the triple knockdown cells. To address this speculation, we examined retrograde transport of a KDELR chimera. The chimera we utilized is a modified version of the original VSVG-KDELR chimera (36, 37), which contains (sequentially from the N terminus) a signal peptide sequence, GFP, the exoplasmic domain of a temperature sensitive (tsO45) form of the vesicular stomatitis virus G protein (VSVG), and KDELR2 (see “Experimental Procedures”). The GFP-VSVG-KDELR chimera accumulates in the ER at the non-permissive temperature (40 °C; Fig. 5Aa) because of misfolding of the VSVG segment and is delivered to the Golgi at the permissive temperature (<32 °C, Fig. 5Ab at 20 °C). However, upon shifting again to the non-permissive temperature, the chimera is redistributed from the Golgi to the ER (Fig. 5Ac; see Refs. 36 and 37) because the misfolded VSVG segment prevents ER exit of the GFP-VSVG-KDELR chimera, which has once returned to the ER from the Golgi. As shown in Fig. 5Ae, the chimera successfully escaped from the ER at the permissive temperature in the triple knockdown cells. However, in a fraction of triple ArfGAP knockdown cells we found that the chimera could not undergo retrograde transport to the ER at the non-permissive temperature (Fig. 5Af; a cell marked by an asterisk) and instead accumulated in the ERGIC-53-positive compartments (Fig. 5B, f-f″; compare the cell marked by an asterisk with cells surrounding it having normal ERGIC-53 distribution), in contrast to the observations of the control cells (Fig. 5A, a-c). Although the retrograde transport of the KDELR chimera was blocked in ∼20% of cells subjected to simultaneous knockdown of the three ArfGAPs (Fig. 5C), almost all the cells exhibiting a stage 1 or stage 2 phenotype of ERGIC-53 showed a considerable retrieval block (Fig. 5D); this is probably because of incomplete depletion of any one of the three ArfGAPs in any given cell. These observations indicate that the Golgi-to-ER retrograde transport of KDELR is blocked in the cells simultaneously depleted of ArfGAP1, ArfGAP2, and ArfGAP3.
FIGURE 5.
Block in Golgi-to-ER transport in cells triple-depleted of ArfGAPs. HeLa cells were treated with siRNAs for LacZ (A, a-c) or ArfGAP1+ArfGAP2+ArfGAP3 (A, d-f, and B, f-f″) and transfected with the expression vector for the EGFP-VSVG-KDELR chimera. After transfection, the cells were incubated at 40 °C overnight (A, a and d), successively incubated at 20 °C for 3 h (A, b and e) and 40 °C for 2 h (A, c and e, and B) in the presence of 50 μg/ml cycloheximide, and processed for staining for ERGIC-53. Note that panels Af and Bf represent the same single image. C, after the final 40 °C incubation, the cells were scored for ER distribution of the EGFP-VSVG-KDELR chimera. The bar graph represents the percentage of cells with ER distribution of the chimera among all cells expressing the chimera. The number (n) is the sum of counted cells in two independent experiments. D, the cells triple-knocked down of ArfGAPs were classified as having normal, stage 1, and stage 2 distributions of ERGIC-53 (for example, see a cell indicated by an asterisk in B). The bar graph represents the percentage of cells with ER distribution of the chimera in each cell population. The number (n) is the sum of counted cells in two independent experiments.
