Abstract
As obligate intracellular parasites, viruses exploit diverse cellular signaling machineries, including the mitogen-activated protein-kinase pathway, during their infections. We have demonstrated previously that the open reading frame 45 (ORF45) of Kaposi sarcoma-associated herpesvirus interacts with p90 ribosomal S6 kinases (RSKs) and strongly stimulates their kinase activities (Kuang, E., Tang, Q., Maul, G. G., and Zhu, F. (2008) J. Virol. 82 ,1838 -1850). Here, we define the mechanism by which ORF45 activates RSKs. We demonstrated that binding of ORF45 to RSK increases the association of extracellular signal-regulated kinase (ERK) with RSK, such that ORF45, RSK, and ERK formed high molecular mass protein complexes. We further demonstrated that the complexes shielded active pERK and pRSK from dephosphorylation. As a result, the complex-associated RSK and ERK were activated and sustained at high levels. Finally, we provide evidence that this mechanism contributes to the sustained activation of ERK and RSK in Kaposi sarcoma-associated herpesvirus lytic replication.
The extracellular signal-regulated kinase (ERK)2 mitogen-activated protein kinase (MAPK) signaling pathway has been implicated in diverse cellular physiological processes including proliferation, survival, growth, differentiation, and motility (1-4) and is also exploited by a variety of viruses such as Kaposi sarcoma-associated herpesvirus (KSHV), human cytomegalovirus, human immunodeficiency virus, respiratory syncytial virus, hepatitis B virus, coxsackie, vaccinia, coronavirus, and influenza virus (5-17). The MAPK kinases relay the extracellular signaling through sequential phosphorylation to an array of cytoplasmic and nuclear substrates to elicit specific responses (1, 2, 18). Phosphorylation of MAPK is reversible. The kinetics of deactivation or duration of signaling dictates diverse biological outcomes (19, 20). For example, sustained but not transient activation of ERK signaling induces the differentiation of PC12 cells into sympathetic-like neurons and transformation of NIH3T3 cells (20-22). During viral infection, a unique biphasic ERK activation has been observed for some viruses (an early transient activation triggered by viral binding or entry and a late sustained activation correlated with viral gene expression), but the responsible viral factors and underlying mechanism for the sustained ERK activation remain largely unknown (5, 8, 13, 23).
The p90 ribosomal S6 kinases (RSKs) are a family of serine/threonine kinases that lie at the terminus of the ERK pathway (1, 24-26). In mammals, four isoforms are known, RSK1 to RSK4. Each one has two catalytically functional kinase domains, the N-terminal kinase domain (NTKD) and C-terminal kinase domain (CTKD) as well as a linker region between the two. The NTKD is responsible for phosphorylation of exogenous substrates, and the CTKD and linker region regulate RSK activation (1, 24, 25). In quiescent cells ERK binds to the docking site in the C terminus of RSK (27-29). Upon mitogen stimulation, ERK is activated by its upstream MAPK/ERK kinase (MEK). The active ERK phosphorylates Thr-359/Ser-363 of RSK in the linker region (amino acid numbers refer to human RSK1) and Thr-573 in the CTKD activation loop. The activated CTKD then phosphorylates Ser-380 in the linker region, creating a docking site for 3-phosphoinositide-dependent protein kinase-1. The 3-phosphoinositide-dependent protein kinase-1 phosphorylates Ser-221 of RSK in the activation loop and activates the NTKD. The activated NTKD autophosphorylates the serine residue near the ERK docking site, causing a transient dissociation of active ERK from RSK (25, 26, 28). The stimulation of quiescent cells by a mitogen such as epidermal growth factor or a phorbol ester such as 12-O-tetradecanoylphorbol-13-acetate (TPA) usually results in a transient RSK activation that lasts less than 30 min. RSKs have been implicated in regulating cell survival, growth, and proliferation. Mutation or aberrant expression of RSK has been implicated in several human diseases including Coffin-Lowry syndrome and prostate and breast cancers (1, 24, 25, 30-32).
KSHV is a human DNA tumor virus etiologically linked to Kaposi sarcoma, primary effusion lymphoma, and a subset of multicentric Castleman disease (33, 34). Infection and reactivation of KSHV activate multiple MAPK pathways (6, 12, 35). Noticeably, the ERK/RSK activation is sustained late during KSHV primary infection and reactivation from latency (5, 6, 12, 23), but the mechanism of the sustained ERK/RSK activation is unclear. Recently, we demonstrated that ORF45, an immediate early and also virion tegument protein of KSHV, interacts with RSK1 and RSK2 and strongly stimulates their kinase activities (23). We also demonstrated that the activation of RSK plays an essential role in KSHV lytic replication (23). In the present study we determined the mechanism of ORF45-induced sustained ERK/RSK activation. We found that ORF45 increases the association of RSK with ERK and protects them from dephosphorylation, causing sustained activation of both ERK and RSK.
EXPERIMENTAL PROCEDURES
Cell Cultures and Reagents—BCBL-1 cells latently infected with KSHV were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and antibiotics. HEK293 cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics. HEK293-teton-ORF45 is a cell line that expresses ORF45 from a tetracycline-inducible promoter. It was generated by cotransfection of HEK293 cells with linearized pTet-On and pTRE2hyg (Clontech, Mountain View, CA) harboring the ORF45 coding sequence followed by G418 and hygromycin selection. A clone with low background expression and high inducibility was used in our study. Anti-HA and anti-FLAG M2 antibodies, anti-FLAG M2 and anti-HA affinity resins, 3× FLAG and HA peptides, TPA, phosphatase inhibitor cocktails I and II, and protein standards for gel filtration chromatography were purchased from Sigma. Antibodies detecting pRSK (Thr-359/Ser-363), pRSK (Ser-380), pRSK (Thr-573), ERK1/2, MEK1/2, pERK1/2 (Thr-202/Tyr-204), pMEK1/2 (Ser-217/221), and SignalSilence® pool ERK1/2 and control siRNAs were purchased from Cell Signaling Technology (Danvers, MA). Rat anti-LANA (latency-associated nuclear antigen) antibody was purchased from Advanced Biotechnologies, Inc. (Columbia, MD). Active ERK2, RSK1, RSK2, and phosphatase 2A (PP2A) proteins and rabbit anti-RSK1 and RSK2 antibodies were purchased from Upstate Biotechnology, Inc. (now part of Millipore, Billerica, MA). ORF45 protein, anti-ORF45, and anti-K8 antibodies have been described previously (36, 37).
Plasmid Constructs—Plasmids pKH3-RSK2, pKH3-RSK1, pCR3.1-ORF45, FLAG-tagged pCMV-ORF45full-length, and derivatives have been described previously (23, 37). FLAG-tagged Rat RSK1-WT and docking-site deletion constructs were kindly provided by James Maller of the University of Colorado. ERK1 and ERK2 constitutive active and kinase-dead mutant constructs were provided by Shou-Jiang Gao of the University of Texas Health Science Center at San Antonio and by Melanie H. Cobb of the University of Texas Southwestern Medical Center at Dallas.
Immunoprecipitation and Western Blot Analysis—Immunoprecipitation and Western blot analysis were performed as previously described (23). Briefly, cells were lysed with whole-cell lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm sodium orthovanadate (Na3VO4), 40 mm β-glycerophosphate, 30 mm sodium fluoride, 10% glycerol, 5 mm EDTA, 1× protease inhibitor mixture (Roche Applied Science), and 1 mm phenylmethylsulfonyl fluoride). For immunoprecipitation with anti-FLAG or anti-HA antibodies, the cell lysates were incubated with EZview red anti-FLAG M2 or anti-HA affinity resin for 4 h or overnight at 4 °C. After washing with the lysis buffer and Tris-buffered saline (TBS: 50 mm Tris-HCl, pH 7.4, 150 mm NaCl), proteins were eluted by incubation with 150 μg/ml 3× FLAG or HA peptide in TBS for 1 h at 4 °C. For Western blot, about 20 μg of proteins was resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked in 5% dried milk in 1× phosphate-buffered saline plus 0.2% Tween 20 and then incubated with diluted primary antibodies for 2 h at room temperature or overnight at 4 °C. Anti-rabbit, anti-rat, or anti-mouse IgG antibodies conjugated to horseradish peroxidase (Pierce) were used as the secondary antibodies. An enhanced chemiluminescence system (Pierce) was used for detection.
siRNA—The strategy for knockdown of gene expression in BCBL-1 cells by a retrovirus-based siRNA vector including target sequences for RSK1 and RSK2 has been described previously (23). The two siRNA sequences for ORF45 are siORF45-1 (AGAGAATGCTTCCAATTGA) and siORF45-5 (GACCCACGGTGATAGATAT). The two for the ERK1 are siERK1-1 (AGCTGGATGACCTACCTAA) and siERK1-2 (GCCATGAGAGATGTCATCA). Because ERK2 is not expressed in BCBL-1 cells,3 only siRNAs against ERK1 were used to knock down ERK1/2 expression. For transient assays, SignalSilence® pool Erk1/2 and control siRNAs from Cell Signaling Technology were transfected into HEK293 cells with Lipofectamine® 2000 (Invitrogen) according to the protocols suggested by the manufacturer. The off-target effects such as activation of interferon were examined and found to be minimal as we previously described (23).
