In 2005, the U.S. Environmental Protection Agency (EPA), with support from the U.S. National Toxicology Program (NTP), funded a project at the National Research Council (NRC) to develop a long-range vision for toxicity testing and a strategic plan for implementing that vision. Both agencies wanted future toxicity testing and assessment paradigms to meet evolving regulatory needs. Challenges include the large numbers of substances that need to be tested and how to incorporate recent advances in molecular toxicology, computational sciences, and information technology; to rely increasingly on human as opposed to animal data; and to offer increased efficiency in design and costs (1–5). In response, the NRC Committee on Toxicity Testing and Assessment of Environmental Agents produced two reports that reviewed current toxicity testing, identified key issues, and developed a vision and implementation strategy to create a major shift in the assessment of chemical hazard and risk (6, 7). Although the NRC reports have laid out a solid theoretical rationale, comprehensive and rigorously gathered data (and comparisons with historical animal data) will determine whether the hypothesized improvements will be realized in practice. For this purpose, NTP, EPA, and the National Institutes of Health Chemical Genomics Center (NCGC) (organizations with expertise in experimental toxicology, computational toxicology, and high-throughput technologies, respectively) have established a collaborative research program.
EPA, NCGC, and NTP Joint Activities
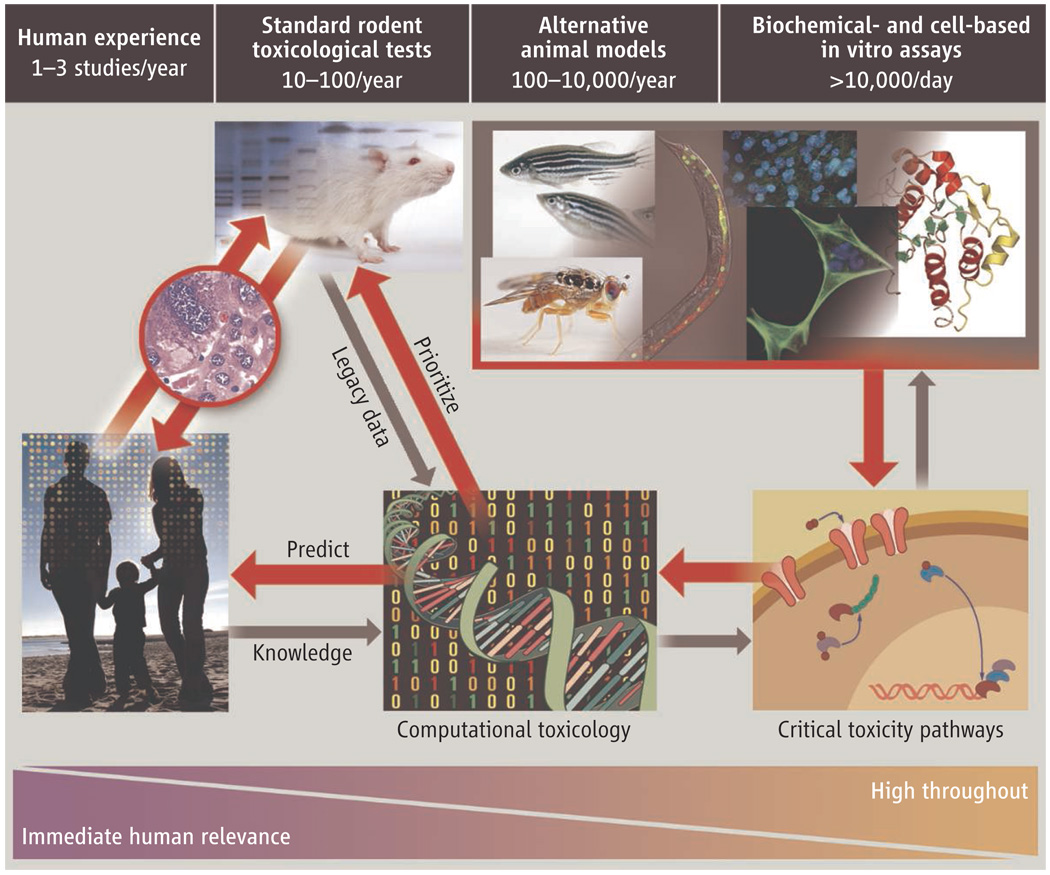
In 2004, the NTP released its vision and roadmap for the 21st century (1), which established initiatives to integrate high-throughput screening (HTS) and other automated screening assays into its testing program. In 2005, the EPA established the National Center for Computational Toxicology (NCCT). Through these initiatives, NTP and EPA, with the NCGC, are promoting the evolution of toxicology from a predominantly observational science at the level of disease-specific models in vivo to a predominantly predictive science focused on broad inclusion of target-specific, mechanism-based, biological observations in vitro (1, 4) (see figure, below).