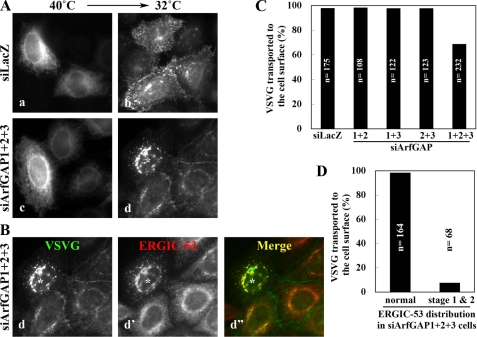
Simultaneous Depletion of ArfGAP1, ArfGAP2, and ArfGAP3 Block Anterograde Transport of VSVG—Because ArfGAP depletion affects the morphology of Golgi compartments, we next assessed the functional integrity of the exocytic pathway through the Golgi. To this end we followed trafficking from the ER to the plasma membrane of a GFP-VSVG tsO45 construct, which is a well characterized marker for the exocytic pathway (48). In control cells the GFP-VSVG construct accumulated in the ER at the non-permissive temperature (40 °C, Fig. 6Aa) was transported to the plasma membrane after temperature shift to 32 °C for 60 min (Fig. 6Ab). In contrast, in a fraction of cells triply knocked down of ArfGAPs (Fig. 6Ad; a cell marked by an asterisk), GFP-VSVG was transported to the Golgi but not farther to the plasma membrane. The epifluorescence of intracellular GFP-VSVG showed a superimposition to the ERGIC-53 staining (Fig. 6B; d-d″), indicating that anterograde transport was blocked at the ERGIC and/or cis-Golgi level. Although ∼30% cells simultaneously knocked down of the three ArfGAPs showed a block in VSVG transport (Fig. 6C), almost all the cells exhibiting the stage 1 or stage 2 phenotype of ERGIC-53 showed the VSVG transport block (Fig. 6D).
FIGURE 6.
Block in anterograde transport through the Golgi in cells triple-depleted of ArfGAPs. HeLa cells were treated with siRNAs for LacZ (A, a and b) or ArfGAP1+ArfGAP2+ArfGAP3 (A, c and d, and B, d-d″) and transfected with the expression vector for EGFP-VSVG. After transfection the cells were incubated at 40 °C overnight (A, a and c), and subsequently at 32 °C for 60 min in the presence of 50 μg/ml cycloheximide (A, b and d, and B). The cells were then stained for ERGIC-53. Note that panels Ad and Bd represent the same single image. C, after the 32 °C incubation, the cells were scored for cell surface delivery of EGFP-VSVG. The bar graph represents the percentage of cells with surface EGFP-VSVG expression among all cells expressing EGFP-VSVG. The number (n) is the sum of counted cells in two independent experiments. D, the cells triple-knocked down of ArfGAPs were classified as having normal, stage 1, and stage 2 distributions of ERGIC-53 (for example, see the cell indicated by an asterisk in B). The bar graph represents the percentage of cells in each cell population with surface expression of EGFP-VSVG. The number (n) is the sum of counted cells in two independent experiments.
Simultaneous Depletion of ArfGAP1, ArfGAP2, and ArfGAP3 Does Not Affect Endocytosis—We then examined whether the triple ArfGAP knockdown affected compartments beyond the Golgi. The triple knockdown had no discernible effects on localization of an endosomal marker, EEA1 (data not shown). Furthermore, as shown in supplemental Fig. S4, internalization of extracellularly applied AlexaFluor488-conjugated transferrin through early endosomes (10 and 30 min) to recycling endosomes (30 and 60 min) was not significantly altered in the stage 1 or stage 2 cells (marked by 1 and 2, respectively, in the middle panels) as compared with the normal phenotype cells (cells with no mark in the middle panels) and the control siRNA-treated cells (top panels). These observations together indicate that the ArfGAP depletion specifically affect the early secretory and retrieval pathways.
Phenotypic Similarity between Cells Depleted of ArfGAPs and Those Depleted of COPI—In the course of the present study we noticed that the phenotypes observed in the triple-ArfGAP-depleted cells resembled those in COPI-depleted cells (49, 50); namely, β-COP depletion by siRNA resulted in redistribution of ERGIC-53 and giantin to large globular structures scattered throughout the cytoplasm. These structures are reminiscent of the stage 2 phenotype of ERGIC-53 distribution observed in the ArfGAP-depleted cells (Fig. 3Ac). As shown in supplemental Fig. S5, we confirmed that the number of cells with stage 1 and stage 2 phenotypes of ERGIC-53 distribution increased in a time-dependent manner (12 and 24 h) upon treatment with β-COP siRNAs. Longer treatment of cells with β-COP siRNAs resulted in cell death (data not shown) as noted previously (49).