ATP Depletion—Serum-starved cells were washed once with Dulbecco's modified Eagle's medium and then treated with 20 ng/ml TPA for 10 min. Then ATP depletion was performed as described previously (38, 39). Briefly, after TPA treatment, cells were incubated with Dulbecco's modified Eagle's medium supplemented with 5 μm rotenone and 10 mm 2-deoxyglucose at 37 °C for the indicated times. Rotenone, an inhibitor of the respiratory chain, blocks the synthesis of ATP, and 2-deoxyglucose is a competitive inhibitor of glycolysis. This treatment leads to a rapid decrease in cytosolic ATP concentration. After incubation, cells were washed with phosphate-buffered saline, lysed with whole-cell lysis buffer, and analyzed by Western blot.
In Vitro Dephosphorylation Assay—Active ERK2, active RSK1, and ORF45 were mixed in a molar ratio of 1:1:5 (150, 200, and 750 ng, respectively) in 25 μl of buffer (50 mm Tris, pH 8.0, 150 mm NaCl) and incubated at 4 °C for 30 min. The reaction mixture was adjusted with 10× calf-intestine alkaline phosphatase (CIAP) buffer (500 mm Tris-HCl, pH 8.5, 1 mm EDTA) or 5× PP2A reaction buffer (50 mm Tris-HCl, pH 7.5, 1 mm EGTA, 1 mm EDTA, 1 mm dithiothreitol, and 100 μg/ml bovine serum albumin); 1 unit CIAP or 0.2 units of PP2A were then added to each reaction. After incubation at 37 °C for 30 min for CIAP or 30 °C for 1 h for PP2A, the reactions were stopped by the addition of 2× Laemmli gel-loading buffer and boiled for 10 min. We also prepared crude cell extracts that contained various phosphatases as described by Wang et al. (40). Briefly, the quiescent HEK293 cells were homogenized in buffer A (50 mm Tris-HCl, pH 7.5, 0.1 mm EGTA, 1 mm dithiothreitol, and 1× protease inhibitor mixture). The lysates were centrifuged at 10,000 × g for 10 min at 4 °C, and supernatant was collected as crude extract that contained various phosphatase activities. Thirty μg of the above supernatant fraction were added to the mixtures of ORF45, active ERK, and active pRSK to a total volume of 100 μl in buffer A. The mixtures were incubated at 30 °C, and the reactions were stopped at different time points as described above.
Gel Filtration Chromatography—Semiconfluent HEK293-teton-ORF45 cells were treated with 500 ng/ml doxycycline for 3 days to induce ORF45 expression. After induction, the cells were harvested, washed with cold phosphate-buffered saline, resuspended in buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 30 mm NaF, 40 mm glycerophosphate, 5 mm EDTA, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin A), and lysed by three rounds of freezing and thawing followed by 10 s of sonication in a micro-ultrasonic cell disrupter. Cell lysates were clarified by ultracentrifuge at 100,000 × g for 1 h at 4 °C. The cell lysates, purified ORF45, ERK2, RSK1, RSK2, or their mixtures were fractionated on a Superdex 200 HR 10/30 gel filtration column with a Beckman System GOLD HPLC system (GMI, Inc., Ramsey, MN). The HPLC running buffer consisted of 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin A. One column volume (24 ml) of eluate was collected in aliquots of 0.5 ml. Fractions were analyzed by Western blot.
RESULTS
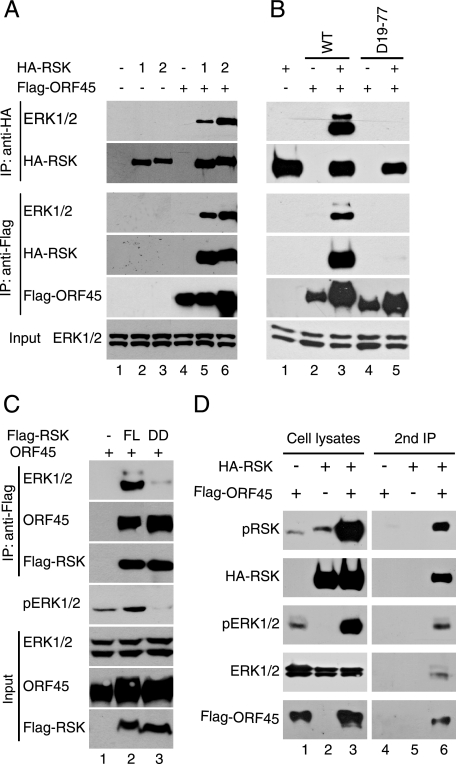
KSHV ORF45 Increases the Association of RSK with ERK—In previous studies we found that KSHV ORF45 interacts with and significantly increases the activities of RSK1 and RSK2 kinase (23). Efficient RSK activation by ERK requires its association with ERK. ERK associates with RSK through the docking site near the C terminus of RSK in quiescent cells and transiently dissociates upon mitogen stimulation (28). Prolonged association with ERK has been noticed to promote sustained RSK activation (28), so we examined whether ORF45 could increase the association of ERK and RSK. As shown in Fig. 1A, we found that ERK was coimmunoprecipitated with HA-RSK1 or HA-RSK2 in the presence of ORF45 (Fig. 1A, lanes 5 and 6), but coprecipitation was barely detectable in the absence of ORF45 expression (lanes 2 and 3), suggesting that ORF45 significantly increases the ERK/RSK interaction. In addition, ERK was coprecipitated with ORF45 in the presence of RSK1 (lane 5) or RSK2 (lane 6) but was nearly not in the absence of RSK overexpression (lane 4). These results suggest that ORF45 interacts with ERK indirectly through RSKs.
FIGURE 1.
KSHV ORF45 forms complexes with RSK and ERK in cells. A, KSHV ORF45 increases association of ERK with RSK. HEK293 cells were transfected with HA-tagged RSK-expressing vectors in the presence (lanes 1-3) or absence (lanes 4-6) of FLAG-ORF45-expressing vectors. At 48 h after transfection, the cells were serum-starved for an additional 24 h. Whole-cell lysates were immunoprecipitated, and the IP complexes were immunoblotted with antibodies as indicated (upper panel). Whole-cell lysates were also analyzed with anti-ERK1/2 to show the inputs in all samples (lower panel). B, the RSK binding site on ORF45 is required for its ability to increase RSK/ERK association. HEK293 cells were cotransfected with FLAG-tagged ORF45 wild-type (WT), Δ19-77 deletion mutant together with pKH3-RSK1 or empty vector. After serum starvation for 24 h, cells were lysed and immunoprecipitated with anti-FLAG or anti-HA affinity resins. The immunocomplexes and whole-cell lysates were immunoblotted with antibodies as indicated. C, the ERK docking site on the C terminus of RSK is required for ORF45·RSK·ERK association. HEK293 cells were transfected with full-length (FL) rat RSK or the ERK docking-site-deletion mutant (DD) and serum-starved for 24 h. Cell lysates were immunoprecipitated with anti-FLAG M2 affinity resin. The immunocomplexes and input whole cell lysates were analyzed by Western blot with antibodies as indicated. D, ORF45, RSK, ERK were present in the same complex as revealed by sequential coimmunoprecipitation assay. HEK293 cells were transfected with FLAG-ORF45 and/or HA-RSK expression vectors for 48 h and then serum-starved for additional 24 h. Cell lysates were immunoprecipitated with anti-FLAG affinity resin and eluted with 3× FLAG peptides. The eluates were then immunoprecipitated with anti-HA resins. Cell lysates and eluates were analyzed by Western blotting with phosphorylation-specific anti-pRSK, anti-pERK antibodies. Membranes were stripped and reprobed with anti-ERK or anti-RSK antibodies for detection of total ERK and RSK.
