Abstract
Circulating cell-free fetal deoxyribonucleic acids (cffDNA) are promising biomarkers with various promising clinical applications. Second and third trimester amniotic fluid (AF) is a rich source of cffDNA. Further improvements to the original protocol for the extraction of cffDNA from AF supernatant resulted in statistically significant higher yields of high quality cffDNA, allowing for a substantial majority of samples to be analyzed with subsequent molecular methods (e.g. comparative genomic hybridization [CGH] micro arrays) to further assess for genetic abnormalities. Several advantages have been realized with the optimized protocol. In addition to an improved yield from a greater proportion of samples as compared to the original protocol, the current method, using large silico-membranes, allows for the extraction of cffDNA from up to ten samples in less than three hours. The replacement of the original lysis buffer eliminates the need for a heating bath during the lysis step, and fewer overall steps are involved in the protocol (e.g. to reduce potential contamination).
The improvements in the yield with the current protocol make it possible to augment current standard of care through the analysis of this previously unappreciated source of genetic material, and furthermore, will allow for exploration of widely unknown genetic, pathophysiological and kinetic issues of cell-free fetal DNA in amniotic fluid.
Keywords: Prenatal diagnosis, cell-free fetal DNA, prental screening, pregnancy
Introduction
Since the detection of cell-free fetal nucleic acids (cffDNA) in maternal plasma and serum, many investigators have focused on its biology and potential role as a genetic marker for non-invasive prenatal diagnosis and screening (1–4). However, these investigations have yet to fully exploit this source of genetic material in maternal body fluids other than plasma and serum. Prenatal cytogenetic diagnosis traditionally relies on the analysis of cultured metaphase cells harvested from amniotic fluid (AF) obtained at amniocentesis. Typically, 10 to 30 mL of AF is removed and intact cells are cultured for cytogenetic analysis and therefore not available for additional molecular techniques, such as PCR amplification or DNA micro array profiling. After removal of the cells, a small amount of the amniotic fluid supernatant is taken to assess levels of cell-free proteins as markers of fetal abnormalities. The remaining supernatant is normally discarded, thus this fraction can yield cffDNA for additional genetic testing to potentially augment current standard of prenatal diagnostic care (5, 5).
In 2001, Bianchi et al. first demonstrated the presence of cell-free fetal DNA (cffDNA) in amniotic fluid as a potential source of genetic material for research and/or clinical applications (5). In that preliminary study we showed that larger quantities (100–200 fold) of cffDNA per milliliter are present in amniotic fluid compared to maternal plasma/serum. However, due to difficulties in extracting sufficient amounts of high quality cffDNA from AF for effective analysis, only a minor proportion of samples could be further analyzed (e.g. with DNA microarrays, in which a minimum of 100 ng of DNA is necessary)(6). Our original protocol was based on known protocols for the extraction of cffDNA from maternal plasma/serum, as specific guidelines for the extraction of this DNA from AF did not exist. Therefore, further investigation was needed to optimize cffDNA extraction from AF supernatant to more fully exploit this promising source of genetic material. Our objective was to develop an approach for the improved isolation of cffDNA from AF supernatant using information regarding DNA extraction from urine, as AF is similarly excreted by the fetal urogenital system during the second and third trimesters.
To improve the yield of extracted cffDNA, the original method (6) using the “Blood and Body fluid” vacuum protocol (Qiagen, Valencia, CA) was changed in the following ways: 1) increasing vacuum extraction pressure from 400 to 800 mbar to allow for maximal absorption of DNA to mini filters. Although this pressure may exceed that available in some laboratories (e.g. using building vacuum pressure), reduced vacuum leads to lower yield of extracted DNA; 2) replacing the volume over-loaded Mini Spin Columns with Maxi Spin Columns (Qiagen). In the original protocol, 10 mL of amniotic fluid was added to 1 mL of protease, 10 mL of AL lysis buffer (Qiagen) and 10 mL of 100% ethanol, which exceeded the volumetric capacity of the mini columns. The use of maxi columns allows for larger starting volumes to be processed and therefore a larger quantity of cffDNA can be obtained as compared to mini columns; and 3) substituting AL buffer with AVL buffer (Qiagen). AL buffer, used in the original protocol, was selected based on prior experience for the isolation of cffDNA from plasma and serum, as there was no information available on the most suitable buffer for extraction of cffDNA from amniotic fluid. However, AVL buffer supplemented with nucleic acid carrier for the extraction of low concentrations of target DNA was selected for the current protocol on the basis of similar qualities between amniotic fluid and urine, a body fluid in which AVL buffer is recommended for DNA extraction.
