Abstract
We demonstrated that mouse spermatozoa cleave their DNA into ~50 kb loop-sized fragments with topoisomerase IIB when treated with MnCl2 and CaCl2 in a process we term sperm chromatin fragmentation (SCF). SCF can be reversed by EDTA. A nuclease then further degrades the DNA in a process we term sperm DNA degradation (SDD). MnCl2 alone could elicit this activity, but CaCl2 had no effect. Here, we demonstrate the existence of a nuclease in the vas deferens that can be activated by EGTA to digest the sperm DNA by SDD. Spermatozoa were extracted with salt and dithiothreitol to remove protamines and then incubated with EGTA. Next, the EGTA was removed and divalent cations were added. We found that Mn+2, Ca+2, or Zn+2 could each activate SDD in spermatozoa but Mg+2 could not. When the reaction was slowed by incubation on ice, EGTA pretreatment followed by incubation in Ca+2 elicited the reversible fragmentation of sperm DNA evident in SCF. When the reactions were then incubated at 37°C they progressed to the more complete degradation of DNA by SDD. EDTA could also be used to activate the nuclease, but required a higher concentration than EGTA. This EGTA-activatable nuclease activity was found in each fraction of the vas deferens plasma: in the spermatozoa, in the surrounding fluid, and in the insoluble components in the fluid. These results suggest that this sperm nuclease is regulated by a mechanism that is sensitive to EGTA, possibly by removing inhibition of a calcium binding protein.
Keywords: Nuclease, Spermatozoa, Vas Deferens, Epididymis, Nuclear Matrix
Introduction
During somatic cell apoptosis, DNA is first degraded by topoisomerase II which cleaves the DNA into ~50 kb loop-sized fragments (Gromova et al., 1995; Li and Liu, 2001). This fragmentation can be reversed by treatment with EDTA and probably occurs on the nuclear matrix at the site of DNA attachment. Subsequently, a set of nucleases that may interact directly with the topoisomerase irreversibly degrades the DNA (Durrieu et al., 2000; Widlak et al., 2000). We have previously demonstrated that there exists a similar mechanism for chromatin degradation in spermatozoa that requires both MnCl2 and CaCl2 for full activity (Shaman et al., 2006). When treated with MnCl2 and CaCl2 mouse spermatozoa cleave their DNA into ~50 kb loop-sized fragments with nuclear topoisomerase IIB (TOPIIB) in a process we term sperm chromatin fragmentation (SCF). Like somatic cell TOPII-induced DNA cleavage, SCF can be reversed by EDTA. A nuclease then further degrades the DNA in a process we term sperm DNA degradation (SDD). MnCl2 alone can elicit this activity, but CaCl2 had no effect. When spermatozoa were extracted with salt and dithiothreitol (DTT), the DNA degradation was more complete. Recently, we demonstrated that the nuclease activity was present in much higher amounts in the vas deferens spermatozoa than in the epididymis (Yamauchi et al., 2007a).
The ability of spermatozoa to thoroughly degrade its own DNA is unexpected because its role is to protect the paternal genome in order to deliver a pristine copy of the father’s DNA to the embryo. To accomplish this, mammalian sperm chromatin is so tightly compacted that it is resistant to nucleases. The histones that organize somatic cell DNA are replaced by protamines (Clarke, 1992; Pogany et al., 1981; Zirkin et al., 1982)], which condense the DNA into tightly packed toroids (Allen et al., 1995; Hud et al., 1995). This highly compact sperm DNA is also organized into loop domains (Risley et al., 1986; Ward et al., 1989), similar to somatic cells (Jackson and Cook, 1986; Pardoll et al., 1980). Each protamine toroid contains about 50 kb (Allen et al., 1995; Hud et al., 1995), and represents a single DNA loop domain (Sotolongo et al., 2003). The protamine toroids are connected by toroid linker chromatin, which attaches the loop domain to the nuclear matrix. This linker is DNAse I sensitive, and may contain residual histones. The toroid linker region is probably the site of sperm nuclease initiation, as well as the initiation of sperm DNA replication after fertilization (Shaman et al., 2007).