We have shown previously that the N-terminal region of ORF45 binds to RSK and that deletion of amino acids 19-77 abolishes its binding (23). In contrast to the full-length ORF45, the Δ19-77 deletion mutant was not coprecipitated with either RSK or ERK (Fig. 1B, bottom panel, lane 5). ERK was coprecipitated with HA-RSK2 in the presence of full-length ORF45 but not the Δ19-77 deletion mutant (Fig. 1B, top panel). So the wild-type ORF45 increased the association between RSK2 and ERK as expected, but the Δ19-77 amino acid mutant did not. The results presented in Fig. 1B were obtained with the HA-RSK2 construct, and identical results were obtained with HA-RSK1 (data not shown). These results further support that ORF45 interacts with ERK indirectly, and their association is bridged by RSK. Interestingly, ERK1 and ERK2 were equally expressed, but ERK2 was retained by RSK preferentially in the presence of ORF45 (Fig. 1, B and C). This pattern could explain our previous observation that ERK2 was preferentially activated in HEK293 cells expressing KSHV ORF45 (23).
RSK interacts with ORF45 through its NTKD (23) and with ERK through its CTKD (27-29), but neither NTKD nor CTKD alone could mediate ORF45/ERK interaction; full-length RSK was required (data not shown). A possible scenario is that binding of ORF45 to RSK stabilizes the transient association between RSK and ERK that normally occurs through the C-terminal ERK docking site. If this were the case, the ERK-docking site would be required for the increased ERK/RSK interaction by ORF45. The coimmunoprecipitation assay showed that disruption of the ERK-docking site (deletion of the last 17 amino acid from the C terminus) diminished the binding of ERK to RSK (Fig. 1C, compare lane 3 to lane 2). Furthermore, the ERK docking site deletion mutant (DD) displayed a dominant inhibitory effect on the ERK activation by ORF45 (Fig. 1C, middle panel, compare lane 3 to lane 2). This result suggests that the increased association of ERK with RSK indeed depends on the ERK docking site. Collectively, the above data demonstrate that binding of ORF45 to the RSK increases the association of ERK with RSK and, therefore, that all three form a complex in the cells.
To determine whether these components exist in the same complex in cells, we coexpressed FLAG-ORF45 and HA-RSK2 in HEK293 cells for a sequential immunoprecipitation analysis. We first immunoprecipitated FLAG-ORF45 and eluted the immunocomplexes with competing FLAG peptides. RSK2 bound to the eluted ORF45 was then immunoprecipitated with anti-HA resin. We found that endogenous ERK as well as pERK and pRSK were present in the RSK2/ORF45 complexes (Fig. 1D), but MEK was not (data not shown). Similar results were obtained with HA-RSK1 construct (data not shown). These results further support that ORF45, ERK, and RSK form complexes that contain active pRSK and pERK in the cell.
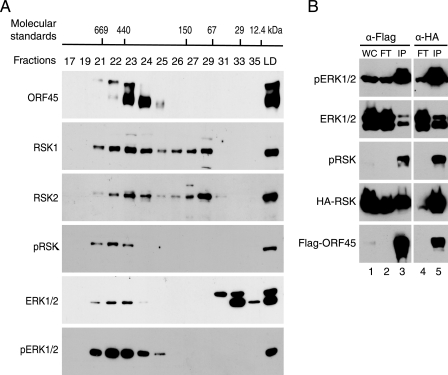
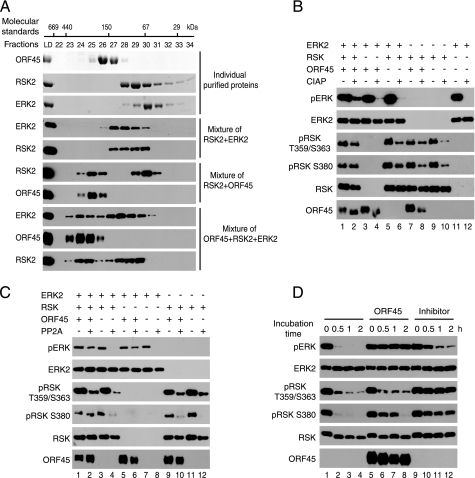
Active ERK Is Associated with the ORF45·RSK Complex—To provide further evidence that ORF45, RSK, and ERK form a complex in the cells, we fractionated the lysates of ORF45-expressing HEK293 cells by gel filtration chromatography and analyzed each fraction by Western blot. As shown in Fig. 2A, we found that: 1) KSHV ORF45 protein (407 amino acids) exists mainly in high molecular weight (Mr) complexes (fractions 21-25), although its predicted Mr is only 43.3 kDa (but its apparent Mr on denaturing SDS-PAGE is about 70 kDa (41, 42); 2) RSK1 and RSK2 (both have apparent Mr of 90 kDa) exist in low Mr (fractions 27-31) and high Mr (fractions 21-25) forms; 3) the high Mr RSK proteins cofractionate with ORF45, supporting the conclusion that RSKs and ORF45 are associated with each other in cells; 4) active pERK and pRSK are only detected in fractions where high Mr ORF45 and RSK reside (fractions 21-24), although the majority of ERK1/2 proteins exist in low Mr, presumably nonphosphorylated, monomer form (fractions 31-35); 5) two forms of ORF45 are detected in these cells; the hypophosphorylated form (higher mobility) cofractionates with the high Mr RSK1 and RSK2 (fractions 22-25), and the hyperphosphorylated ORF45 (lower mobility) appears larger in size and cofractionates with pRSK, pERK (fractions 21-23). Overall, these results support ORF45, RSK, and ERK complex formation in which active pRSK and pERK are highly enriched.
FIGURE 2.
ORF45 forms complexes with active RSK and ERK. A, lysate of HEK293-teton-ORF45 cell lines overexpressing ORF45 was fractionated on a Superdex 200 HR 10/30 column. Fractions and input were analyzed by Western blotting with antibodies as indicated. The molecular masses of protein standards, thyroglobulin (669 kDa), apoferritin (440 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (67 kDa), carbonic anhydrase (29 kDa), and cytochrome C (12.4 kDa, were marked at the approximate fractions at the top. LD, load. B, active pERK and pRSK are greatly enriched in the ORF45·RSK·ERK complex. HEK293 cells were transfected with FLAG-ORF45 and HA-RSK expression vectors for 48 h. Cell lysates were immunoprecipitated with anti-FLAG or anti-HA immunoaffinity resins. The input whole-cell lysates (WC), flow-through (FT), and IP complexes (IP) were analyzed by Western blot with antibodies as indicated.
To confirm further that the active pERK and pRSK are in complexes with ORF45, we assessed the phosphorylation status of the ORF45-associated ERK and RSK. Lysates of HEK293 cells transfected with FLAG-ORF45 and HA-RSK2 expression plasmids were immunoprecipitated (IP) with anti-FLAG affinity resins. When the IP complexes and the flow-through were analyzed by Western blot, we found that a small portion of ERK was coprecipitated with FLAG-ORF45 but that the majority of ERK remained in the flow-through (Fig. 2B, compare ERK signals of lanes 3 and 2), confirming the above observation that most ERK proteins in cells were detected in low Mr fractions (Fig. 2A). HA-RSK2 in the cells was efficiently coprecipitated with FLAG-ORF45 (Fig. 2B, compare the RSK signals of lanes 3 and 2). The RSK and ERK in the IP complexes contain significantly higher levels of active pRSK and pERK than does the flow-through (Fig. 2B, compare the pERK and pRSK signals of lanes 3 and 2). Considering the lower amount of total ERK and RSK in the immunoprecipitated complexes than in the flow-through, we conclude that the phosphorylated forms, pERK and pRSK, are highly enriched in the ORF45·RSK·ERK complexes. When anti-HA affinity resins were used to immunoprecipitate HA-RSK2 from the same lysates, they pulled down ORF45 efficiently (Fig. 2B, lane 5). As with IP complexes with anti-FLAG, only a small portion of total ERK in the cells (Fig. 2B, compare ERK signals of lanes 5 and 4) is associated with RSK2, but that is highly phosphorylated (compare pERK signals of lanes 5 and 4). This experiment indicates that ORF45, RSK, and ERK form complexes that retain high levels of pRSK and pERK.
