Abstract
Recently, Salmonella spp. were shown to induce apoptosis in infected macrophages. The mechanism responsible for this process is unknown. In this report, we establish that the Inv-Spa type III secretion apparatus target invasin SipB is necessary and sufficient for the induction of apoptosis. Purified SipB microinjected into macrophages led to cell death. Binding studies show that SipB associates with the proapoptotic protease caspase-1. This interaction results in the activation of caspase-1, as seen in its proteolytic maturation and the processing of its substrate interleukin-1β. Caspase-1 activity is essential for the cytotoxicity. Functional inhibition of caspase-1 activity by acetyl-Tyr-Val-Ala-Asp-chloromethyl ketone blocks macrophage cytotoxicity, and macrophages lacking caspase-1 are not susceptible to Salmonella-induced apoptosis. Taken together, the data demonstrate that SipB functions as an analog of the Shigella invasin IpaB.
The enteric, Gram-negative bacteria Salmonellae are a significant cause of morbidity and mortality among human populations, especially those in regions of the world lacking adequate sanitation and health delivery systems. Salmonella typhimurium and Salmonella typhi are two serotypes of particular interest to researchers because they account for the most prevalent and the most severe diseases, respectively, among genus members. S. typhimurium is a leading cause of gastroenteritis, an acute localized inflammation of the intestine. It is generally self-limiting and without complication provided supportive therapy is available. Typhoid fever, a more lethal yet less common systemic disease mainly caused by S. typhi, is marked by protracted fever and a variable clinical presentation and course depending on the organ systems affected (1).
Both serotypes are acquired through the ingestion of contaminated food or water. After passage through the upper gastrointestinal tract, Salmonellae infect the host at the level of the ileum or the colon (2). The bacteria cross the intestinal epithelial barrier by translocating through the M cells that are positioned among the epithelial cells overlying lymphoid follicles (3, 4). Salmonella subvert the normal function of M cells to sample luminal antigens to invade the host. After reaching the lamina propria of the intestinal wall, Salmonella encounter resident tissue macrophages that intimately associate with the M cells. Rather than being destroyed by these professional phagocytes after internalization, Salmonella survives intracellularly (5). In vitro studies have documented that infection of macrophages by several Salmonella spp., including S. typhimurium and S. typhi, leads to the death of the phagocyte by apoptosis (6–8).
Shigella flexneri, the etiological agent of bacillary dysentery, is an enteric pathogen that induces macrophage apoptosis in vitro and in vivo (9–11). The secreted invasin IpaB is necessary and sufficient for this process (12, 13). According to the current model, cytoplasmically distributed IpaB binds to the cysteine protease caspase-1 [interleukin 1-β (IL-1β) converting enzyme] (14), leading to its activation (15). Caspase-1 activity is crucial for the induction of Shigella-induced macrophage apoptosis (13, 16).
A novel set of Salmonella virulence genes located in an operon at centisome 63 of the chromosome were identified through mutagenesis studies (17–20). The operon was denoted sip (Salmonella invasion protein), and contains five genes: sipE, sipB, sipC, sipD, and sipA. Not only are the proteins encoded by each of these genes homologous to corresponding gene products of the ipa operon of Shigella, the genes of the operons are organized in the same order (17). In particular, SipB and IpaB share 55% identity over the C-terminal 500 amino acids of the proteins (17). The suggestion of functional similarity between these two proteins is supported by the ability of SipB to complement a S. flexneri ipaB mutant for HeLa cell invasion and escape from the phagosome in macrophages (17).
To this point, the only pathogenic function tentatively ascribed to SipB is derived from studies involving the translocation of proteins via the Inv-Spa type III secretion apparatus in sipB mutants. Several effector proteins (such as SptP, SopE, and AvrA) can be secreted into bacterial supernatants, but they are not delivered to the eukaryotic cytoplasm by Salmonella strains containing this mutation (21–23). These results suggest that SipB may be involved in the translocation process.
In this study, we investigated the potential role of SipB in Salmonella-induced macrophage apoptosis and the potential involvement of caspase-1 in this process. We report that SipB is necessary for Salmonella-induced macrophage apoptosis to occur, and that this protein functions by directly interacting with the essential proapoptotic enzyme caspase-1 in a manner that activates this protease.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions.
The strains of bacteria used in this study were as follows. Wild-type, pathogenic Salmonella strains were S. typhi Ty2, S. typhimurium C5 (17), and S. typhimurium SL1344 (24). S. typhi Ty233 and S. typhimurium ST100 are insertion mutants in sipB of the parental strains (17). BJ66 is a noninvasive isogenic mutant strain of SL1344 containing an insertion in orgA (25). SF620 is an avirulent derivative of the S. flexneri wild-type strain M90T, which contains a nonpolar deletion mutant of the ipaB gene (26).