Despite the unexpected presence of nucleases in fully mature spermatozoa, several laboratories have reported their existence. At least two groups have proposed that nucleases prevent spermatozoa from absorbing foreign DNA (Carballada and Esponda, 2001; Maione et al., 1997). This could explain their presence in mature spermatozoa – their function is to protect the paternal DNA. On the other hand, several groups have suggested that spermatozoa can undergo apoptosis or apoptotic-like events (Anzar et al., 2002; Baccetti et al., 1996; Blanco-Rodriguez and Martinez-Garcia, 1998; Lin et al., 1999; Sakkas et al., 1999). In the hamster epididymis, defective spermatozoa are wrapped in a “death cocoon” which may protect the healthy spermatozoa in the same region from destruction by enzymes released from dying spermatozoa (Olson et al., 2004). We have demonstrated that when spermatozoa are induced to activate the initial DNA fragmentation through SCF, and then injected into mouse oocytes, the paternal DNA is degraded six hours later at the initiation of DNA synthesis (Yamauchi et al., 2007a; Yamauchi et al., 2007b). These reports suggest that mammalian spermatozoa are associated with a variety of nucleases that may serve different functions.
Here we show that a nuclease with activity similar to that in SDD can be activated with CaCl2 alone if spermatozoa are pretreated with ethylene glycol tetraacetic acid (EGTA). The sequence of this reaction is important because while EGTA is required to activate the nuclease, it must subsequently be removed and CaCl2 then added before DNA degradation occurs. Nucleases exist that are dependent on MgCl2 and CaCl2 (Kyprianou et al., 1988; Yakovlev et al., 2000), and there are other nucleases that are active in the presence of EDTA (Pohlman et al., 1993; Przykorska et al., 2004). But as far as we are aware, there are none that are first activated by EGTA and then require CaCl2. This sequence implies a specific regulation of the nuclease, and of SCF and SDD.
Materials and Methods
Animals
Male B6D2F1 (C57BL/6J X DBA/2) mice were obtained from National Cancer Institute (Raleigh, NC). Mice were fed ad libitum and kept in standard housing in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and those prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Resources National Research Council (DHEF publication no. [NIH] 80-23, revised 1985). The protocol for animal handling and the treatment procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Hawaii.
Sperm Chromatin Fragmentation and Sperm DNA Degradation
Plasma from the caudal epididymides and vas deferens of ~ 9 week old mice was extracted separately and collected in 25 mM Tris, pH 7.4, 150 mM KCl buffer (TKB) on ice. The final concentration of spermatozoa was roughly 108 spermatozoa/ml. The suspension was mixed with agarose to a final concentration of 1% and poured into molds to make 5 mm thick plugs which were used for all assays unless noted otherwise. Each plug contained 75 μl of sperm suspension.
For the sperm chromatin fragmentation (SCF) assay, half of the plugs were pre-incubated in 30 mM EGTA for 15 minutes at room temperature, and then all plugs were incubated in TKB with no cations, 10 mM MnCl2, 10 mM CaCl2, or 10 mM of each cation at 37°C for one hour. To test for the reversibility of the fragmentation, plugs were then incubated in 50 mM ethylenediamine tetraacetic acid (EDTA) for 30 min at 37°C.
The EGTA-sperm DNA degradation (EGTA-SDD) assay involves the complete degradation of the sperm chromatin. Agarose plugs of sperm suspension were first incubated in 2 M NaCl, 2 mM DTT for 1 hour at 37°C in order to remove the protamines. The plugs were then incubated in 30 mM EGTA for 15 minutes at room temperature, and finally all plugs were incubated in TKB or TKB with 10 mM MnCl2 and/or CaCl2 at 37°C for one hour. The reactions were stopped by incubating the plugs in digestion buffer (10 mM Tris, 5 mM EDTA, pH 7.8, 100 mM NaCl, 0.5% SDS, and 20 mM DTT) at 55°C for at least 30 minutes before they are placed in a 1% agarose gel for Field Inversion Gel Electrophoresis (FIGE).