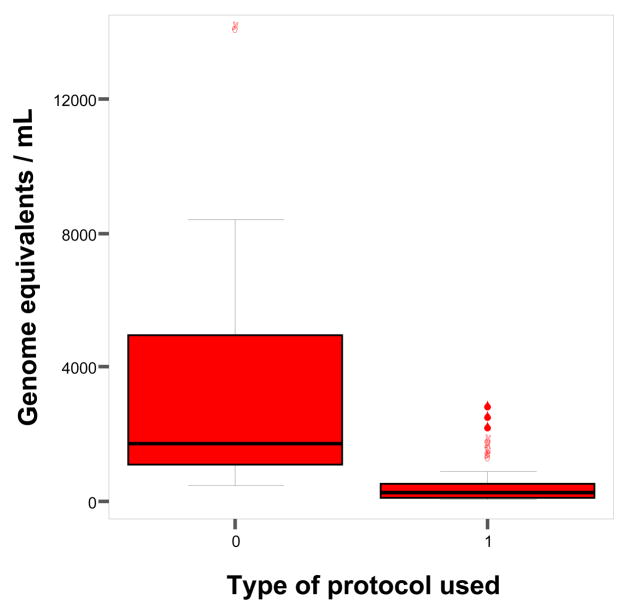
From euploid singleton pregnancies (n = 29), the median amount of GAPDH DNA extracted from 10 mL of amniotic fluid with the new protocol was 1700 genomic equivalents (GE)/mL (25th, 75th percentiles: 1071, 4938 GE/mL) compared to 246 GE/mL (93, 523.5 GE/mL) using the original protocol (6,7)(p<0.0001, see Figure 1).
Figure 1.
Comparison of the yield of cell free fetal DNA, as measured by real-time PCR amplification of the GAPDH locus, extracted from amniotic fluid supernatant from euploid singleton pregnancies. 0: new protocol (7), 1: original protocol (6). The lines inside the boxes denote medians. The box indicates 25th and 75th percentiles. The whiskers denote the 10th and the 90th percentiles. Symbols indicate data points outside the 10th and 90th percentiles.
Several advantages have been realized with the protocol developed here. In addition to an improved yield from a greater proportion of samples as compared to the original protocol, the current protocol allows for the extraction of cffDNA from up to ten samples in less than three hours. The replacement of AL buffer with AVL buffer eliminates the need for a heating bath during the lysis step, and fewer overall steps are involved in the protocol (e.g. to reduce potential contamination). However, the cost of cffDNA extraction from a 10 mL AF supernatant sample using the new protocol is about 10 fold higher compared to the original protocol (~$39 and ~$ 4 per sample, respectively), although the advantage of the new protocol with respect to improved DNA yield justifies this higher cost per sample.
For clinical applications, one major advantage of using the amniotic fluid supernatant is its availability without interfering with current standard of care or compromising fetal health.
Materials and Methods
For each sample (10 mL) you will need:
Three 50 mL polypropylene tubes (e.g. BLUE MAX Falcon, cat. Number 352070)
One 2 mL microcentrifuge tube (e.g. Fischer Scientific, package of 500, 02-681-258) for storage of the final eluted sample
One to three 0.5 mL tubes for storage of the final eluted sample
Sterile pipette tips (1000 μL, 200 μL, 20 μL)
Sterile 10 mL or 20 mL pipettes
-
Buffer AVL (155 mL) Viral Lysis Buffer and 4.2 mg Carrier RNA/DNA for 250 reactions (Qiagen, Valencia, CA, Cat. No. 19073)
After arrival, lysis buffer can be stored at room temperature (15–25°C)
After addition of RNA/DNA carrier, lysis buffer should be stored at 4°C.