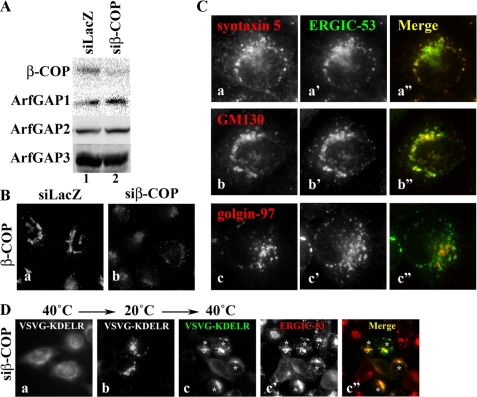
Efficient depletion of β-COP by RNAi was confirmed by immunoblot and immunofluorescence analyses (Fig. 7, A and B). As observed for cells with triple ArfGAP depletion (Fig. 4), the β-COP-depleted cells with the stage 1 phenotype showed significant colocalization of ERGIC-53 and cis-Golgi markers (syntaxin 5 and GM130) on punctate ERGIC-like structures (Fig. 7C, a-a″ and b-b″) and juxtaposition of ERGIC-53 and golgin-97, a trans-Golgi marker (Fig. 7C, c-c″). Furthermore, retrieval of the VSVG-KDELR chimera from the Golgi to the ER was blocked in the β-COP-depleted cells, and the chimera was accumulated in the ERGIC-53-positive structures (Fig. 7D). Thus, cells with triple ArfGAP depletion and those with COPI depletion showed similar phenotypes, both with regard to protein localization and protein transport.
FIGURE 7.
Cells depleted of COPI show phenotypes similar to those of cells triple-depleted of ArfGAPs. HeLa cells were treated with siRNAs for LacZ (A, lane 1, and Ba) or β-COP (A, lane 2, Bb, C, and D) for 24 h. A, cell extracts were prepared and subjected to immunoblot analysis using antibody against β-COP, ArfGAP1, ArfGAP2, or ArfGAP3. B, the cells were immunostained for β-COP. C, β-COP-depleted cells were doubly stained for either syntaxin 5 (a), GM130 (b), or golgin-97 (c) and ERGIC-53 (a′-c′). D, β-COP-depleted cells were subjected to retrograde transport assay as described under the legend for Fig. 5, A and B. Asterisks indicate cells showing stage 1 and 2 distribution of ERGIC-53.
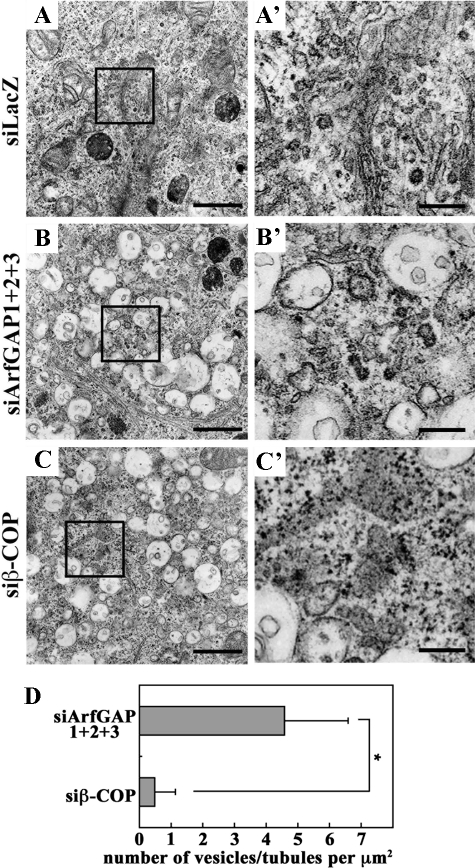
We then examined the similarity in the morphological changes between the cells triple-depleted of ArfGAPs and those depleted of β-COP at the electron microscopic level. Although typical Golgi stacks were observed in control cells (Fig. 8A), they were poorly observed in cells triple-depleted of ArfGAPs (Fig. 8B) and in those depleted of β-COP (Fig. 8C). Instead, both in the ArfGAP and β-COP knockdown cells, clusters of vacuoles were often found in the perinuclear region. It is noteworthy that the vacuoles often contained internal membranes, some of which were continuous with their outermost membranes. We also noticed that the triple ArfGAP knockdown cells possessed many vesicular or tubular structures in the vicinity of the vacuoles, often closely associated with them (Fig. 8B′); in contrast, such structures were barely detectable in the β-COP knockdown cells (Fig. 8, C′ and D; see “Discussion”).