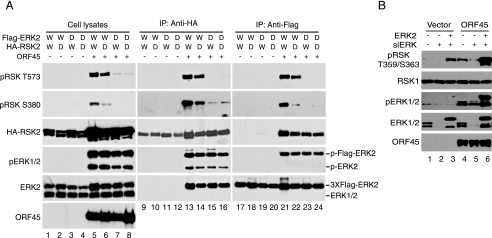
Activation of RSK by KSHV ORF45 Requires ERK Activity—We noticed previously that activation of RSK1 and RSK2 by ORF45 is correlated with the activation of ERK but not further upstream MEK kinase (23), in agreement with our observation that active pERK but not pMEK was detected in the ORF45·RSK·ERK complex (data not shown). The next experiment was designed to determine whether ERK kinase activity is required for the RSK activation by ORF45. We used an ERK kinase-dead (KD) mutant to determine whether ERK activity is required for RSK activation by ORF45. We cotransfected the wild-type (Fig. 3A, lanes 1 and 2 and lanes 5 and 6) or FLAG-ERK-KD (lanes 3 and 4 and lanes 7 and 8) and the wild-type (lanes 1 and 3 and lanes 5 and 7) or HA-RSK2-KD (lanes 2 and 4 and lanes 6 and 8) with (lanes 5-8) or without (lanes 1-4) ORF45 expression vectors into HEK293 cells. We then analyzed the levels of RSK and ERK phosphorylation in the transfected cells by Western blot. The overall levels of RSK and ERK phosphorylation were significantly higher in the presence of ORF45 (Fig. 3A, compare lanes 5-8 to lanes 1-4), as we have reported previously (23). In the presence of ORF45, the levels of RSK phosphorylation were obviously lower in cells transfected with the ERK-KD construct (Fig. 3A, compare lanes 7 and 8 to lanes 5 and 6). The lower phosphorylation of RSK was evident not only on Thr-573 (thought to be phosphorylated by ERK according to the current model; see below) but also on Ser-380 (thought to be autophosphorylated by RSK itself), suggesting that ERK activity is indeed required for the activation of RSK by ORF45. Interestingly, in the presence of ORF45, ERK phosphorylation showed no difference in cells transfected with wild-type or RSK2-KD constructs (Fig. 3A, compare the pERK signals of lanes 5 and 6). The dramatically reduced autophosphorylation on Ser-380 (Fig. 3, lane 6) suggested RSK2-KD was not functionally activated, but this RSK2-KD increased ERK2 phosphorylation as well as did the wild type-RSK2, and its phosphorylation at Thr-573 by ERK was comparable with that of wild-type RSK2 (Fig. 3A, compare lane 6 to lane 5), indicating that RSK kinase activity is not absolutely required for the activation of ERK by ORF45.
FIGURE 3.
ERK kinase activity is required for RSK activation by KSHV ORF45. A, HEK293 cells were cotransfected with HA-tagged wild-type RSK2 (W) or RSK2 kinase-dead mutant (D), FLAG-tagged wild-type ERK2 (W) or ERK2 kinase-dead mutant (D), and ORF45 expression vectors in combination as indicated. At 48 h after transfection, the cells were serum-starved for an additional 24 h. Whole-cell lysates were prepared and immunoprecipitated with anti-HA or anti-FLAG affinity resins. Whole-cell lysates (left panel), the IP complexes of anti-HA (middle panel), and anti-FLAG (right panel) were analyzed by Western blotting with antibodies as indicated. The signals of exogenous 3× FLAG-ERK2 and endogenous ERK2 are marked at the right. B, knockdown of endogenous ERK reduces activation of RSK by ORF45. HEK293 cells were transfected with SignalSilence® siRNA against ERK1/2 or control siRNA (both purchased from Cell Signaling Technology) in the presence or absence of ORF45 expression vector. After 24 h of starvation, whole-cell lysates were prepared and analyzed by Western blotting with the antibodies as indicated.
Because the majority of activated pERKs and pRSKs are associated with ORF45-containing complexes (Fig. 2), we also determined the levels of phosphorylation of the complex-associated RSKs and ERKs. The HA-RSK was precipitated with anti-HA resin, and the immunocomplexes were analyzed for total ERKs and RSKs as well as their phosphorylated forms. In the presence of ORF45, the ERK-KD mutant bound to RSK, as did the wild-type ERK (compare the ERK signals of lanes 15 and 16 to those of lanes 13 and 14), but caused a drastically lower level of RSK phosphorylation (compare pRSK signals of lanes 15 and 16 to those of lanes 13 and 14). Analysis of the levels of pRSK in the immunocomplexes of FLAG-ERK revealed similar results (lanes 17-24), confirming that ERK activity is necessary for activation of RSK by ORF45. If ERK activity is required, we reasoned that knockdown of endogenous ERK would reduce the activation of RSK by ORF45. As shown as in Fig. 3B, ERK siRNA dramatically reduced expression of ERK1/2 (compare lanes 2 and 5 to lanes 1 and 4) and significantly inhibited ORF45-induced RSK phosphorylation. The effect was specific to ablation of ERK because ectopic expression of exogenous rat ERK2 restored RSK activation by ORF45 (lanes 3 and 6). In conclusion, the above experiments suggest that the ERK activity is required for activation of RSK by ORF45 presumably in protein complexes in which an RSK molecule seems to act as scaffold.
ORF45 Shields pERK and pRSK from Dephosphorylation—Next we explored how RSK and ERK maintain high levels of phosphorylation when they form complexes with ORF45. We first determined whether ORF45 could activate RSK when they were mixed together in vitro. We purified recombinant ORF45 proteins from either bacteria or baculovirus expression systems. We mixed purified ORF45, RSK2, and ERK2 with various combinations and found no significant RSK activation, although these proteins formed complexes as revealed by gel-filtration chromatography (see below and data not shown). Although different ones could be drawn, we favor the interpretation that ORF45 does not activate RSK directly or by physical interactions. Its failure to do so is not surprising because ORF45 contains no conserved kinase domain or NTP binding motif.
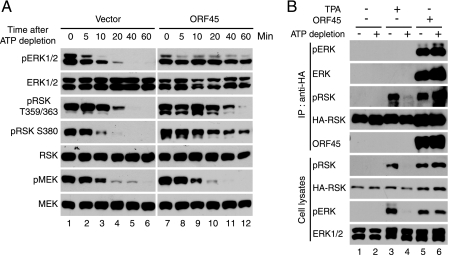
The phosphorylation levels of ERK and RSK in cells are determined by two processes; that is, their phosphorylation by upstream kinases and their dephosphorylation by various phosphatases. Because the upstream kinase MEK is not activated in the RSK activation by ORF45 (23), we asked whether ORF45 affects dephosphorylation of pRSK and pERK. We used the technique of ATP depletion, which has been successfully used to examine the rate of dephosphorylation of p38 and JNK (Jun N-terminal kinase) after various stress treatments or viral infection (38, 39). The ORF45 and mock-transfected cells were serum-starved for 24 h, then treated with TPA for 10 min and subjected to ATP depletion. Cell extracts were harvested at 0, 5, 10, 20, 40, and 60 min after the addition of ATP-depleting reagents. Subsequent Western blot analysis demonstrated that the amount of phosphorylated ERK and RSK decreased rapidly and became undetectable 20 min after ATP depletion in the absence of ORF45 (Fig. 4A). The rate of pERK and pRSK dephosphorylation was significantly lower in cells that expressed ORF45, and the phosphorylated forms of pERK and pRSK were visible even after 60 min (Fig. 4A, compare lanes 10-12 to lanes 4-6). The ORF45·RSK·ERK complexes remained intact after ATP depletion because both ORF45 and ERK were efficiently coimmunoprecipitated with HA-RSK (Fig. 4B). These results indicate that ORF45 and its induced protein complexes inhibit dephosphorylation of active pERK and pRSK. In contrast, MEK dephosphorylation by ATP depletion was not affected by ORF45, a finding that supports our previous observation that MEK is not directly involved in RSK activation by ORF45 (23).
FIGURE 4.