Figure 1. Transforming toxicology.
The studies we propose will test whether high-throughput and computational toxicology approaches can yield data predictive of results from animal toxicity studies, will allow prioritization of chemicals for further testing, and can assist in prediction of risk to humans.
Toxicity pathways
In vitro and in vivo tools are being used to identify cellular responses after chemical exposure expected to result in adverse health effects (7). HTS methods are a primary means of discovery for drug development, and screening of >100,000 compounds per day is routine (8). However, drug-discovery HTS methods traditionally test compounds at one concentration, usually between 2 and 10 µM, and tolerate high false-negative rates. In contrast, in the EPA, NCGC, and NTP combined effort, all compounds are tested at as many as 15 concentrations, generally ranging from ∼5 nM to ∼100 µM, to generate a concentration-response curve (9). This approach is highly reproducible, produces significantly lower false-positive and false-negative rates than the traditional HTS methods (9), and facilitates multiassay comparisons. Finally, an informatics platform has been built to compare results among HTS screens; this is being expanded to allow comparisons with historical toxicologic NTP and EPA data (http://ncgc.nih.gov/pub/openhts). HTS data collected by EPA and NTP, as well as by the NCGC and other Molecular Libraries Initiative centers (http://mli.nih.gov/), are being made publicly available through Webbased databases [e.g., PubChem (http://pubchem.ncbi.nlm.nih.gov)]. In addition, efforts are under way to link HTS data to historical toxicological test results, including creating relational databases with controlled ontologies, annotation of the chemical entity, and public availability of information at the chemical and biological level needed to interpret the HTS data. EPA’s DSSTox (Distributed Structure Searchable Toxicity) effort is one example of a quality-controlled, structure-searchable database of chemicals that is linked to physicochemical and toxicological data (www.epa.gov/ncct/dsstox/).
At present, more than 2800 NTP and EPA compounds are under study at the NCGC in over 50 biochemical- and cell-based assays. Results from the first study, in which 1408 NTP compounds were tested for their ability to induce cytotoxicity in 13 rodent and human cell types, have been published (10). Some compounds were cytotoxic across all cell types and species, whereas others were more selective. This work demonstrates that titration-based HTS can produce high-quality in vitro toxicity data on thousands of compounds simultaneously and illustrates the complexities of selecting the most appropriate cell types and assay end points. Additional compounds, end points, and assay variables will need to be evaluated to generate a data set sufficiently robust for predicting a given in vivo toxic response.
In 2007, the EPA launched ToxCast (www.epa.gov/ncct/toxcast) to evaluate HTS assays as tools for prioritizing compounds for traditional toxicity testing (5). In its first phase, ToxCast is profiling over 300 wellcharacterized toxicants (primarily pesticides) across more than 400 end points. These end points include biochemical assays of protein function, cell-based transcriptional reporter assays, multicell interaction assays, transcriptomics on primary cell cultures, and developmental assays in zebrafish embryos. Almost all of the compounds being examined in phase 1 of ToxCast have been tested in traditional toxicology tests. ToxRefDB, a relational database being created to house this information, will contain the results of nearly $1 billion worth of toxicity studies in animals.
Another approach to identifying toxicity pathways is exploring the genetic diversity of animal and human responses to known toxicants. The NCGC is evaluating the differential sensitivity of human cell lines obtained from the International HapMap Project (http://www.hapmap.org/) to the 2800 compounds provided by NTP and EPA. Similarly, NTP has established a Host Susceptibility Program to investigate the genetic basis for differences in disease response using various mouse strains (http://ntp.niehs.nih.gov/go/32132); cell lines derived from these animals will be evaluated at the NCGC for differential sensitivity to the compounds tested in the HapMap cell lines. The resulting collective data sets will be used to develop bioactivity profiles that are predictive of the phenotypes observed in standard toxicological assays and to identify biological pathways which, when perturbed, lead to toxicities. The ultimate goal is to establish in vitro “signatures” of in vivo rodent and human toxicity. To assist, computational methods are being developed that can simulate in silico the biology of a given organ system.