FIGURE 8.
Electron microscopic analyses of cells triple-depleted of ArfGAPs and those depleted of β-COP. HeLa cells treated for 120 h with siRNAs for LacZ (A) or ArfGAP1 + ArfGAP2 + ArfGAP3 (B) or for 48 h with siRNAs for β-COP (C) were processed for electron microscopic analysis. Boxed regions are enlarged and shown in A′, B′, and C′. Bars:5 μm in A-C; 1 μm in A′-C′. D, the number of vesicular and tubular structures surrounded by vacuoles was counted. *, p < 0.001 (n = 40).
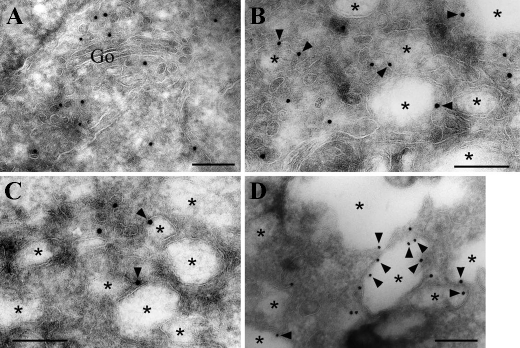
We then performed immunoelectron microscopic analysis of the triple ArfGAP knockdown cells using anti-β-COP antibody. In the control cells (Fig. 9A), β-COP was localized mainly on vesicles around the Golgi stacks, a subset of which might possibly represent the cis-Golgi compartments. In striking contrast, β-COP signals were found mainly on the rims of vacuoles (arrowheads) and on vesicles in regions surrounded by the vacuoles in the triple ArfGAP knockdown cells (Fig. 9, B-D). These observations suggest that the vacuolar structures are accumulated through perturbation of COPI function caused by the ArfGAP depletion.
FIGURE 9.
Association of β-COP with vacuolar structures in cells triple-depleted of ArfGAPs. HeLa cells treated for 120 h with siRNAs for LacZ (A) or ArfGAP1+ArfGAP2+ArfGAP3 (B-D) were processed for immunoelectron microscopic analysis. Ultrathin cryosections were stained with polyclonal anti-β-COP antibody (1:10) followed by goat anti-rabbit IgG conjugated with 10-nm colloidal gold. Note that β-COP is localized on vesicles near the Golgi (Go) in the control cells (A), whereas it is found on rims (arrowhead) of the vacuoles (asterisks) and vesicular structures in the triple knockdown cells (B-D). Bars, 0.2 μm.
DISCUSSION
Although ArfGAP1 has been shown to be responsible for COPI-mediated trafficking between the Golgi and ER, the roles of ArfGAP2 and ArfGAP3 have remained poorly understood. In the present study we observed no significant phenotypic changes in cells depleted of any one of the three ArfGAPs or any two in pairwise combinations. However, simultaneous depletion of the three ArfGAPs has considerable impacts on Golgi morphology and trafficking between the Golgi and ER. Specifically, in the triple ArfGAP knockdown cells, we observe accumulation of cis-Golgi proteins, including COPI, in ERGIC-like structures and a concomitant block in retrograde transport from the Golgi to the ER. A substantial elevation in the level of GTP-bound ARFs in the triple knockdown cells supports the hypothesis that these phenotypic changes are indeed caused by depletion of these ArfGAPs. These observations indicate that the three ArfGAPs, at least in part, have overlapping functions; the data are in line with previous findings that neither deletion of GCS1 nor GLO3 has an effect on yeast growth, whereas deletion of both genes is lethal (7). In the context of the overlapping ArfGAP functions, it is interesting to note that when any one of the three ArfGAPs (or any pairwise combination) was depleted, the level of the other ArfGAP(s) was disposed to increase (see supplemental Fig. S1A), although we have not further addressed this issue in the present study. Thus, the ArfGAPs could compensate for each other under conditions where the level of any one of the ArfGAPs is somehow decreased.