To determine if the nuclease responsible for the EGTA-SDD DNA degradation was located in the sperm cell, as opposed to in the surrounding fluid, we ran the EGTA-SDD assay using isolated sperm cells centrifuged at 700 × g and washed five times with TKB, and finally resuspended in TKB. This solution was then made into agarose plugs. The EGTA-SDD assay, as described above, was then carried out on these plugs instead of the plugs with the neat, sperm-containing, plasma.
Plasmid-Based Nuclease Assay
Plasma from the vas deferens of ~ 9 week old mice was extracted and collected in 25 mM Tris and 150 mM KCl buffer (TKB) on ice. This was centrifuged with a small tabletop centrifuge (Sigma-Aldrich Micro Centrifuge 100 VAC) for 2 minutes at 1,500 × g to separate the plasma (supernatant) from the sperm cells (pellet). The supernatant was aspirated and ultra-centrifuged at 36,000 × g (20,000 rpm, in an Allegra 64R table top ultracentrifuge from Beckman/Colter using an F2402 rotor) for 30 minutes at 4°C, using an Allegra™ 64R Centrifuge (Beckman/Coulter). The supernatant (Luminal Supernatant) and the pellet (Luminal Pellet) were resuspended in TKB and both used in this assay. The sperm cell pellet was then washed three times with TKB at room temperature and resuspended in TKB. Triton X-100 (TX), a non-ionic detergent, was added to the sperm-TKB solution to a final concentration of 0.25%. This was centrifuged at 700 × g, and the supernatant (Sperm Extract) used for this assay. Each solution – the Luminal Supernatant, Luminal Pellet, and the Sperm Extract – was pre-treated with nothing or with 30 mM EGTA for 15 minutes at room temperature. Then, 3 ul of each solution (with or without EGTA pre-treatment) was added to 20 ul of TKB, containing 1.25 μg of plasmid DNA, and either no cations, 10 mM MnCl2, 10 mM CaCl2, or 10 mM of each cation for one hour at 37°C. The solutions were applied to a 1% agarose gel and were electrophoresed.
Results
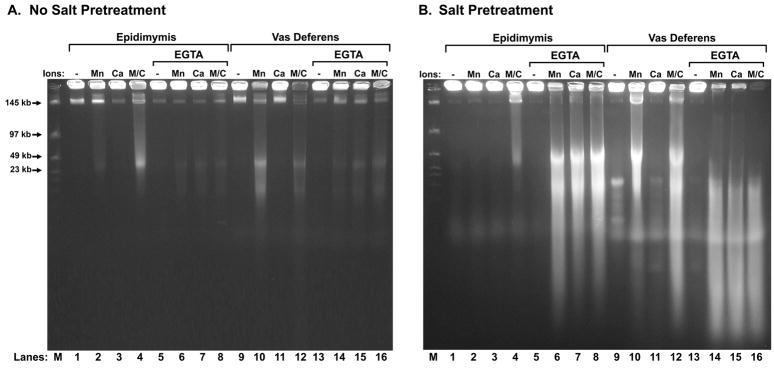
The epididymides and vas deferens sperm have a nuclease that is activated by EGTA, that requires CaCl2 and salt extraction
As we have recently reported (Yamauchi et al., 2007a), the sperm chromatin fragmentation (SCF) nuclease activity in the vas deferens is much greater than in the epididymides. Spermatozoa treated with both MnCl2 and CaCl2 digest their DNA to about 50 kb (Fig. 1A, lanes 4 and 12). There is some SCF activity detectable with only MnCl2 in vas deferens (Fig. 1A, lane 10), but CaCl2 alone has no effect on DNA degradation in spermatozoa isolated from either the epididymis or vas deferens (Fig. 1A, lanes 3 and 11). We next extracted the plugs with salt and DTT first to remove the protamines, exposing the sperm DNA, and while this alone did increase the nuclease digestion with MnCl2 and CaCl2 (Fig 1B, lanes 4 and 12), CaCl2 still had no effect on the reaction by itself (Fig. 1B, lanes 3 and 11). However, when the plugs were incubated with EGTA just after the salt treatment, all combinations of cations induced marked increase in DNA degradation, in both epididymides and in vas deferens (Fig. 1B, lanes 6 – 8 and 14 – 16). CaCl2 alone had just as strong an effect as MnCl2 or as MnCl2 and CaCl2 combined. This activation of the nuclease by Ca2+ alone, after EGTA treatment, is the hallmark of EGTA-SDD. In fact, this is the first time we detected any nuclease activity in SDD with CaCl2, alone. EGTA alone after salt had no effect (Fig. 1B, lanes 5 and 13), indicating that the nuclease still required a divalent cation to digest DNA. The same assay was tested with EDTA in place of EGTA, and we found that EDTA can also be used, but EGTA works at a slightly lower concentration (Fig. 2).