S. typhi sipB was amplified by PCR and a FLAG epitope tag (N-DYKDDDDK-C) was included at the 3′ end of sipB. The PCR product was ligated into the vector pUC19 using PstI and XbaI sites (pFSipB).
Salmonella strains were grown overnight without agitation in LB medium supplemented with 139 mM NaCl at 37°C. The day of the assay, bacteria were subcultured into LB broth + NaCl and grown for 3 hr at 37°C. The Shigella strains were grown with aeration in tryptic soy broth at 37°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 10 μg/ml.
Cytotoxicity Assays.
Murine macrophages were seeded onto a 96-well plate at a density of 2 × 104 cells per well and incubated overnight at 37°C. Before infection with the bacteria, the medium was replaced with serum-free RPMI 1640 medium. Infection of cells with Shigella and Salmonella was done as described (27), by spinning bacteria of 100:1 multiplicity of infection (moi), unless otherwise stated, onto the cells (600 × g for 10 min) followed by incubation at 37°C. After 30 min of incubation, gentamycin (90 μg/ml) was added to kill extracellular bacteria. At the indicated time points during the infections, culture supernatants were collected for analysis as the experimental release samples. Cytotoxicity was quantified colorimetrically with the CytoTox96 lactate dehydrogenase (LDH)-release kit (Promega). The percentage of cytotoxicity was calculated with the formula: 100 × [(experimental release − spontaneous release)/(total release − spontaneous release)], in which spontaneous release is the amount of LDH activity in the supernatant of uninfected cells and total release is the activity in macrophage lysates.
Caspase-1 Inhibitor Studies.
RAW264.7 macrophages were incubated with 50 μM acetyl-Tyr-Val-Ala-Asp-chloromethyl ketone (Ac-YVAD-CMK; Bachem Bioscience, King of Prussia, PA) 1 hr before infection. Infection of the macrophages was as described earlier. Culture supernatants were assayed for the release of LDH at 4 hr postinfection.
Microinjection Assay.
The microinjection experiments were performed as described (28). Briefly, peritoneal macrophages were isolated from mice and plated onto glass coverslips in RPMI 1640 medium (GIBCO/BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum, 2 mM glutamine, and 50 μg/ml each penicillin/streptomycin (complete medium). The cells were incubated at 37°C for 3–8 hr then washed with PBS. Samples were loaded into glass capillary micropipets made with an automatic P80/PC micropipet puller (Sutter Instruments.). A specific area on the coverslip was scratched to localize the injection area, and a monolayer of macrophages in this area was microinjected (0.3–0.7 × 10−11 ml per cell) with coded samples (750 μg protein/ml) by using an Eppendorf microinjection system. After microinjection, cells were incubated at 37°C for 4–6 hr and living cells were stained with 1 μM propidium iodide (PI)/PBS. Every microinjected cell was identified and scored for PI uptake into the nucleus. No background killing within the macrophage monolayer was observed because of the mechanical manipulations from the injection equipment, as controlled by injecting PBS. The glutathione S-transferase (GST) sample controlled for the presence of this peptide in the GST fusion proteins.
Immunocytochemistry.
Indirect immunofluorescence studies were performed as follows: RAW264.7 macrophages were seeded onto 22-cm glass coverslips in 35-mm tissue culture dishes at a density of 5 × 105 per dish and incubated overnight at 37°C. Cells were infected by spinning bacteria at 100:1 moi (600 × g for 10 min) onto them and then incubation at 37°C. At the time point, cells were washed with PBS, fixed with 4% paraformaldehyde/PBS, quenched with 0.1 M glycine/PBS, permeabilized with cold acetone, blocked with normal goat serum, and then incubated with anti-FLAG M2 Antibody (Eastman Kodak) + RNase A (1 mg/ml). Cells were then blocked again, incubated with an anti-mouse secondary antibody conjugated to fluorescein, and lastly, incubated with PI before mounting on glass slides for analysis by confocal microscopy (Molecular Dynamics). For immunoelectron microscopy, RAW264.7 macrophages were infected with S. typhi Ty233/pFSipB or Ty233/pSipB for 40 min followed by fixation with a 2% formaldehyde and 1% glutaraldehyde mixture for 35 min on ice. The samples were dehydrated by incubations in a graded ethanol series and then infiltrated with graded lowicryl white series. The samples were incubated at 55°C on a rotator until polymerization at 50°C. Sections (70 nm) were blocked with 2% BSA and then incubated with the anti-FLAG mAb (1:250) for 1 hr. Samples were then incubated with anti-mouse IgG conjugated to 10-nm gold particles (BBInternational) for 30 min followed by stainings with 1% uranyl acetate for 3 min and lead citrate for 30 sec. Samples were analyzed on a CM12 Philips Transmission electron microscope. Images are presented as the original and a version modified by using photoshop (Adobe Systems, Mountain View, CA) in which the background intensity has been decreased and individual gold particles are highlighted.