The other notable finding in the present study is that the phenotypes observed in cells with triple ArfGAP depletion closely resemble those in β-COP-depleted cells, which show accumulation of cis-Golgi proteins in ERGIC-like structures and a block in Golgi-to-ER retrograde transport. Moreover, at the electron microscopic level, both the triple ArfGAP- and β-COP-depleted cells accumulate characteristic vacuolar structures. Based on these observations, it is likely that phenotypic changes induced by the triple ArfGAP knockdown result largely, if not completely, from perturbation of COPI functions. In this context it is worth mentioning that numerous vesicles and tubules associated with vacuoles are present in triple-ArfGAP-depleted cells but are barely detectable in β-COP-depleted cells (Fig. 8D). Because β-COP is found on these vesicles/tubules as well as on rims of the vacuoles by immunoelectron microscopy, it is likely that a fraction of the vesicles represents COPI-coated vesicles. These observations are consistent with early in vitro reconstitution experiments suggesting that COPI vesicles can be formed in the absence of ArfGAPs (9). However, more recent studies have indicated that ArfGAPs positively regulate COPI-coated vesicle formation by coupling cargo selection and vesicle uncoating through ARF inactivation (18-20). In the triple-ArfGAP-depleted cells, COPI-coated vesicles might be formed with very low efficiency because of substantial, but not complete depletion of ArfGAPs by RNAi. Once formed, however, COPI-coated vesicles might be accumulated for the inefficient uncoating process.
A recent study (27) reported that ArfGAP2 and ArfGAP3, but not ArfGAP1, associate with COPI-coated vesicles in the presence of GTPγS in vitro, in good agreement with data pertaining to yeast Gcs1 and Glo3 (6). Furthermore, while this manuscript was in preparation two independent groups (51, 52) showed biochemically that although ArfGAP1 is recruited directly onto membranes via its membrane curvature-sensing motif as previously shown (23, 24), ArfGAP2 and ArfGAP3 are recruited onto membranes in the presence of coatomer and ARF, and their GAP activities are highly stimulated by coatomer. These studies suggest differential roles of ArfGAP1 and ArfGAP2/ArfGAP3; ArfGAP2 and ArfGAP3 are COPI coat-dependent ARFGAPs, whereas ArfGAP1 has a more general role.
How can we reconcile our cell biological data and yeast genetic data, suggesting overlapping functions of ArfGAP1-(Gcs1) and ArfGAP2/ArfGAP3(Glo3) in COPI-mediated trafficking, with biochemical data suggesting differential roles? The most likely explanation is that ArfGAP2 and ArfGAP3 regulate COPI-dependent trafficking, whereas ArfGAP1 is involved in both COPI-dependent and -independent processes and shares COPI-independent functions, such as clathrin/AP-1-mediated transport, with other ARF-GAPs. For example, AGAP2 and SMAP2 have been suggested to function in AP-1-dependent transport (53, 54). Another intriguing possibility is that formation of COPI-coated vesicles is optimized by the concerted actions of ArfGAP1, which senses membrane curvature (23, 24), and ArfGAP2/ArfGAP3, which interact with coatomer (27, 51) even though vesicle formation requires a single ArfGAP.
Finally, it remains possible that ArfGAP2 and ArfGAP3 favor distinct populations of COPI-coated vesicles, as the appendage domain of γ-COP (γ1-COP) has been reported to bind strongly to ArfGAP2 but only weakly to ArfGAP3 (55). In mammals there are two isoforms of γ-COP (γ1 and γ2) and two isoforms of ζ-COP (ζ1 and ζ2) (56) which are incorporated into coatomer in any combination (57) and show differential localization within the Golgi apparatus (58). Our future studies will, therefore, focus on potential differences between ArfGAP2 and ArfGAP3 functions.