QIAamp DNA Blood Maxi Kit (10), for 10 DNA maxipreps, (Qiagen Cat. No. 51192)
-
Alternatively: QIAamp DNA Blood Maxi Kit (50), for 50 DNA maxipreps (Qiagen Cat. No. 51194)
Maxi columns should be stored at 4°C to maintain constant high yields
Wash solutions (AW1 and AW2) are supplied in the QIAamp DNA Blood Maxi Kit. Storage at room temperature (15–25°C)
AE elution buffer is supplied in the QIAamp DNA Blood Maxi Kit. Store at room temperature (15–25°C).
Ethanol 100%, store at room temperature (15–25°C).
Notes
All steps of the protocol should be performed at room temperature. During the procedure, work quickly.
Wash buffer AW1 und AW2 are supplied. Add volumes of ethanol (100%), as indicated on the bottles
Methods
Prepare tubes for extraction: for each sample (10 mL) you will need three 50 mL screw cap tube (polypropylene), one 2 mL microcentrifuge tube and one to three 0.5 mL tubes. Place tube rack, pipettes and tubes into the ultraviolet (UV) cross-linker for 20 minutes, leaving caps open.
If amniotic fluid (AF) samples are frozen, thaw at 37° C and equilibrate to room temperature (see note 1).
Vortex thawed sample briefly.
Check Buffer AVL for precipitate, and, if necessary, incubate at 80°C until precipitate is dissolved. Add 1 mL of buffer AVL to one tube of lyophilized Carrier RNA/DNA. Dissolve Carrier RNA/DNA thoroughly. Transfer to AVL bottle, and mix thoroughly before using AVL buffer for the first time (see note 2).
Pipette 20 mL (or 400% of AF volume) of AVL/Carrier RNA/DNA in each of two 50 mL tubes.
Add 5 mL of AF into each of the 50 mL tubes. Mix by pulse-vortexing for 15 sec (see note 3).
Incubate at room temperature (15–25°C) for 10 minutes.
Briefly centrifuge the two 50 mL tubes at 1200 rpm to remove residual droplets from the cap.
Add 20 mL (or same amount as AVL) of 100% ethanol to each sample. Mix by pulse-vortexing for 15 seconds.
Briefly centrifuge the 50 mL tubes at 1200 rpm (Sorvall RT) to remove residual droplets from the cap.
Apply 15 mL (of total 90 mL) from step 10 to the QIAamp Maxi column, placed in a 50 mL centrifugation tube. Do not moisten the rim of the QIAamp maxi column. Close the cap and centrifuge at 1850 × g (3000 rpm) (Sorvall RT) for 3 minutes (see note 4).
-
Remove the QIAamp maxi column, discard the filtrate, and place the QIAamp maxi column back into the 50 mL centrifugation tube. Load 15 mL of the solution from step 10 onto the QIAamp maxi column. Close the cap and centrifuge at 1850 × g (3000 rpm) (Sorvall RT) for 3 minutes. Note: wipe off any spillage from the thread of the 50 mL centrifugation tube before reinserting the QIAamp maxi column. Do not wet the rim of the QIAamp maxi column.
Repeat step 12 until the whole volume passed through the Maxi column.
Remove the QIAamp maxi column, discard the filtrate, and place the QIAamp maxi column back into the 50 mL centrifugation tube. Note: wipe off any spillage from the thread of the 50 mL centrifugation tube before reinserting the QIAamp maxi column.
Carefully, without moistening the rim, add 5 mL Buffer AW1 to the QIAamp maxi column. Close the cap and centrifuge at 3715 × g (4150 rpm) (Sorvall RT) for 5 minutes. Do not discard the flow-through at this stage and continue directly with step 10.
Carefully, without moistening the rim, add 5 mL Buffer AW2 to the QIAamp maxi column. Close the cap and centrifuge at 3715 × g (4150 rpm) (Sorvall RT) for 25 minutes (see note 5).