The liver is the most frequent target of more than 500 orally consumed environmental chemicals, based on an analysis of the distribution of critical effects in rodents by organ, in the EPA Integrated Risk Information System (IRIS) (www.epa.gov/iris). The goal of the recently initiated Virtual Liver project (www.epa.gov/ncct/virtual_liver.html) is to develop models for predicting liver injury due to chronic chemical exposure by simulating the dynamics of perturbed molecular pathways, their linkage with adaptive or adverse processes leading to alterations of cell state, and integration of the responses into a physiological tissue model. When completed, the Virtual Liver Web portal and accompanying query tools will provide a framework for incorporation of mechanistic information on hepatic toxicity pathways and for characterizing interactions spatially and across the various cells types that comprise liver tissue.
Targeted testing
The NRC committee report (7) proposes in vitro testing as the principal approach with the support of in vivo assays to fill knowledge gaps, including tests conducted in nonmammalian species or genetically engineered animal models. A goal would be to use genetically engineered in vitro cell systems, microchip-based genomic technologies, and computer-based predictive toxicology models to address uncertainties.
Dose-response and extrapolation models
Dose-response data and extrapolation models encompass both pharmacokinetics (the relation between exposure and internal dose to tissues and organs) and pharmacodynamics (the relation between internal dose and toxic effect). This knowledge can aid in predicting the consequences of exposure at other dose levels and life stages, in other species, or in susceptible individuals. Physiologically based pharmacokinetic (PBPK) models provide a quantitative simulation of the biological processes of absorption, distribution, metabolism, and elimination of a substance in animals or humans. PBPK models are being created to evaluate exposure-response relation for critical target organ effects (11). These can be combined with models that measure changes in cells in target tissues under different test substance concentrations. This will help to determine the likelihood of adverse effects from “low-dose” exposure, as well as to assess variation among individuals in specific susceptible groups.
Making It Happen
Elements of this ongoing collaboration and coordination include sharing of databases and analytical tools, cataloging critical toxicity pathways for key target organ toxicities, sponsoring workshops to broaden scientific input into the strategy and directions, outreach to the international community, scientific training of end users of the new technologies, and support for activities related to the requirements for national and international regulatory acceptance of the new approaches. It is a research program that, if successful, will eventually lead to new approaches for safety assessment and a marked refinement and reduction of animal use in toxicology. The collective budget for activities across the three agencies that are directly related to this collaboration has not yet been established. Future budgets and the pace of further development will depend on the demonstration of success in our initial efforts. As our research strategy develops, we welcome the participation of other public and private partners.
Footnotes
We propose a shift from primarily in vivo animal studies to in vitro assays, in vivo assays with lower organisms, and computational modeling for toxicity assessments.
References and Notes
- 1.NTP. Research Triangle Park, NC: NTP, NIEHS; A National Toxicology Program for the 21st Century: A Roadmap for the Future. 2004 ( http://ntp.niehs.nih.gov/go/vision)
- 2.NTP. Research Triangle Park, NC: NTP, NIEHS; Current Directions and Evolving Strategies: Good Science for Good Decisions. 2004 ( http://ntp.niehs.nih.gov/ntp/Main_Pages/PUBS/2004CurrentDirections.pdf)
- 3.Bucher JR, Portier C. Toxicol. Sci. 2004;82:363. doi: 10.1093/toxsci/kfh293. [DOI] [PubMed] [Google Scholar]
- 4.Kavlock R, et al. Toxicol. Sci. in press published online 7 December 2007, 10:1093/toxsci/kfm297. [Google Scholar]
- 5.Dix D, et al. Toxicol. Sci. 2007;95:5. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 6.NRC Toxicity Testing for Assessment of Environmental Agents: Interim Report. Washington, DC: National Academies Press; 2006. [Google Scholar]
- 7.NRC Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. [DOI] [PubMed] [Google Scholar]
- 8.Pereira DA, Williams JA. Br. J. Pharmacol. 2007;152:53. doi: 10.1038/sj.bjp.0707373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inglese J, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11473. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia M, et al. Environ. Health Perspect. in press published online 22 November 2007, 10.1289/ehp.10727. [Google Scholar]
- 11.Chiu WA, et al. J. Appl. Toxicol. 2007;27:218. doi: 10.1002/jat.1225. [DOI] [PubMed] [Google Scholar]
- 12.We thank R. Tice (NIEHS), R. Kavlock (EPA), and C. Austin (NCGC and NHGRI), without whose contributions this manuscript would not have been possible. This research was supported in part by the Intramural Research programs of the NHGRI, EPA, and NIEHS.