KSHV ORF45 protects pERK and pRSK from dephosphorylation. A, ATP depletion. ORF45-transfected HEK293 or control cells were serum-starved for 24 h and then treated with TPA for 10 min. ATP depletion was initiated by addition of rotenone and 2-deoxyglucose as described under “Experimental Procedures.” The cells were harvested at the indicated times after depletion. Cell lysates were analyzed by Western blotting for determination of phosphorylation status and total amounts of MEK, ERK1/2, and RSK. B, ATP depletion does not affect complex formation of ORF45, RSK, and ERK. HEK293 cells were cotransfected with pKH3-RSK plus ORF45-expressing or empty vectors as controls. After 24 h of serum starvation, cells were treated with TPA, then depleted of ATP as indicated. After 30 min of ATP depletion, cell lysates were prepared and immunoprecipitated with anti-HA affinity resins. The immunocomplexes (upper panel) and whole-cell lysates (lower panel) were analyzed for total amounts of ERK1/2 and RSK1 and their phosphorylated forms.
Next, we determined whether ORF45 is able to prevent dephosphorylation in vitro. Purified ORF45, RSK, and ERK formed higher Mr complexes, as revealed by distinct chromatography profiles of their mixtures, than did individual proteins. The triple complexes were consistently eluted earlier than dual complexes and than individual proteins (Fig. 5A). Next we determined the status of dephosphorylation of ERK and RSK in triple complex. Purified ORF45, active RSK1 or RSK2 (only data with RSK1 were shown), and active ERK2 were mixed and incubated for 30 min for complex formation. The mixtures were then subjected to limited dephosphorylation by incubation with CIAP for 30 min. The phosphorylation status of ERK and RSK was then determined. The increased mobility of phosphoprotein ORF45 on SDS-PAGE gel in CIAP-treated samples attested to authentic CIAP activity (Fig. 5B, bottom row, compare lanes 2 and 1, lanes 4 and 3, or lanes 8 and 7). Dephosphorylation of pERK is inhibited when both ORF45 and RSK are present but not if either one is absent (Fig. 5B, top row). We next used protein PP2A and obtained similar results (Fig. 5C). In both cases the protection by ORF45 seems more obvious for pERK than for pRSK, suggesting pERK is directly protected by the triple complexes. The protection of pRSK by ORF45 could be indirect and through active phosphorylation by pERK. Such an event might not take place under the above in vitro conditions because ATP was not included in the dephosphorylation reactions. To prove the protection of pERK from dephosphorylation by ORF45 does not result from a particular phosphatase, we also used crude cell extracts that contain various cellular phosphatases. As shown in Fig. 5D, pERK was completely protected, and pRSK was also protected, but to a lesser extent, from dephosphorylation (lanes 5-8). As a control, phosphatase inhibitor cocktails inhibited dephosphorylations of pERK2 and pRSK (lanes 9-12). The protection by ORF45 of pERK was stronger than that of pRSK, in comparison to the protection by phosphatase inhibitors respectively, again suggesting pERK is the target of protection by ORF45-containing triple complexes. Collectively, both in vivo and in vitro experiments support that ORF45 complexes protect pERK from dephosphorylation.
FIGURE 5.
KSHV ORF45 protects pERK from dephosphorylation in vitro. A, formation of a triple complex of ORF45, RSK, and ERK in vitro. Purified ERK2, RSK2, and ORF45 were mixed in different combinations and incubated at 4 °C for 30 min. The mixtures were then fractionated on a Superdex 200 HR 10/30 column as described under “Experimental Procedures.” Fractions and input load were analyzed by SDS-PAGE followed by Coomassie staining (top three panels) or Western blotting with antibodies as indicated. ORF45 protects pERK from in vitro dephosphorylation by CIAP (B), PP2A (C), and cellular phosphatases in the crude cell extract (D). Active ERK2, active RSK, and ORF45 were mixed and incubated at 4 °C for 30 min. The reaction mixtures were adjusted to ideal buffer conditions before the additions of CIAP and PP2A or extract of HEK293 cells in the presence or absence of phosphatase inhibitor mixture, respectively. After incubation at 37 °C for 30 min (for CIAP), 30 °C for 1 h (for PP2A), or 30 °C for the indicated time (for crude cellular phosphatase extract), the reactions were stopped by the addition of 2× Laemmli gel-loading buffer and boiled for 10 min before immunoblotting with the indicated antibodies.
ERK, RSK, and ORF45 Are All Necessary for the Sustained Activation of ERK and RSK in KSHV Lytic Replication—We have demonstrated that ORF45 forms complexes with RSK and ERK, retains active pRSK and pERK in the complexes, and protects them from dephosphorylation, resulting in sustained activation of ERK and RSK. We next determined whether such a mechanism plays a role to the sustained RSK and ERK activation observed during KSHV lytic replication (23).
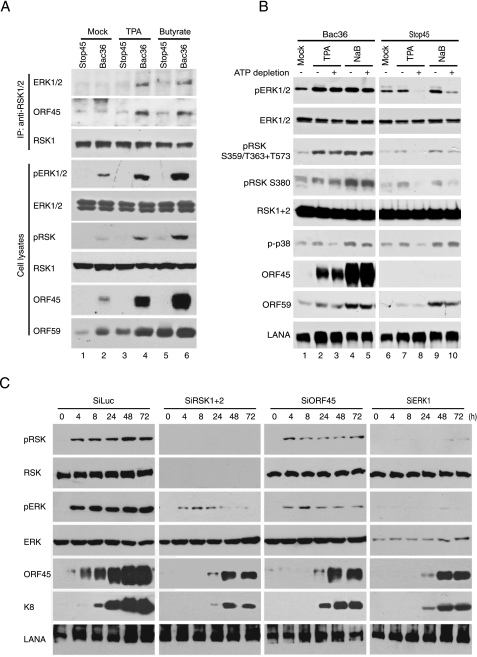
We first determined whether loss of ORF45 affected the association between RSK and ERK in KSHV-infected cells. We transfected 293T cells with the wild-type Bac36 and ORF45-null Bac-stop45 DNAs to establish KSHV latently infected Bac36-293T and Bac-stop45-293T cells as previously described (43). We then treated both types of cells with either TPA or sodium butyrate for 3 days to induce KSHV lytic replication. After induction with TPA, a protein kinase C agonist that induces KSHV lytic replication through activating ERK MAPK signaling (35, 44, 45), a lower level of lytic gene expression was observed in Bac-stop45-293T than in Bac36-293T cells (Fig. 6A, compare the ORF59 signals of lanes 3 and 4). The difference became negligible after induction with sodium butyrate, a histone deacetyltransferase inhibitor that presumably induces KSHV lytic cycle by a different mechanism (Fig. 6A, see the ORF59 signals of lanes 5 and 6, and data not shown). No obvious difference in expression of several KSHV lytic genes was observed between Bac-stop45-293T and Bac36-293T cells when they were induced with a combination of TPA and sodium butyrate (43). The lysates of TPA- or sodium butyrate-induced cells were immunoprecipitated with anti-RSK antibody, and immunocomplexes were then analyzed for the presence of ERK. As shown in Fig. 6A, the association between ERK and RSK was evident in Bac36-293T cells (lanes 4 and 6) but was much weaker in Bac-Stop45 293T cells no matter what inducing agent was used (lanes 3 and 5). Importantly, such association was correlated with the activation of RSK and ERK, as demonstrated by higher levels of pRSK and pERK signals in Bac36-293T and lower ones in Bac-stop45 cells (compare lanes 4 and 3 and lanes 6 and 5). This experiment demonstrated that loss of ORF45 reduced ERK/RSK interaction and abolished sustained activation of ERK/RSK signaling. At least in the case of sodium butyrate induction, we observed no obvious defect on expression of other KSHV lytic genes (Fig. 6A and data not shown), so we conclude that ORF45 plays a critical role in the sustained activation of RSK and ERK by increasing their association during KSHV lytic replication.
FIGURE 6.