Maturation Assays.
C57BL/6 mice were injected i.p. with 1 ml of thioglycolate medium (GIBCO/BRL) 4 days before they were killed. Macrophages were harvested by peritoneal lavage with cold, sterile PBS, washed in RPMI 1640 medium, plated at a density of 5 × 105 onto 24-well plates in RPMI complete medium, and then incubated at 37°C for 3 hr. The adherent cells were washed three times with media and then activated overnight by culture media containing 1 μg/ml phenol-water purified S. flexneri 1A lipopolysaccharide (LPS) (Sigma). The cells were then washed with serum-free media before infection with indicated bacteria. Gentamycin (90 μg/ml) was added to cell cultures infected for >30 min. Lysates of infected cells were cleared by centrifugation, and then analyzed by SDS/PAGE followed by transfer to nitrocellulose and immunoblotting (29). Caspase-1 and IL-1β maturation was determined by immunoblotting with a rabbit anti-mouse caspase-1 (p45) antibody (R10311, a kind gift of Doug Miller, Merck Laboratories, Rahaway, NJ) or a goat anti-mouse IL-1β primary antibody (R&D Systems, Minneapolis, MN).
Infections of Caspase-1 −/− Macrophages.
Caspase-1 knockout mice (kind gift of John Mudgett, Merck Laboratories) and Balb/c mice were injected with 3% thioglycolate 4 days before harvesting peritoneal macrophages. Approximately 5 × 104 peritoneal macrophages were seeded into each well of a 96-well dish and infected as described above with a 10:1 moi for 8 hr. Cytotox96 assays were then performed on the culture supernatants.
Binding Assays.
Cell-free assay. To produce GST-SipB, PCR-amplified sipB was ligated into the BamHI and XbaI sites of the expression vector pGEX-KG (30) to generate the fusion protein. Strain SF620 containing pGEX-KG/SipB or pGEX-KG alone was induced for 3 hr by isopropyl β-d-thiogalactoside at 37°C. Bacterial cultures were collected by centrifugation and resuspended in PBS with 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, and 10 μg/ml leupeptin. Bacteria were lysed by French press, and the lysates were incubated with glutathione-Sepharose 4B beads (Pharmacia Biotech). After washing, the GST-SipB or GST was eluted from the beads by glutathione, and then dialyzed and concentrated before use in the assay. J774A.1 cells (109 cells per binding reaction) were collected, washed, and lysed by incubation in lysis buffer (PBS with 1% Triton X-100, 5 mM EDTA, 0.1% 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, and 10 μg/ml leupeptin). Lysates were spun at 160,000 × g for 1 hr at 4°C, then cleared overnight by incubation with glutathione-Sepharose beads at 4°C. Either purified GST-SipB or GST (25 μg) was added to an equal aliquot of cell lysate and incubated overnight at 4°C, before washing with 60 bed volumes of 1% Triton X-100 in PBS. Proteins were eluted from the beads by boiling in SDS/PAGE sample buffer, separated by SDS/PAGE, transferred to nitrocellulose, and analyzed by immunoblotting with an anti-mouse caspase-1 antibody.
Infected, intact cell assay.
Approximately 8 × 107 J774A.1 cells were infected as described above with either SF620/pFSipB or SF620/pSipB for 40 min at 37°C. Cells were washed with cold PBS three times, scraped, and collected by centrifugation. Cells were incubated in lysis buffer mentioned above, then spun at 2,700 × g for 30 min at 4°C. The lysates were cleared overnight by IgG-agarose beads before being incubated with IgG-agarose beads preloaded with the anti-FLAG M2 antibody. After washes with 40-bed volumes of 1% Triton X-100 in PBS, proteins were eluted and processed for immunoblotting as described above.
RESULTS
Salmonella-Induced Macrophage Cytotoxicity Is SipB-Dependent.
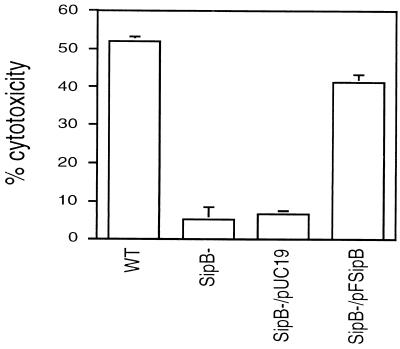
The capacity of S. typhimurium and S. typhi to induce apoptosis in macrophages was first recognized by in vitro assays using two murine macrophage cell lines, J774A.1 and RAW264.7, and bone marrow-derived macrophages as target cells (6–8). Cell death was dependent on time and moi. We sought to determine the role of SipB in this process. Fig. 1 shows the results of a cell death assay for macrophages infected for 10 hr by wild-type S. typhi, the sipB mutant Ty233, or the mutant transformed by either vector alone or vector containing sipB. Cytotoxicity was determined by measuring the release of the cytoplasmic enzyme LDH. Wild-type S. typhi killed 52% of the macrophages, whereas loss of sipB expression led to an abrogation of cytotoxicity. The dying cells demonstrated the classical cellular and nuclear morphologic alterations of apoptosis (31) (data not shown). Adding sipB in trans led to a restoration of killing, thereby demonstrating that the cytotoxic defect was caused by the loss of SipB and not to a polar effect of the mutation on other members of the operon. Similar results were seen when RAW264.7 cells, murine peritoneal macrophages, and phorbol 12-myristate 13-acetate-differentiated THP-1 human monocytic cells were tested in cytotoxicity assays (data not shown). Our results are in agreement with the report of Chen et al. (7), in which a sipB mutant was found to be noncytotoxic.