In the present study we have established a system in which ArfGAP1, ArfGAP2, and ArfGAP3 are simultaneously knocked down. In this system phenotypic changes induced by the triple knockdown can be recovered by exogenous expression of any one of these ArfGAPs. The system will be helpful in future studies aimed at understanding both the common and specific functions of these ArfGAPs.
Supplementary Material
Acknowledgments
We thank Hans-Peter Hauri, Nobuhiro Nakamura, Yoshio Misumi, Jennifer Lippincott-Schwartz, and Victor W. Hsu for kindly providing materials. We also thank Atsuko Yabashi and Katsuyuki Kanno for excellent techniques in electron microscopy.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Japan Society for Promotion of Science, the Targeted Proteins Research Program, the International Young Scientists Career Development Organization (ICDO) of Kyoto University, the Uehara Memorial Foundation, the NOVARTIS Foundation (Japan) for the Promotion of Science, and the Hayashi Memorial Foundation for Female Natural Scientists.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5.
Footnotes
The abbreviations used are: ARF, ADP-ribosylation factor; COPI, coat protein I; ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartment; GAP, GTPase-activating protein; KDELR, KDEL receptor; siRNA, small interfering RNA; RNAi, RNA interference; VSVG, vesicular stomatitis virus G protein; GTPγS, guanosine 5′-3-O-(thio)triphosphate; GFP, green fluorescent protein; EGFP, enhanced GFP; HA, hemagglutinin; si-, small interfering-.
References
- 1.Nickel, W., and Wieland, F. T. (1998) Histochem. Cell Biol. 109 477-486 [DOI] [PubMed] [Google Scholar]
- 2.Donaldson, J. G., Honda, A., and Weigert, R. (2005) Biochim. Biophys. Acta 1744 364-373 [DOI] [PubMed] [Google Scholar]
- 3.Gillingham, A. K., and Munro, S. (2007) Annu. Rev. Cell Dev. Biol. 23 579-611 [DOI] [PubMed] [Google Scholar]
- 4.Shin, H.-W., and Nakayama, K. (2004) J. Biochem. (Tokyo) 136 761-767 [DOI] [PubMed] [Google Scholar]
- 5.Nie, Z., and Randazzo, P. A. (2006) J. Cell Sci. 119 1203-1211 [DOI] [PubMed] [Google Scholar]
- 6.Lewis, S. M., Poon, P. P., Singer, R. A., Johnston, G. C., and Spang, A. (2004) Mol. Biol. Cell 15 4064-4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poon, P. P., Cassel, D., Spang, A., Rotman, M., Pick, E., Singer, R. A., and Johnston, G. C. (1999) EMBO J. 18 555-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cukierman, E., Huber, I., Rotman, M., and Cassel, D. (1995) Science 270 1999-2002 [DOI] [PubMed] [Google Scholar]
- 9.Tanigawa, G., Orci, L., Amherdt, M., Ravazzola, M., Helms, J. B., and Rothman, J. E. (1993) J. Cell Biol. 123 1365-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teal, S. B., Hsu, V. W., Peters, P. J., Klausner, R. D., and Donaldson, J. G. (1994) J. Biol. Chem. 269 3135-3138 [PubMed] [Google Scholar]
- 11.Aoe, T., Cukierman, E., Lee, A., Cassel, D., Peters, P. J., and Hsu, V. W. (1997) EMBO J. 16 7305-7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhard, C., Schweikert, M., Wieland, F. T., and Nickel, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8253-8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szafer, E., Rotman, M., and Cassel, D. (2001) J. Biol. Chem. 276 47834-47839 [DOI] [PubMed] [Google Scholar]
- 14.Goldberg, J. (1999) Cell 96 893-902 [DOI] [PubMed] [Google Scholar]