ORF45, RSK, and ERK are all required for the sustained activation of RSK and ERK during KSHV reactivation. A, loss of ORF45 abolishes RSK/ERK association and activation during KSHV lytic replication. Stable wild-type Bac36-293T and ORF45-null Bac-stop45-293T cells were treated with TPA or sodium butyrate for 72 h. Cell lysates were immunoprecipitated with rabbit polyclonal anti-RSK1/2 antibodies, and the immunocomplexes were analyzed by Western blotting with mouse monoclonal anti-ERK1/2 antibodies. Cell lysates were also analyzed by Western blotting with phosphorylation-specific antibodies against pRSK and pERK as well as antibodies against KSHV lytic gene products ORF45 and ORF59. Membranes were stripped and reprobed with anti-ERK or anti-RSK antibodies for determination of total ERK and RSK. B, ORF45 protects pRSK and pERK from dephosphorylation during KSHV lytic replication. Stable wild-type Bac36-293T and ORF45-null Bac-stop45-293T cells were treated with TPA and sodium butyrate for 72 h and then subjected to ATP depletion as described under “Experiment Procedures.” The cells were harvested 30 min after depletion. Cell lysates were analyzed by Western blotting with antibodies as indicated. LANA, latency-associated nuclear antigen. C, knockdown of ORF45·RSK·ERK triplex component by siRNA drastically reduced sustained activation of ERK and RSK signaling during KSHV lytic replication. BCBL cells were transduced with retrovirus-based siRNAs of ORF45, RSK1/2, ERK, and luciferase as a control. The cells were induced with TPA for various lengths of time as indicated. Cell lysates were then prepared and analyzed by Western blotting with the specified antibodies.
We next determined whether ORF45 could protect active pERK and pRSK from dephosphorylation during KSHV lytic replication. Bac36-293T and Bac-stop45-293T cells were induced with TPA or sodium butyrate for 3 days and then subjected to ATP depletion as described under “Experimental Procedures,” As shown in Fig. 6B, the levels of ERK and RSK phosphorylation were not affected in Bac36-293T cells (Fig. 6B, compare lane 3 to lane 2 and lane 5 to lane 4) but were dramatically diminished in Bac-stop45 cells by ATP depletion (compare lane 8 to lane 7 and lane 10 to lane 9), suggesting that ORF45 protects pERK and pRSK from dephosphorylation. In contrast, ORF45 seems to have no effect on dephosphorylation of phospho-p38 after ATP depletion. These results provide evidence that ORF45 plays a crucial role in sustained activation of ERK and RSK by protecting them from dephosphorylation in KSHV lytically infected cells.
We have shown that ORF45 forms complexes with RSK and ERK and activates both of them. The activation of ERK by ORF45 requires RSK, and RSK activation by ORF45 requires ERK. Furthermore, we determined whether both RSK and ERK are essential for sustained activation of ERK and RSK during KSHV lytic replication. The expression of ORF45, ERK, or RSK in BCBL-1 cells was ablated with the appropriate specific siRNAs as described under “Experimental Procedures.” The status of ERK and RSK phosphorylation was determined at different times after TPA treatment (Fig. 6C). Sustained ERK and RSK phosphorylation up to 72 h was observed in control siRNA-luc-treated cells. Ablation of either ORF45, ERK, or RSK resulted in a decreased level of ERK and RSK phosphorylation and much short duration, suggesting that ERK, RSK, and ORF45 are all necessary for the sustained ERK/RSK signaling in the KSHV lytic cycle.
DISCUSSION
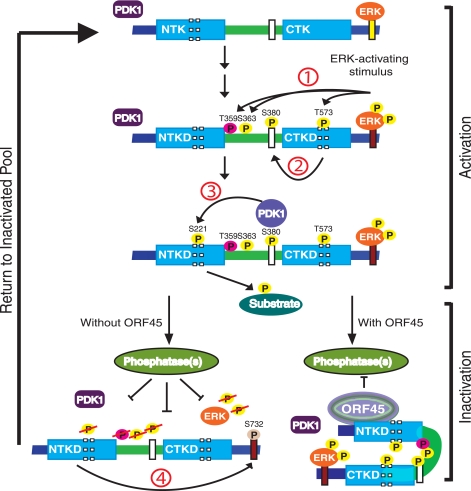
The aim of this study was to elucidate the mechanism by which KSHV ORF45 activates the sustained ERK and RSK during KSHV lytic replication. We found that KSHV ORF45 increases RSK/ERK association and that all three form multiprotein complexes in which activated pRSK and pERK are highly enriched. With the aid of kinase-dead ERK and RSK mutants, we found that ERK activity is required for RSK activation by ORF45, but RSK kinase activity is not required ERK activation under the same conditions, suggesting that RSK serves as a scaffold that bridges ORF45 and ERK to form complexes. Endogenous ERK activity seems to be required for ORF45-induced RSK activation because knockdown of ERK expression by siRNAs reduces the level of activation. Conceivably, the residual ERK activity allows phosphorylation of RSK to initiate its activation. In the absence of ORF45, activated pRSK autophosphorylates itself at Ser-732 (corresponding to Ser-749 in avian RSK1), near the ERK docking site, causing dissociation of pRSK from pERK. Subsequently, both pRSK and pERK are quickly dephosphorylated and, thus, inactivated by cellular phosphatases, but when RSK is bound by KSHV ORF45, the otherwise transient interaction between pRSK and pERK becomes more stable, so they form multiprotein complexes. Such complexes seem to shield the active pERK and pRSK from cellular-phosphatase attack. As a result, their activities are sustained at high levels (Fig. 7).
FIGURE 7.
Schematic diagram of KSHV ORF45-mediated sustained activation of ERK and RSK. The current model suggests that RSK activation consists of multistep ordered phosphorylation events (24-26, 60). First, ERK bound to the C-terminal docking site phosphorylates Thr-573 in the activation loop of CTKD and Thr-359/Ser-363 in the linker region upon upstream signal stimulation. Second, phosphorylation of Thr-573 activated CTKD and enabled it to autophosphorylate Ser-380 in the hydrophobic motif. Third, phosphorylation of Ser-380 generated a docking site for 3-phosphoinositide-dependent protein kinase 1, and allowed it to phosphorylate Ser-221 in the activation loop of NTKD and activate RSK completely. The fully activated RSK relies on the NTKD to phosphorylate various exogenous substrates. Finally, the NTKD also phosphorylates Ser-732 (corresponding to Ser-749 in avian RSK1) in the ERK docking site, causing dissociation of pERK from pRSK. Subsequently, dissociated pRSK and pERK were dephosphorylated by cellular phosphatases. Binding of ORF45 to RSK prevents dissociation between ERK and RSK, and all three, therefore, form multiprotein complexes. Through possible conformational reorganization, the complexes could shield active pERK and pRSK from phosphatase attack, so they are accumulated, resulting in sustained activation of RSK and ERK.
The interaction of ERK with RSK is critical for RSK activation. In the current model ERK binds to RSK in quiescent cells (27-29). Upon stimulation by a mitogen such as epidermal growth factor or TPA, ERK is activated and subsequently dissociated from activated RSK, resulting in usually transient RSK activation (Fig. 6 and Ref. 28). Different cellular mechanisms that regulate such interaction and, thus, RSK activation have been reported. Recently a cellular protein, PEA-15, has been reported to interact with both RSK2 and ERK as a scaffold, regulating RSK2 activation in a concentration-dependent manner (46). Another recent study demonstrates that phosphorylation of tyrosine 529 on RSK2 (amino acid number refers to murine RSK2) by fibroblast growth factor receptor 3 promotes the interaction between ERK and RSK2 and subsequent phosphorylation of RSK2 by ERK (47). Interestingly, both mechanisms seem to be specific to RSK2 (46, 47). In contrast, we found that ORF45 activates both RSK1 and RSK2; thus, a different mechanism seems to be involved in the action of ORF45. Binding of ORF45 to the N-terminal domain of RSK reinforces its interaction with ERK, which depends on the ERK docking site near the C terminus, but the exact mechanism is unclear. A conformational reorganization could be involved (Fig. 7), and further structural-functional analysis is needed to reveal the mechanism in detail.
Repeated attempts failed to demonstrate that ORF45 activates RSK directly in vitro. A lead prompted us to examine the dephosphorylation process. Reversible phosphorylation plays a key role in cellular signal transduction. The levels of ERK and RSK phosphorylation in cells are determined by both phosphorylation and dephosphorylation processes. Numerous phosphatases such as PP2A, dual-specificity phosphatases, MAPK phosphatase-1, and phosphatase-3 have been implicated in the regulation of ERK signaling (48-51). Because of the critical role of dephosphorylation processes in the regulation of MAPK signal strength and duration, viruses evolve various mechanisms to suppress it. For example, SV40 small T antigen interacts with and inhibits protein phosphatase PP2A to allow accumulation of phosphorylated MKK1/2 and ERK (52); human cytomegalovirus infection causes sustained activation of both ERK and p38 by virally induced mechanisms that appear to involve the inhibition of a cellular phosphatase activity, but the details remain elusive, and the viral factors responsible are unknown (9, 38).