Figure 1.
SipB dependence of the cytotoxicity of Salmonella on the J774A.1 macrophage cell line. Cells were infected at an moi of 100:1 for 10 hr and then processed for a LDH-release assay. The wild-type S. typhi (WT) was cytotoxic to J774A.1 cells. An insertion mutant of sipB (SipB-) did not retain the cytotoxic phenotype, which can be complemented by sipB in trans (SipB−/pFSipB). The negative control (SipB−/pUC19) did not restore the killing activity. The means and standard deviations of three experiments are shown.
SipB Is Sufficient to Cause Apoptosis.
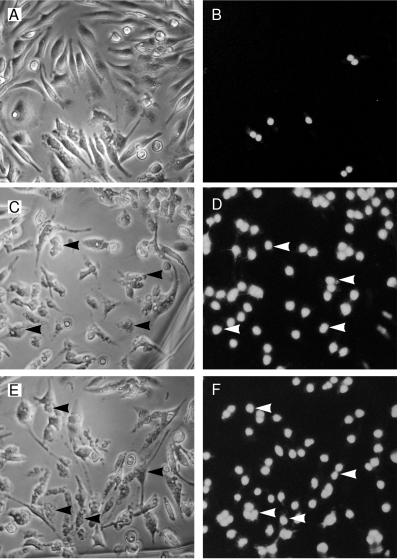
We performed microinjection studies to test whether SipB is directly involved in promoting apoptosis or indirectly involved by acting as a “translocase” (21, 22). At least 600 peritoneal macrophages for each sample were microinjected with 750 μg/ml of either purified GST–SipB fusion protein, purified GST–IpaB, or GST alone as a negative control. Cell death was quantified by PI uptake into the nucleus of the cells and confirmed by morphological alterations consistent with apoptosis. Both GST–SipB and GST–IpaB killed macrophages efficiently (76 ± 5% and 85 ± 5%, respectively), whereas, injection of GST at the same concentration yielded background levels of cytotoxicity (24 ± 4%). Fig. 2 (C and E) shows cells with characteristic apoptotic morphology such as cell rounding, shrinkage, and vacuolization. These results provide evidence of a direct role for SipB in the induction of apoptosis.
Figure 2.
Macrophages are killed after microinjection of SipB. Every macrophage in the field was microinjected with purified GST (A and B, same field), GST–IpaB (C and D, same field), or GST–SipB (E and F, same field) and then stained with PI in PBS without fixation. Apoptotic cells were then scored for PI uptake and cellular morphology. Many more cells are positive for PI uptake and apoptotic cellular alterations in GST–SipB (E and F) and GST–IpaB (C and D) injected cells than found in GST-injected cells (A and B). Arrowheads highlight some of the cells undergoing apoptosis. See text for quantification of the levels of apoptosis.
SipB Is Localized to the Cell Cytoplasm in Salmonella-Infected Macrophages.
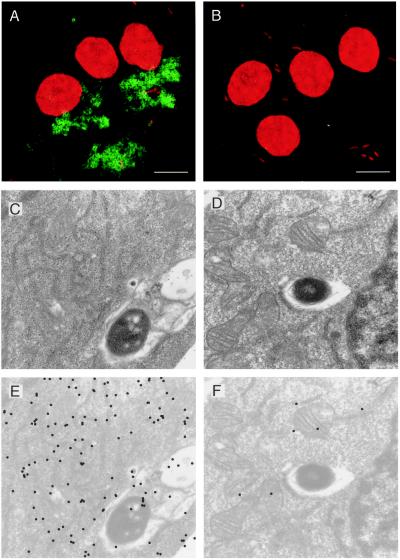
As a candidate effector molecule for inducing apoptosis, we investigated the subcellular localization of SipB. During the course of infection in macrophages, Salmonellae are maintained in an endosomal compartment; the bacteria never gain direct access to the cytoplasm. Indirect immunofluorescence was performed to determine whether SipB exists outside the confines of the phagosome during infection by S. typhi. By using PCR, we engineered a version of sipB that encodes the FLAG octapeptide epitope tag at the amino terminus of the protein (FSipB). The FSipB protein could then be detected by the monoclonal antibody against FLAG (M2Ab). FSipB is secreted by S. typhi in vitro and can complement S. typhi lacking endogenous SipB (Ty233) in macrophage cytotoxicity assays (data not shown). Macrophages were infected with either Ty233 as a negative control or Ty233 transformed with the FSipB construct (Ty233/pFSipB). After 45 min of infection, samples were processed for indirect immunofluorescence, and the slides were analyzed by confocal microscopy (Fig. 3 A and B). The bacteria and nuclei are visible in red because of staining by the DNA-binding dye PI. FSipB was detected by a fluorescein-labeled secondary antibody and appears green in the images. As seen in Fig. 3A, macrophages infected with Ty233/pFSipB contain a green fluorescence pattern that extends beyond the phagosomal boundary and encompasses a large portion of the cytoplasm. We noticed that SipB was not evenly distributed throughout the cytoplasm. In fact, even at later time points, this pattern of SipB dispersal persists (data not shown). There appears to be a restriction or exclusion of some nature occurring. Control cells infected with Ty233 (Fig. 3B) were not stained by the anti-FLAG antibody.