Figure 1. EGTA Stimulates Sperm DNA Degradation after Salt Treatment.
(A) Total plasma from the epididymis (lanes 1–8) or vas deferens (lanes 9–16) was embedded in agarose plugs and then treated with or without 30 mM EGTA, as indicated. All plugs were then treated with or without 10 mM MnCl2 and/or with or without 10 mM CaCl2 as indicated, and as described in Methods. The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE). (B) As in (A) except that all plugs were treated with 2 M NaCl, 2 mM DTT prior to EGTA and divalent cation treatment.
Figure 2. Concentration Dependence of EGTA and EDTA in SDD.
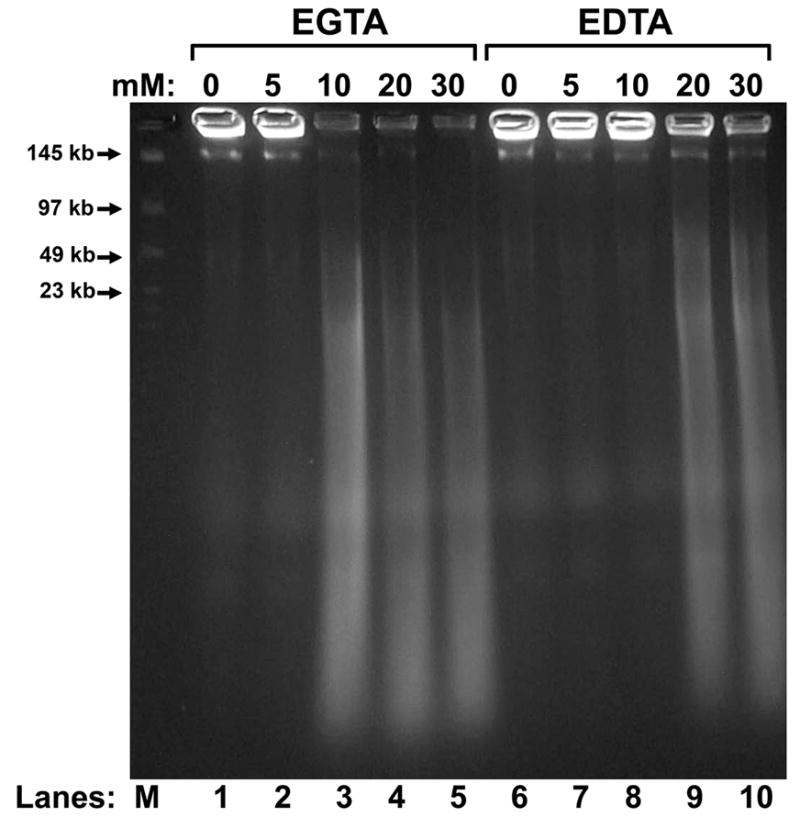
Total plasma from the vas deferens was embedded in agarose plugs. These were then extracted with 2 M NaCl and 2 mM DTT, and then treated with 0 to 30 mM EGTA (lanes 1 – 5) or EDTA (lanes 6 – 10), and finally with 10 mM CaCl2, alone. The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE).
These data demonstrate that the epididymal and vas deferens plasma contain a nuclease that is activated by treatment with EGTA, but that then requires the addition a divalent cation for DNA digestion.