The ATP depletion experiment indicates that ORF45 protects pERK and pRSK from dephosphorylation in vivo (Fig. 4A). The in vitro dephosphorylation assays suggest that the pERK is protected from dephosphorylation when both ORF45 and RSK are present but not when either one is absent (Fig. 5, B and C). The dependence on RSK suggests that tertiary structure is required for this protection. Possibly, the complex of ORF45, RSK, and ERK provides a physical shield that fends off cellular phosphatases, resulting in an extended activation of ERK and RSK in the complexes. This conjecture is supported by the observations that most of the activated pERK and pRSK in cells is associated with the ORF45-containing complexes (Fig. 2, A and B). These observations also explain ORF45 causing sustained activation of both ERK and RSK even though RSK negatively regulates ERK activation in general (53, 54). Interestingly, in these in vitro assays pERK seemed to be better protected than pRSK by ORF45-induced complexes. We hypothesize that pERK is the primary target of protection, whereas pRSK is protected indirectly through active phosphorylation by pERK. Conceivably, such a phosphorylation event would occur in cells but might not happen in the in vitro dephosphorylation assays or ATP depletion experiments because of absence of ATP. This model explains why both RSK and ERK are activated and sustained in the ORF45-expressing cells. In conclusion, our data suggest that ORF45 activates both RSK and ERK largely by increasing their association and decreasing their dephosphorylation. Because the structural natures and components of ORF45·RSK complexes are largely unknown, the mechanistic details remain to be determined. In addition to inhibition of dephosphorylation, other mechanisms not mutually exclusive are also possible.
Note that almost all ORF45 proteins in cells reside in the high Mr complexes (Fig. 2A). The exact components of such complexes have not been fully characterized, but RSK and ERK are probably among them. Because ORF45 is a virion tegument protein, it is expected to interact with other virion proteins in various stages of the viral life cycle. Indeed, a recent virion-wide interaction screening has revealed that ORF45 interacts with capsid protein ORF62, tegument proteins ORF63, and ORF64 (55). ORF45 has also been found to interact with RTA in another viral genome-wide interaction screening (56). Furthermore, KSHV ORF45 is known to interact with other cellular proteins such as IRF-7 and SIAH-1 (37, 57). Therefore, KSHV ORF45 may form different complexes in cells in different situations, so further characterization of the natures and components of such complexes is important to revealing not only the mechanism of sustained ERK/RSK activation by ORF45 but also the functional consequences of such activation in the KSHV life cycle.
Here we have described a novel mechanism of extended activation of both ERK and RSK by KSHV ORF45. Similar high molecular mass ORF45·RSK complexes are detectable in TPA-induced BCBL-1 cells (data not shown), suggesting that such a mechanism could be the cause of sustained ERK/RSK signaling activities in KSHV lytically infected cells. Indeed, the association between ERK and RSK became weaker in ORF45-null KSHV-infected cells, making pERK/pRSK more vulnerable to dephosphorylation by cellular phosphatase(s) (Fig. 6, A and B). Furthermore, knockdown of expression of any one of ORF45, RSK, or ERK by siRNA reduced the level of their phosphorylation and abolished the sustained activation of RSK and ERK (Fig. 6C). Although the sustained activation of the ERK signaling cascade has been noted in several viruses, its exact role in the viral life cycle is not well understood (5, 8, 13). We previously demonstrated that ablation of RSK expression or inhibition of RSK activation by specific inhibitors leads to lower levels of lytic gene expression in TPA-induced BCBL-1 cells, suggesting an essential role of RSK in KSHV lytic replication (23). Besides playing a role in the viral life cycle, sustained ERK/RSK activation might be involved in viral pathogenesis because of the critical role of ERK MAPK signaling in regulation of cell growth, survival, and proliferation (1, 4, 24-26). Although lytically infected cells are thought to die within a few days, spontaneous lytic replications of KSHV are crucial for KSHV pathobiology because they facilitate dissemination of the virus and cause expression of viral lytic oncoproteins that are involved in paracrine regulation of tumor growth. Conceivably, extension of survival of spontaneously reactivated cells would prolong and increase paracrine regulation and would, thus, have a significant effect on KSHV pathogenesis. The ERK/RSK MAPK signaling cascade has been known to promote cell survival and proliferation, so the ORF45·RSK·ERK axis may lead to prolonged cell survival and, thus, contribute to KSHV viral pathogenesis. Because of the essential role of RSK-dependent signaling in KSHV lytic replication and possible pathogenesis (23), development of specific RSK inhibitors may represent a promising and more specific antiviral strategy for treatment of KSHV-related diseases (32, 58, 59).
Acknowledgments
We are grateful to Drs. Melanie H. Cobb, Shou-Jiang Gao, Deborah A. Lannigan, and James Maller for kindly providing reagents. We thank Drs. Betty Gaffney and Hengli Tang for reading the manuscript and Dr. Anne B. Thistle for excellent editorial assistance. We also thank Hongying Deng for generating HEK293-tet-ORF45 cell line and all members of the Zhu laboratory for suggestions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DE016680. This work was also supported by a Bankhead-Coley bridge grant, Florida State University set-up funds, and a planning grant (to F. Z.).
Footnotes
The abbreviations used are: ERK, extracellular signal-regulated kinase; Bac, bacterial artificial chromosome; BCBL, body cavity-based lymphoma; CIAP, calf intestine alkaline phosphatase; CTKD, C-terminal kinase domain; HPLC, high performance liquid chromatography; KSHV, Kaposi sarcoma-associated herpesvirus; MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; NTKD, N-terminal kinase domain; ORF45, open reading frame 45; pERK, phosphorylated ERK; RSK, p90 ribosomal S6 kinase; pRSK, phosphorylated RSK; RPMI, Roswell Park Memorial Institute; PP2A, phosphatase 2A; Tet, tetracycline; TPA, 12-O-tetradecanoylphorbol-13-acetate; HA, hemagglutinin; siRNA, small interfering RNA; IP, immunoprecipitated; KD, kinase dead.