Figure 3.
Subcellular localization of SipB in Salmonella-infected macrophages. RAW264.7 cells infected with S. typhi Ty233 transformed by pFSipB (A, C, and E), pSipB (D and F), or vector alone (B) for 40 min. Indirect immunofluorescence (A and B): Both macrophage nuclei and the bacteria were stained with the DNA binding dye PI. FSipB was detected by using a fluorescein-labeled secondary antibody (green) and shown to be dispersed in the cytoplasm of infected macrophages. Immunoelectron microscopy: Gold particles show that the majority of FSipB is localized in the cytoplasm of the macrophage (C and E). There appears to be an association between FSipB and endomembranes, most notably endoplasmic reticulum. (E) Computer enhancement of signal found in C. SipB lacking the FLAG epitope was not detected (D and F). (F) Computer enhancement of signal found in D. (Bars = 10 μm.)
To address the uneven distribution of SipB and determine its localization with a higher degree of resolution, we performed immunoelectron microscopy. We used the Ty233/pFSipB strain (Fig. 3C) and Ty233 transformed with SipB not containing the FLAG epitope (Ty233/pSipB) as a negative control (Fig. 3D). Macrophages were infected for 45 min with these two strains and processed for immunoelectron microscopy. To visualize the ImmunoGold particles, in Fig.3 C and D the background intensity of these images was decreased and the gold particles were labeled by using Adobe photoshop to yield Fig. 3 E and F, respectively. The distribution of ImmunoGold particles representing FSipB localization concurs with the immunofluorescence images. There are gold particles associated with the bacterium and contained within the phagosome. However, a majority of the signal is found in the cytoplasm. Through a review of numerous images, we noticed that cytoplasmic SipB generally associated with intracellular membranes, predominantly what appears to be endoplasmic reticulum. As expected, sections of cells infected with the negative control Ty233/pSipB were not labeled (Fig. 3 D and F).
SipB Binds Directly to Caspase-1.
The localization studies indicate that SipB is within the macrophage cytoplasm shortly after infection by Salmonella. Thus, SipB shares the same subcellular compartment as the proapoptotic caspase-1 (32). To determine whether SipB interacts directly with caspase-1, we undertook both cell-free and intact cell binding studies. Caspase-1 is expressed in macrophages as a 45-kDa precursor form, which then requires proteolytic processing to eventually yield the 10- and 20-kDa subunits of mature caspase-1 (33).
For the cell-free binding assay, we generated a GST–SipB fusion protein to use as a ligand for affinity purifications. Macrophage cytoplasmic lysates were incubated with either GST–SipB or GST alone and then applied to glutathione-agarose beads. Immunoblotting of the fraction bound by GST–SipB revealed two proteins recognized by an anti-caspase-1 antibody (Fig. 4A). The molecular masses of these proteins were 45 and 33 kDa, matching the sizes of the precursor form of caspase-1 and an intermediate cleavage product. The interaction between SipB and caspase-1 was specific because the anti-caspase-1 antibody did not detect any proteins in the GST bound fraction.
Figure 4.
Binding assays to determine if SipB binds to caspase-1. (A) Western blot analysis of GST (lane 1) and GST–SipB (lane 2) affinity-purified proteins from J774A.1 cell lysates resolved on 15% SDS/PAGE gel with anti-mouse caspase-1 rabbit serum. GST–SipB specifically bound to two proteins of 45 and 33 kDa recognized by the anti-caspase-1 antibody. These proteins are the precursor of caspase-1 and one of its intermediate forms. (B) Western blot analysis of proteins coimmunoprecipitated by anti-FLAG antibody from J774A.1 cells infected for 40 min with either SF620/pSipB (lane 2) or SF620/pFSipB (lane 3). Included as control is lysate from uninfected equivalent number of cells (lane 1). Samples were resolved on 15% SDS/PAGE and immunoblotted with an anti-caspase-1 antibody. The precursor form of caspase-1 (p45) was coimmunoprecipitated with FSipB, demonstrating that SipB binds to caspase-1 during infection of macrophages. No proteins immunoprecipitated from the lysate of cells exposed to SipB lacking the FLAG epitope were recognized by the anti-caspase-1 antibody.