Cation Specificity of EGTA-SDD
We next tested the cation specificity for the EGTA-activated nuclease. Our results in Fig. 1 suggested that EGTA-SDD can be activated with either MnCl2 or CaCl2, so we tested two additional divalent cations for activity. We chose Mg+2 because many nucleases use this cation as a cofactor, and found that the EGTA-SDD nuclease could not use this cation at all (Fig. 3, lanes 2 and 7). We also tested Zn+2 as it has been shown to inhibit many nucleases (Giannakis et al., 1991; Widlak and Garrard, 2001), and found that in this case, it appeared to serve as a cofactor for DNA digestion by the EGTA-SDD nuclease (Fig. 3, lanes 5 and 10).
Figure 3. Ion Specificity of EGTA-SDD.
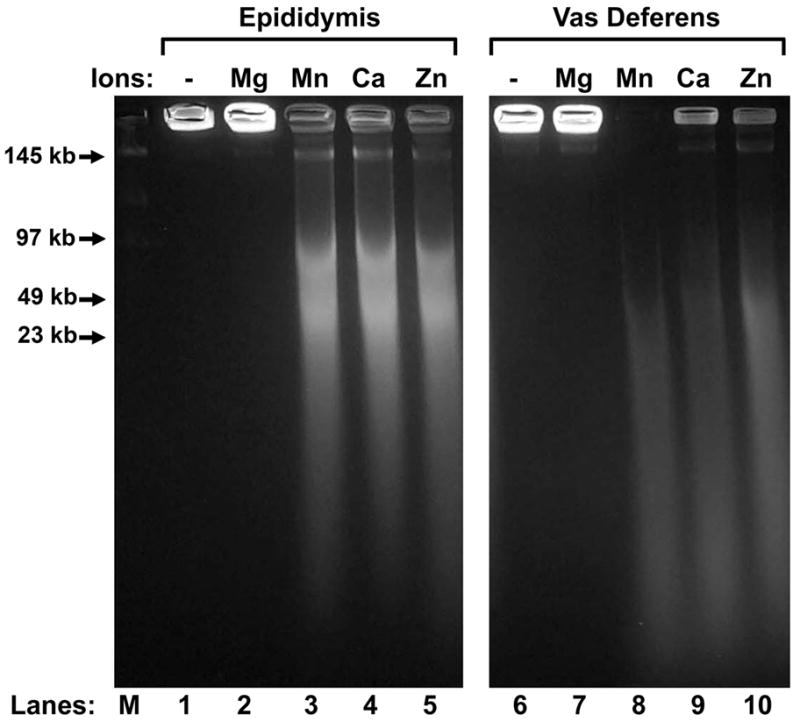
Epididymal (lanes 1 – 5) or vas deferens (lanes 6 – 10) total plasma was embedded in agarose plugs, extracted with extracted with 2 M NaCl, 2 mM DTT, and then treated with 30 mM EGTA. The plugs were then placed in either TKB alone, or 10 mM MgCl2, MnCl2, CaCl2, or ZnCl2. The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE).
Even though Ca+2 appeared to work as a cofactor for the EGTA-activated nuclease, Mn+2 was more active than Ca+2. In the EGTA-SDD assay, even as low as 2 mM CaCl2 had some effect after EGTA, but without EGTA, even the highest concentration we tested, 10 mM, had no effect (Fig. 4A). MnCl2 was much more active in initiating the nuclease activity. With EGTA, MnCl2 digested the sperm chromatin completely even with only 1 mM(Fig. 4B). Without EGTA, MnCl2 initiates partial chromatin digestion even at a concentration of only 1 mM, reminiscent of SCF. Under these conditions, the sperm DNA was all fragmented to loop-sized fragments (Fig. 4B, lane 7). These data indicate that MnCl2 can activate the nuclease even without EGTA, but that EGTA stimulates the nuclease with MnCl2 much more. It also suggests that the EGTA-activated nuclease is much more specific for MnCl2 than CaCl2.
Figure 4. Comparison of the Relative Activities of EGTA-SDD with Ca+2 and Mn+2.