Discard the 50 mL centrifugation tube containing the filtrate, and place the QIAamp maxi column in a new 50 mL centrifugation tube. Note: wipe off any spillage from the thread of the 50 mL centrifugation tube before reinserting the QIAamp maxi column.
Add 1 mL of Buffer AE (15–25°C). Pipet directly to the membrane of the QIAamp maxi column and close the cap. Incubate at room temperature for 5 minutes and centrifuge at 3715 × g (4150 rpm) for 10 minutes.
For maximum yield: Pipet 1 mL of fresh Buffer AE (15–25°C). Pipet directly to the membrane of the QIAamp maxi column and close the cap. Incubate at room temperature for 5 minutes and centrifuge at 3715 × g (4150 rpm) (Sorvall RT) for 10 minutes. Note: Less than 1 mL will be eluted from the column, but this has no effect on DNA yield.
Transfer the elutant to a 2 mL and additional 500 microliter microcentrifuge tubes for further analysis (e.g. PCR, electrophoresis).
Analyze samples immediately, store at 4° C for a maximum of 2 weeks, or freeze indefinitely (at −80°C).
Notes
Samples can be thawed in approximately 15 minutes, using a water bath (37°C).
AVL buffer that is stored at 4°C after addition of carrier RNA/DNA usually shows small crystalloid precipitates at the bottom of the bottle. Dissolve them by putting the bottle in a warm water bath and vortexing the buffer quickly.
It is important to mix the sample thoroughly with the lysis buffer to avoid any clotting of the silico-membrane.
Use an appropriate centrifuge that allows centrifugation of 50 mL tubes at 3700–4000 g (e.g. Sorval RT). If the solution has not completely passed through the membrane, centrifuge at a slightly higher speed.
Please allow the silico-membranes to dry completely. A reduction of the centrifugation time is not recommended.
Acknowledgments
This work was supported by NIH grant R01HD42053 and the Swiss National Fund (PBBSB-108590). The authors would like to thank Helene Stroh, Inga Peter, and Janet Cowan with her team of the Tufts-New England Medical Center, Boston, for their advice.
References
- 1.Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Zhong XY, Laivuori H, Livingston JC, Ylikorkala O, Sibai BM, Holzgreve W, Hahn S. Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am J Obstet Gynecol. 2001;184:414–9. doi: 10.1067/mob.2001.109594. [DOI] [PubMed] [Google Scholar]
- 3.Wataganara T, LeShane ES, Farina A, Messerlian GM, Lee T, Canick JA, Bianchi DW. Maternal serum cell-free fetal DNA levels are increased in cases of trisomy 13, but not trisomy 18. Hum Genet. 2003;112:204–8. doi: 10.1007/s00439-002-0853-9. [DOI] [PubMed] [Google Scholar]
- 4.Farina A, LeShane ES, Lambert-Messerlian GM, Canick JA, Lee T, Neveux LM. Valuation of fetal cell free DNA as a second-trimester marker of Down syndrome pregnancy. Clin Chem. 2003;49:239–42. doi: 10.1373/49.2.239. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi DW, LeShane E, Cowan JM. Large amounts of cell-free fetal DNA are present in amniotic fluid. Clin Chem. 2001;47:1867–9. [PubMed] [Google Scholar]
- 6.Larrabee PB, Johnson KL, Pestova E, Lucas M, Wilber K, Le Shane E, Tantravahi U, Cowan JM, Bianchi DW. Microarray analysis of cell-free fetal DNA in amniotic fluid: a prenatal molecular karyotype. Am J Hum Genet. 2004;75:485–91. doi: 10.1086/423288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapaire O, Stroh H, Peter I, Cowan JM, Tantravahi U, O’Brien B, Bianchi DW, Johnson KL. Larger filters and change of lysis buffer significantly improve the quantity of extracted cell-free DNA from amniotic fluid. Clin Chem 2006. 2006;52:156–157. doi: 10.1373/clinchem.2005.058420. [DOI] [PubMed] [Google Scholar]