To test whether SipB binds to caspase-1 during an actual cellular infection, we infected J774A.1 macrophages with the Shigella strain SF620 transformed to express either FSipB or SipB. We used this mutant strain because it induces apoptosis more synchronously than the Salmonella strains, probably because it delivers SipB more efficiently to the population of macrophages. After 40 min of infection the cells were lysed and the cytoplasmic fractions were probed with IgG-agarose beads loaded with the anti-FLAG antibody. The resulting immunoprecipitates were analyzed by immunoblotting with the anti-caspase-1 antibody (Fig. 4B). As expected, caspase-1 was not precipitated from the lysate of cells infected with SF620 expressing SipB lacking the FLAG epitope. However, FSipB did bind to the 45 kDa precursor form of caspase-1, thereby showing that the binding we observed in vitro also occurs during an infection of macrophages.
Caspase-1 Activation by SipB Is Necessary for Cytotoxicity.
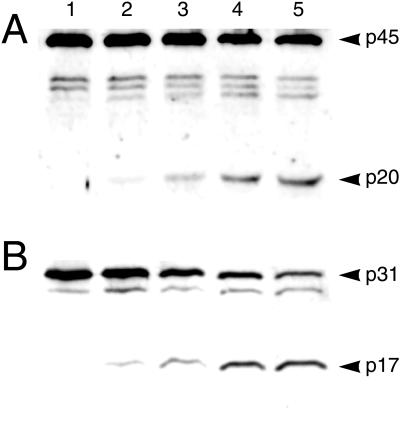
The biological relevance of the interaction between SipB and caspase-1 was then examined. We tested whether exposure to SipB during an infection by Salmonella leads to the catalytic processing of the 45-kDa precursor form of caspase-1 to the 20- and 10-kDa subunits of the mature enzyme. The lysates of LPS-activated peritoneal macrophages infected with S. typhimurium were analyzed by immunoblotting with an anti-caspase-1 antibody (Fig. 5A). LPS activation of the macrophages did not cause an up-regulation in the expression of caspase-1. Maturation of caspase-1 to the 20-kDa form was detected within 10 min of infection by wild-type S. typhimurium and the levels of this mature form continued to increase throughout the course of the observed infection. Note that the smaller 10-kDa subunit cannot be detected by this antibody and so is not visible in the immunoblot. As expected, infection for 70 min with the noncytotoxic sipB insertion mutant ST100 did not result in processing of the enzyme.
Figure 5.
Caspase-1 is activated during infection of macrophages infected with Salmonella. (A and B) LPS-activated murine peritoneal macrophages were infected with S. typhimurium C5. After 10 min (lane 2), 25 min (lane 3), 40 min (lane 4), and 70 min (lane 5) of infection, cell lysates were resolved on an 15% SDS/PAGE gel and immunoblotted with either an anti-caspase-1 antibody (A) or an anti-IL-1β antibody (B). As a control, macrophages were infected with the sipB mutant ST100 for 70 min (lane 1 in each gel) and processed in parallel. Both caspase-1 and its specific substrate IL-1β undergo catalytic maturation within 10 min of infection with mature forms accumulating throughout the course of infection observed.
A test for the induction of caspase-1 biological activity is to determine the maturation status of one of its known substrates, IL-1β. This proinflammatory cytokine exists as a precursor form of 31 kDa, which can be specifically cleaved into the mature, biologically active 17-kDa form. Activation of peritoneal macrophages with LPS treatment leads to an up-regulation of IL-1β precursor expression to levels detectable by immunoblotting with an anti-IL-1β antibody. As shown in Fig. 5B, infection with wild-type S. typhimurium results in the cleavage of IL-1β to its mature form, whereas IL-1β was not cleaved after infection with ST100. Thus, caspase-1 is activated by Salmonella infection.
Caspase-1 Is Directly Involved in Salmonella-Induced Apoptosis.
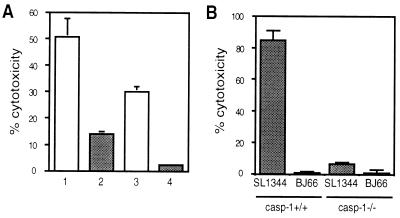
The activation of caspase-1 was found to be important in Salmonella-induced macrophage apoptosis. Caspase-1 activity can be irreversibly blocked by the inhibitor Ac-YVAD-CMK that has a high affinity for this protease. As shown in Fig. 6A, pretreatment of macrophages with Ac-YVAD-CMK before infection by wild-type S. typhimurium led to a significant decrease in killing to either 7% or 27% of wild-type cytotoxicity depending on the moi used for the infection. Incubation of the cells with inhibitor alone did not alter their survival rate (data not shown).