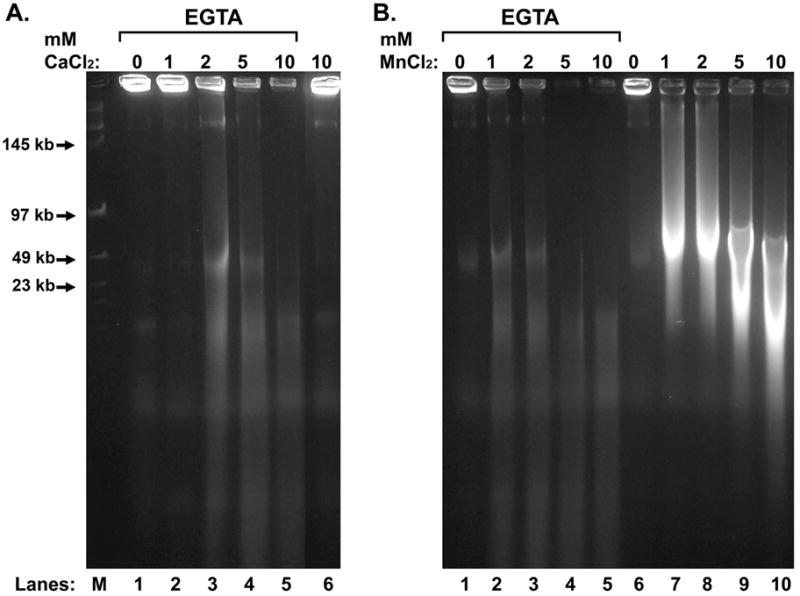
Vas deferens total plasma was embedded in agarose plugs, extracted with 2 M NaCl and 2 mM DTT, and then treated with or without 30 mM EGTA, as indicated. The plugs were then placed in either TKB alone, or various concentrations of CaCl2 (A) or MgCl2 (B). The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE).
EGTA Activated Nuclease is Present in the Sperm Cell, and in the Luminal Fluid Compartments
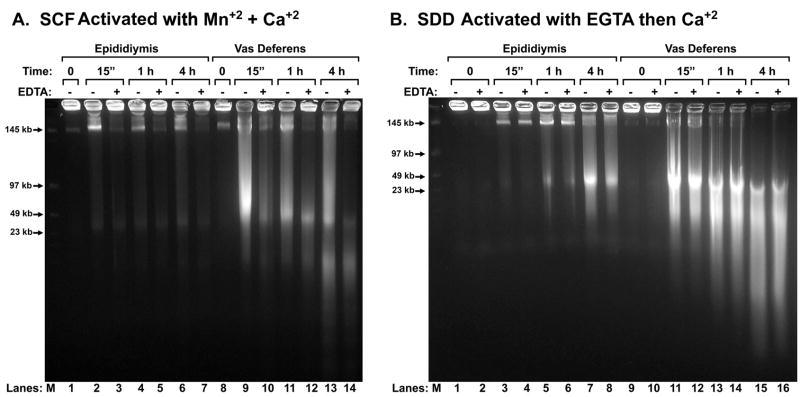
In the assays described above, the entire luminal plasma was used. This includes the sperm cells as well as the fluids and vesicles that surround the spermatozoa. We next tested whether the EGTA activated nuclease was present within each of these three components. We first isolated sperm cells by washing them with repeated centrifugation and then we tested whether MnCl2 and CaCl2 could activate the degradation of DNA in these isolated spermatozoa by SCF. We had previously reported that isolated spermatozoa did not have the ability to degrade their DNA (Shaman et al., 2006), but this was before we discovered that spermatozoa from the vas deferens have a much higher SCF activity than epididymal spermatozoa (Yamauchi et al., 2007a). In our previous report, we used a mixture of spermatozoa from both sources. Here, we found that isolated epididymal spermatozoa were not able to degrade their DNA when incubated with MnCl2 and CaCl2 (Fig. 5A, lanes 1 – 7), but spermatozoa isolated from the vas deferens did (Fig. 5A, lanes 8 – 14). We then isolated spermatozoa from both organs and tested for their ability to digest DNA after EGTA and subsequent CaCl2 treatment, we found that both epididymal and vas deferens spermatozoa degraded their DNA, although once again the latter exhibited much greater degradation (Fig. 5B).