- 15.Lanoix, J., Ouwendijk, J., Lin, C.-C., Stark, A., Love, H. D., Ostermann, J., and Nilsson, T. (1999) EMBO J. 18 4935-4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickel, W., Malsam, J., Gorgas, K., Ravazzola, M., Helms, J. B., and Wieland, F. T. (1998) J. Cell Sci. 111 3081-3090 [DOI] [PubMed] [Google Scholar]
- 17.Pepperkok, R., Whitney, J. A., Gomez, M., and Kreis, T. E. (2000) J. Cell Sci. 113 135-144 [DOI] [PubMed] [Google Scholar]
- 18.Yang, J.-S., Lee, S. Y., Gao, M., Bourgoin, S., Randazzo, P. A., Premont, R. T., and Hsu, V. W. (2002) J. Cell Biol. 159 69-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, S. Y., Yang, J.-S., Hong, W., Premont, R. T., and Hsu, V. W. (2005) J. Cell Biol. 168 281-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanoix, J., Ouwendijk, J., Stark, A., Szafer, E., Cassel, D., Dejgaard, K., Weiss, M., and Nilsson, T. (2001) J. Cell Biol. 155 1199-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, W., Duden, R., Phair, R. D., and Lippincott-Schwartz, J. (2005) J. Cell Biol. 168 1053-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majoul, I., Straub, M., Hell, S. W., Duden, R., and Söling, H.-D. (2001) Dev. Cell 1 139-153 [DOI] [PubMed] [Google Scholar]
- 23.Bigay, J., Casella, J.-F., Drin, G., Mesmin, B., and Antonny, B. (2005) EMBO J. 24 2244-2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigay, J., Gounon, P., Robineau, S., and Antonny, B. (2003) Nature 426 563-566 [DOI] [PubMed] [Google Scholar]
- 25.Kahn, R. A., Bruford, E., Inoue, H., Logsdon, J. M., Jr., Nie, Z., Premont, R. T., Randazzo, P. A., Satake, M., Theibert, A. B., Zapp, M. L., and Cassel, D. (2008) J. Cell Biol. 182 1039-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yahara, N., Sato, K., and Nakano, A. (2006) J. Cell Sci. 119 2604-2612 [DOI] [PubMed] [Google Scholar]
- 27.Frigerio, G., Grimsey, N., Dale, M., Majoul, I., and Duden, R. (2007) Traffic 8 1644-1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, X., Zhang, C., Xing, G., Chen, Q., and He, F. (2001) FEBS Lett. 490 79-83 [DOI] [PubMed] [Google Scholar]
- 29.Zhang, C., Yu, Y., Zhang, S., Liu, M., Xing, G., Wei, H., Bi, J., Liu, X., Zhou, G., Dong, C., Hu, Z., Zhang, Y., Luo, L., Wu, C., Zhao, S., and He, F. (2000) Genomics 63 400-408 [DOI] [PubMed] [Google Scholar]
- 30.Schweizer, A., Fransen, J. A., Bächi, T., Ginsel, L., and Hauri, H.-P. (1988) J. Cell Biol. 107 1643-1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimura, S., Yamamoto, A., Misumi, Y., Sohda, M., Barr, F. A., Fujii, G., Shakoori, A., Ohno, H., Mihara, K., and Nakamura, N. (2004) J. Biochem. (Tokyo) 135 201-216 [DOI] [PubMed] [Google Scholar]
- 32.Sohda, M., Misumi, Y., Yoshimura, S., Nakamura, N., Fusano, T., Ogata, S., Sakisaka, S., and Ikehara, Y. (2007) Traffic 8 270-284 [DOI] [PubMed] [Google Scholar]
- 33.Kasai, K., Shin, H.-W., Shinotsuka, C., Murakami, K., and Nakayama, K. (1999) J. Biochem. (Tokyo) 125 780-789 [DOI] [PubMed] [Google Scholar]
- 34.Waguri, S., Dewitte, F., Le Borgne, R., Rouillé, Y., Uchiyama, Y., Dubremetz, J.-F., and Hoflack, B. (2003) Mol. Biol. Cell 14 142-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin, H.-W., Kobayashi, H., Kitamura, M., Waguri, S., Suganuma, T., Uchiyama, Y., and Nakayama, K. (2005) J. Cell Sci. 118 4039-4048 [DOI] [PubMed] [Google Scholar]