Figure 6.
Caspase-1 is essential for the induction of apoptosis. (A) Protection by YVAD of RAW264.7 macrophages from Salmonella-induced cell death. At 1 hr before infection with S. typhimurium SL1344 at an moi of either 100:1 (1 and 2) or 50:1 (3 and 4), cells were preincubated with 50 μM Ac-YVAD-CMK (2 and 4) or media alone (1 and 3). The presence of the caspase-1 inhibitor led to a decrease in cytotoxicity. (B) Caspase-1 is necessary for Salmonella-induced cytotoxicity. Peritoneal macrophages from either wild-type or caspase-1 −/− mice were infected for 8 hr with S. typhimurium SL1344 or BJ66. SL1344 killed the macrophages from the wild-type mice, whereas neither SL1344 nor BJ66 were able to kill macrophages lacking caspase-1. The means and SDs of three experiments are shown.
To determine whether caspase-1 is essential for the cytotoxicity observed, we infected peritoneal macrophages derived from wild-type and caspase-1 knockout mice with either S. typhimurium SL1344 or the noninvasive derivative BJ66. After 8 hr of infection, SL1344, but not BJ66, killed ≈85% of the macrophages harvested from wild-type mice, whereas peritoneal macrophages lacking caspase-1 were insensitive to Salmonella-induced cytotoxicity (Fig. 6B). Therefore, caspase-1 activity is necessary for the process of apoptosis to occur in response to Salmonella infection.
DISCUSSION
We have identified the secreted protein SipB as the effector molecule responsible for the induction of apoptosis in macrophages infected by S. typhimurium and S. typhi. Macrophages undergo cell death when microinjected with purified SipB, demonstrating that this molecule is sufficient to induce apoptosis. Cytoplasmically-distributed SipB directly engages a key component of the macrophage’s apoptotic machinery—caspase-1. It is this interaction that presumably leads to the activation of caspase-1 (Fig. 5A), thereby promoting as yet undefined downstream events necessary for apoptosis. As seen in the knockout cytotoxicity studies (Fig. 6B), caspase-1 activity is necessary and therefore central to the apoptotic pathway engaged. In addition, IL-1β undergoes maturation to its biologically active form (Fig. 5B) and could initiate an inflammatory response in vivo. Thus, SipB is not only a sequence homolog to IpaB of Shigella, it is an analog as well. Whether SipB also functions as a translocase for other virulence factors remains to be determined.
SipB and IpaB apparently induce apoptosis through similar mechanisms. However, the method used by the corresponding bacteria to deliver these proteins to the cellular compartment in which they act is quite different. After internalization by phagocytosis, Shigella quickly escapes from the phagosome and directly secretes IpaB into the cytoplasm. In contrast, Salmonella are maintained within the phagosome and therefore apart from the macrophage cytoplasm. Our studies (Figs. 3 A–F) and those of Collazo and Galán (22) clearly show that SipB gains access to the cytoplasm during the infection; however, Collazo and Galán report a homogeneous distribution of SipB throughout the cytoplasm, whereas we observed a restricted cytoplasmic pattern. Regardless, SipB is most likely to either be translocated across the phagosomal membrane barrier directly or inserted into the phagosomal membrane and then released by a vesiculation event to achieve a cytoplasmic distribution. The apparent preferential association of SipB with endomembranes, such as endoplasmic reticulum, supports the latter supposition. In fact, the immunofluorescence studies of Garcia-del Portillo et al. (34) demonstrated that LPS is dispersed throughout the cytoplasm of Salmonella-infected macrophages by a vesiculation of the phagosome. Possibly, SipB uses this mode of transport as well.
The importance of the induction of apoptosis in the pathogenesis of salmonellosis is still undefined. Richter-Dahlfors et al. (35) demonstrated that macrophage apoptosis occurs in the livers of mice infected with S. typhimurium. In their studies, they did not comment on other potential sites of apoptosis such as the intestinal wall, spleen and gall bladder. Thus, we do not know whether programmed cell death is more widespread. In the case of shigellosis, animal models have demonstrated that the release of mature, biologically active IL-1β, presumably by the apoptosis of macrophages, is of central importance for the promotion of the acute inflammation that ensues (36). In salmonellosis, a role for macrophage apoptosis and the release of IL-1β is less clear given the different nature of the intestinal inflammation and the potential for systemic infection. In vivo studies are underway to determine the extent and significance of apoptosis in general and IL-1β release in particular for the development and ultimate resolution of salmonellosis.