E. Kuang and F. Zhu, unpublished data.
References
- 1.Roux, P. P., and Blenis, J. (2004) Microbiol. Mol. Biol. Rev. 68 320-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Z., Gibson, T. B., Robinson, F., Silvestro, L., Pearson, G., Xu, B., Wright, A., Vanderbilt, C., and Cobb, M. H. (2001) Chem. Rev. 1012449 -2476 [DOI] [PubMed] [Google Scholar]
- 3.Raman, M., Chen, W., and Cobb, M. H. (2007) Oncogene 263100 -3112 [DOI] [PubMed] [Google Scholar]
- 4.McCubrey, J. A., Steelman, L. S., Chappell, W. H., Abrams, S. L., Wong, E. W., Chang, F., Lehmann, B., Terrian, D. M., Milella, M., Tafuri, A., Stivala, F., Libra, M., Basecke, J., Evangelisti, C., Martelli, A. M., and Franklin, R. A. (2007) Biochim. Biophys. Acta 17731263 -1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadagopan, S., Sharma-Walia, N., Veettil, M. V., Raghu, H., Sivakumar, R., Bottero, V., and Chandran, B. (2007) J. Virol. 813949 -3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma-Walia, N., Krishnan, H. H., Naranatt, P. P., Zeng, L., Smith, M. S., and Chandran, B. (2005) J. Virol. 7910308 -10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacque, J. M., Mann, A., Enslen, H., Sharova, N., Brichacek, B., Davis, R. J., and Stevenson, M. (1998) EMBO J. 172607 -2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pleschka, S., Wolff, T., Ehrhardt, C., Hobom, G., Planz, O., Rapp, U. R., and Ludwig, S. (2001) Nat. Cell Biol. 3301 -305 [DOI] [PubMed] [Google Scholar]
- 9.Rodems, S. M., and Spector, D. H. (1998) J. Virol. 729173 -9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang, X., and Gabuzda, D. (1999) J. Virol. 733460 -3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie, J., Pan, H., Yoo, S., and Gao, S. J. (2005) J. Virol. 7915027 -15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan, H., Xie, J., Ye, F., and Gao, S. J. (2006) J. Virol. 805371 -5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo, H., Yanagawa, B., Zhang, J., Luo, Z., Zhang, M., Esfandiarei, M., Carthy, C., Wilson, J. E., Yang, D., and McManus, B. M. (2002) J. Virol. 763365 -3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade, A. A., Silva, P. N., Pereira, A. C., De Sousa, L. P., Ferreira, P. C., Gazzinelli, R. T., Kroon, E. G., Ropert, C., and Bonjardim, C. A. (2004) Biochem. J. 381437 -446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai, Y., Liu, Y., and Zhang, X. (2007) J. Virol. 81446 -456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong, X., San Juan, H., Behera, A., Peeples, M. E., Wu, J., Lockey, R. F., and Mohapatra, S. S. (2004) FEBS Lett. 55933 -38 [DOI] [PubMed] [Google Scholar]
- 17.Nijhara, R., Jana, S. S., Goswami, S. K., Rana, A., Majumdar, S. S., Kumar, V., and Sarkar, D. P. (2001) J. Virol. 7510348 -10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon, S., and Seger, R. (2006) Growth Factors 2421 -44 [DOI] [PubMed] [Google Scholar]
- 19.Murphy, L. O., and Blenis, J. (2006) Trends Biochem. Sci. 31268 -275 [DOI] [PubMed] [Google Scholar]
- 20.Marshall, C. J. (1995) Cell 80 179-185 [DOI] [PubMed] [Google Scholar]
- 21.Cowley, S., Paterson, H., Kemp, P., and Marshall, C. J. (1994) Cell 77 841-852 [DOI] [PubMed] [Google Scholar]
- 22.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F., and Ahn, N. G. (1994) Science 265966 -970 [DOI] [PubMed] [Google Scholar]
- 23.Kuang, E., Tang, Q., Maul, G. G., and Zhu, F. (2008) J. Virol. 821838 -1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauge, C., and Frodin, M. (2006) J. Cell Sci. 1193021 -3023 [DOI] [PubMed] [Google Scholar]
- 25.Carriere, A., Ray, H., Blenis, J., and Roux, P. P. (2008) Front. Biosci. 134258 -4275 [DOI] [PubMed] [Google Scholar]
- 26.Anjum, R., and Blenis, J. (2008) Nat. Rev. Mol. Cell Biol. 9747 -758 [DOI] [PubMed] [Google Scholar]
- 27.Gavin, A. C., and Nebreda, A. R. (1999) Curr. Biol. 9281 -284 [DOI] [PubMed] [Google Scholar]
- 28.Roux, P. P., Richards, S. A., and Blenis, J. (2003) Mol. Cell. Biol. 234796 -4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, J. A., Poteet-Smith, C. E., Malarkey, K., and Sturgill, T. W. (1999) J. Biol. Chem. 2742893 -2898 [DOI] [PubMed] [Google Scholar]
- 30.Trivier, E., De Cesare, D., Jacquot, S., Pannetier, S., Zackai, E., Young, I., Mandel, J. L., Sassone-Corsi, P., and Hanauer, A. (1996) Nature 384567 -570 [DOI] [PubMed] [Google Scholar]
- 31.Clark, D. E., Errington, T. M., Smith, J. A., Frierson, H. F., Jr., Weber, M. J., and Lannigan, D. A. (2005) Cancer Res. 653108 -3116 [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. A., Poteet-Smith, C. E., Xu, Y., Errington, T. M., Hecht, S. M., and Lannigan, D. A. (2005) Cancer Res. 651027 -1034 [PubMed] [Google Scholar]
- 33.Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M., and Moore, P. S. (1994) Science 2661865 -1869 [DOI] [PubMed] [Google Scholar]
- 34.Ganem, D. (2007) in Fields Virology (Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., and Straus, S. E., eds) 5th Ed., pp2847 -2888, Lippincott Williams & Wilkins, Philadelphia, PA
- 35.Xie, J., Ajibade, A. O., Ye, F., Kuhne, K., and Gao, S. J. (2008) Virology 371139 -154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, F. X., Chong, J. M., Wu, L., and Yuan, Y. (2005) J. Virol. 79800 -811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, F. X., King, S. M., Smith, E. J., Levy, D. E., and Yuan, Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 995573 -5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, R. A., Huong, S. M., and Huang, E. S. (2000) J. Virol. 741158 -1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meriin, A. B., Yaglom, J. A., Gabai, V. L., Zon, L., Ganiatsas, S., Mosser, D. D., Zon, L., and Sherman, M. Y. (1999) Mol. Cell. Biol. 192547 -2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, P. Y., Liu, P., Weng, J., Sontag, E., and Anderson, R. G. (2003) EMBO J. 222658 -2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, F. X., Cusano, T., and Yuan, Y. (1999) J. Virol. 735556 -5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, F. X., and Yuan, Y. (2003) J. Virol. 774221 -4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu, F. X., Li, X., Zhou, F., Gao, S. J., and Yuan, Y. (2006) J. Virol. 8012187 -12196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen, A., Brodie, C., and Sarid, R. (2006) J. Gen. Virol. 87795 -802 [DOI] [PubMed] [Google Scholar]
- 45.Ford, P. W., Bryan, B. A., Dyson, O. F., Weidner, D. A., Chintalgattu, V., and Akula, S. M. (2006) J. Gen. Virol. 871139 -1144 [DOI] [PubMed] [Google Scholar]
- 46.Vaidyanathan, H., Opoku-Ansah, J., Pastorino, S., Renganathan, H., Matter, M. L., and Ramos, J. W. (2007) Proc. Natl. Acad. Sci. U. S. A. 10419837 -19842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang, S., Dong, S., Gu, T. L., Guo, A., Cohen, M. S., Lonial, S., Khoury, H. J., Fabbro, D., Gilliland, D. G., Bergsagel, P. L., Taunton, J., Polakiewicz, R. D., and Chen, J. (2007) Cancer Cell 12201 -214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, Y., Shepherd, E. G., and Nelin, L. D. (2007) Nat. Rev. Immunol. 7202 -212 [DOI] [PubMed] [Google Scholar]
- 49.Jeffrey, K. L., Camps, M., Rommel, C., and Mackay, C. R. (2007) Nat. Rev. Drug Discov. 6 391-403 [DOI] [PubMed] [Google Scholar]
- 50.Zhou, B., Wang, Z. X., Zhao, Y., Brautigan, D. L., and Zhang, Z. Y. (2002) J. Biol. Chem. 27731818 -31825 [DOI] [PubMed] [Google Scholar]
- 51.Blanco-Aparicio, C., Torres, J., and Pulido, R. (1999) J. Cell Biol. 1471129 -1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sontag, E., Fedorov, S., Kamibayashi, C., Robbins, D., Cobb, M., and Mumby, M. (1993) Cell 75 887-897 [DOI] [PubMed] [Google Scholar]
- 53.Dufresne, S. D., Bjorbaek, C., El-Haschimi, K., Zhao, Y., Aschenbach, W. G., Moller, D. E., and Goodyear, L. J. (2001) Mol. Cell. Biol. 2181 -87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim, M., Lee, J. H., Koh, H., Lee, S. Y., Jang, C., Chung, C. J., Sung, J. H., Blenis, J., and Chung, J. (2006) EMBO J. 253056 -3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozen, R., Sathish, N., Li, Y., and Yuan, Y. (2008) J. Virol. 824742 -4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uetz, P., Dong, Y. A., Zeretzke, C., Atzler, C., Baiker, A., Berger, B., Rajagopala, S., Roupelieva, M., Rose, D., Fossum, E., and Haas, J. (2006) Science 311239 -242 [DOI] [PubMed] [Google Scholar]
- 57.Abada, R., Dreyfuss-Grossman, T., Herman-Bachinsky, Y., Geva, H., Masa, S. R., and Sarid, R. (2008) J. Virol. 822230 -2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen, M. S., Zhang, C., Shokat, K. M., and Taunton, J. (2005) Science 3081318 -1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapkota, G. P., Cummings, L., Newell, F. S., Armstrong, C., Bain, J., Frodin, M., Grauert, M., Hoffmann, M., Schnapp, G., Steegmaier, M., Cohen, P., and Alessi, D. R. (2007) Biochem. J. 401 29-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cuadrado, A., and Nebreda, A. R. (2007) Cancer Cell 12187 -189 [DOI] [PubMed] [Google Scholar]