- 36.Yang, J.-S., Lee, S. Y., Spanó, S., Gad, H., Zhang, L., Nie, Z., Bonazzi, M., Corda, D., Luini, A., and Hsu, V. W. (2005) EMBO J. 24 4133-4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole, N. B., Ellenberg, J., Song, J., DiEuliis, D., and Lippincott-Schwartz, J. (1998) J. Cell Biol. 140 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin, H.-W., Morinaga, N., Noda, M., and Nakayama, K. (2004) Mol. Biol. Cell 15 5283-5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishizaki, R., Shin, H.-W., Iguchi-Ariga, S. M. M., Ariga, H., and Nakayama, K. (2006) Genes Cells 11 949-959 [DOI] [PubMed] [Google Scholar]
- 40.Shin, H.-W., Shinotsuka, C., and Nakayama, K. (2005) Methods Enzymol. 404 206-215 [DOI] [PubMed] [Google Scholar]
- 41.Takatsu, H., Yoshino, K., Toda, K., and Nakayama, K. (2002) Biochem. J. 365 369-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waguri, S., Kohmura, M., Gotow, T., Watanabe, T., Ohsawa, Y., Kominami, E., and Uchiyama, Y. (1999) Arch. Histol. Cytol. 62 423-434 [DOI] [PubMed] [Google Scholar]
- 43.Appenzeller-Herzog, C., and Hauri, H.-P. (2006) J. Cell Sci. 119 2173-2183 [DOI] [PubMed] [Google Scholar]
- 44.Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T. E., and Warren, G. (1995) J. Cell Biol. 131 1715-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gleeson, P. A., Anderson, T. J., Stow, J. L., Griffiths, G., Toh, B.-H., and Matheson, F. (1996) J. Cell Sci. 109 2811-2821 [DOI] [PubMed] [Google Scholar]
- 46.Cole, N. B., Sciaky, N., Marotta, A., Song, J., and Lippincott-Schwartz, J. (1996) Mol. Biol. Cell 7 631-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santy, L. C., and Casanova, J. E. (2001) J. Cell Biol. 154 599-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Presley, J. F., Cole, N. B., Schroer, T. A., Hirschberg, K., Zaal, K. J. M., and Lippincott-Schwartz, J. (1997) Nature 389 81-85 [DOI] [PubMed] [Google Scholar]
- 49.Styers, M. L., O'Connor, A. K., Grabski, R., Cormet-Boyaka, E., and Sztul, E. (2008) Am. J. Physiol. Cell Physiol. 294 1485-1498 [DOI] [PubMed] [Google Scholar]
- 50.Guo, Y., Punj, V., Sengupta, D., and Linstedt, A. D. (2008) Mol. Biol. Cell 19 2830-2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weimer, C., Beck, R., Eckert, P., Reckmann, I., Moelleken, J., Brügger, B., and Wieland, F. (2008) J. Cell Biol. 183 725-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kliouchnikov, L., Bigay, J., Mesmin, B., Parnis, A., Rawet, M., Goldfeder, N., Antonny, B., and Cassel, D. (2009) Mol. Biol. Cell 20 859-869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nie, Z., Fei, J., Premont, R. T., and Randazzo, P. A. (2005) J. Cell Sci. 118 3555-3566 [DOI] [PubMed] [Google Scholar]
- 54.Natsume, W., Tanabe, K., Kon, S., Yoshida, N., Watanabe, T., Torii, T., and Satake, M. (2006) Mol. Biol. Cell 17 2592-2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watson, P. J., Frigerio, G., Collins, B. M., Duden, R., and Owen, D. J. (2004) Traffic 5 79-88 [DOI] [PubMed] [Google Scholar]
- 56.Futatsumori, M., Kasai, K., Takatsu, H., Shin, H.-W., and Nakayama, K. (2000) J. Biochem. (Tokyo) 128 793-801 [DOI] [PubMed] [Google Scholar]
- 57.Wegmann, D., Hess, P., Baier, C., Wieland, F. T., and Reinhard, C. (2004) Mol. Cell. Biol. 24 1070-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moelleken, J., Malsam, J., Betts, M. J., Movafeghi, A., Reckmann, I., Meissner, I., Hellwig, A., Russell, R. B., Söllner, T., Brügger, B., and Wieland, F. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4425-4430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.