Acknowledgments
We thank Dr. Doug Miller for the anti-caspase-1 antibody, Dr. Michel Popoff for providing the Salmonella strains Ty2, Ty233, C5, and ST100, Adam B. Hittelman for his technical assistance, and Antonios O. Aliprantis for his careful reading and helpful comments. This work was supported by National Institutes of Health Grant AI42780 to A.Z., by Public Health Service Grant R01 AI26195, and in part by the Defense Advanced Research Projects Agency to S.F.
ABBREVIATIONS
- Ac-YVAD-CMK
acetyl-Tyr-Val-Ala-Asp-chloromethyl ketone
- LPS
lipopolysaccharide
- IL
interleukin
- PI
propidium iodide
- GST
glutathione S-transferase
- moi
multiplicity of infection
- LDH
lactate dehydrogenase
References
- 1.Keusch G T. In: Harrison’s Principles of Internal Medicine. 13th Ed. Isselbacher K J, Braunwald E, Wilson J D, Martin J B, Fauci A S, Kasper D L, editors. New York: McGraw–Hill; 1994. pp. 671–676. [Google Scholar]
- 2.Finlay B B. Curr Top Microbiol Immunol. 1994;192:163–185. doi: 10.1007/978-3-642-78624-2_8. [DOI] [PubMed] [Google Scholar]
- 3.Jones B D, Ghori N, Falkow S. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark M A, Jepson M A, Simmons N L, Hirst B H. Res Microbiol. 1994;145:543–553. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 5.Fields P I, Swanson R V, Haidaris C G, Heffron F. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monack D M, Raupach B, Hromockyj A E, Falkow S. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L M, Kaniga K, Galán J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 8.Lindgren S W, Stojiljkovic I, Heffron F. Proc Natl Acad Sci USA. 1996;93:4197–4201. doi: 10.1073/pnas.93.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zychlinsky A, Prévost M-C, Sansonetti P J. Nature (London) 1992;358:167–168. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
- 10.Zychlinsky A, Thirumalai K, Arondel J, Cantey J R, Aliprantis A O, Sansonetti P J. Infect Immun. 1996;64:5357–5365. doi: 10.1128/iai.64.12.5357-5365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam D, Veress B, Bardhan P K, Lindberg A A, Christensson B. Infect Immun. 1997;65:739–749. doi: 10.1128/iai.65.2.739-749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zychlinsky A, Kenny B, Ménard R, Prévost M-C, Holland I B, Sansonetti P J. Mol Microbiol. 1994;11:619–627. doi: 10.1111/j.1365-2958.1994.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 14.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W W, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 15.Hilbi H, Chen Y, Thirumalai K, Zychlinsky A. Infect Immun. 1997;65:5165–5170. doi: 10.1128/iai.65.12.5165-5170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilbi H, Moss J E, Hersh D, Chen Y, Arondel J, Banerjee S, Flavel R A, Yuan J, Sansonetti P J, Zychlinsky A. J Biol Chem. 1998;273:32895–32900. doi: 10.1074/jbc.273.49.32895. [DOI] [PubMed] [Google Scholar]
- 17.Hermant D, Ménard R, Arricau N, Parsot C, Popoff M Y. Mol Microbiol. 1995;17:781–789. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaniga K, Trollinger D, Galán J E. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga K, Tucker S C, Trollinger D, Galán J E. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegues D A, Hantman M J, Behlau I, Miller S I. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 21.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 22.Collazo C M, Galán J E. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 23.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 24.Hoiseth S K, Stocker B A. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 25.Jones B D, Falkow S. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ménard R, Sansonetti P J, Parsot C. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clerc P L, Ryter A, Mounier J, Sansonetti P J. Infect Immun. 1987;55:521–527. doi: 10.1128/iai.55.3.521-527.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith M R, Muegge K, Keller J R, Kung H-F, Young H A, Durum S K. J Immunol. 1990;144:1777–1782. [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 30.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 31.Arends M J, Wyllie A H. Int Rev Exp Pathol. 1991;32:223–254. doi: 10.1016/b978-0-12-364932-4.50010-1. [DOI] [PubMed] [Google Scholar]
- 32.Thirumalai K, Kim K-S, Zychlinsky A. Infect Immun. 1997;65:787–793. doi: 10.1128/iai.65.2.787-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whyte M. Trends Cell Biol. 1996;6:245–248. doi: 10.1016/0962-8924(96)20025-x. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-del Portillo F, Stein M A, Finlay B B. Infect Immun. 1997;65:24–34. doi: 10.1128/iai.65.1.24-34.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter-Dahlfors A, Buchan A M J, Finlay B B. J Exp Med. 1997;186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sansonetti P J, Arondel J, Cavaillon J M, Huerre M. J Clin Invest. 1995;96:884–892. doi: 10.1172/JCI118135. [DOI] [PMC free article] [PubMed] [Google Scholar]