Figure 5. Mn+2/Ca+2 SCF and EGTA-SDD in Isolated Spermatozoa.
(A) Spermatozoa were washed of fluid and treated with 10 mM MnCl2 and 10 mM CaCl2 for 0 to 4 hrs, as indicated. Half of the plugs were then treated with EDTA to test for the ability of the DNA fragmentation to religate. The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE). (B) Spermatozoa from caudal epididymides or vas deferens were separated from the luminal fluid components by repeated low-speed centrifugation. The spermatozoa were then embedded in agarose plugs. The agarose plugs were extracted with 2 M NaCl and 2 mM DTT, and then treated with 30 mM EGTA. The plugs were then incubated with 10 mM CaCl2, alone, for 0 to 4 hrs, as indicated. Half of the plugs were then treated with EDTA to test for the ability of the DNA fragmentation to religate. The reactions were stopped with SDS, and the DNA analyzed by field inversion gel electrophoresis (FIGE).
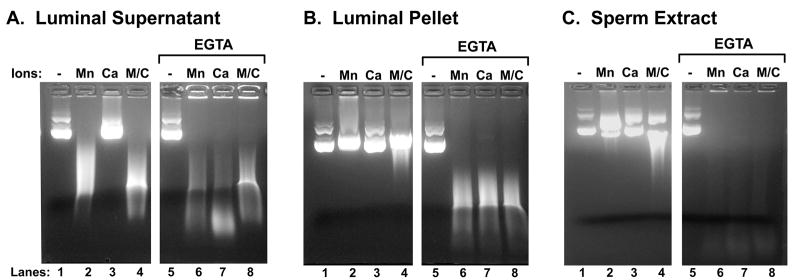
To test for nuclease activity in the luminal fluid from which the spermatozoa were isolated, we developed an assay to screen for the nuclease using a plasmid, rather than sperm chromatin, as the nucleic acid target (Shaman et al., 2006). Spermatozoa were isolated and the subsequent vas deferens fluid was separated into two fractions: a soluble, extracellular portion, the “fluid supernatant” and an insoluble, high-speed pellet, the “fluid pellet”. Spermatozoa were extracted with 0.25% TX and then centrifuged; the supernatant of this treatment, “sperm extract,” was used in the plasmid-based nuclease assay. We found that an EGTA-activatable nuclease was in all three fractions. None of these required salt or DTT, suggesting that salt extraction is only required to expose the sperm chromatin in SDD, not to enhance the EGTA activation mechanism. The existence of the nuclease in the fluid pellet suggests that it is present in vesicles. This is supported by the fact that TX was required to release the nuclease from the pellet. We also found a nuclease in the luminal fluid that could digest the plasmid DNA in the presence of MnCl2 or in MnCl2 and CaCl2, even without EGTA pretreatment (Fig. 6A, lanes 2 and 4). This Mn+2/Ca+2 nuclease activity was not present in either the luminal pellet (Fig. 6B) or the sperm extract (Fig. 6C).
Figure 6. A Nuclease that is Activated by EGTA and then Requires a Divalent Cation Exists in the Vas Deferens Luminal Fluid and Extractable Component of the Sperm Cell.
(A) Luminal fluid was prepared from the vas deferens as described in the Methods. One sixth volume of the luminal fluid was added to the nuclease assay buffer containing either no cation (lane 1), 10 mM MnCl2 (lane 2), 10 mM CaCl2 (lane 3), or both 10 mM MnCl2 and 10 mM CaCl2 (lane 4). Alternatively, the fluid was treated with 30 mM EGTA before one sixth volume was placed into the nuclease buffer under the same conditions (lanes 5 – 8). (B) As in (A), except that the high-speed, luminal pellet was used. (C) as in (A) except the sperm extract was used. All samples were analyzed by conventional agarose electrophoresis.
EGTA-SDD Progresses through a Reversible Step
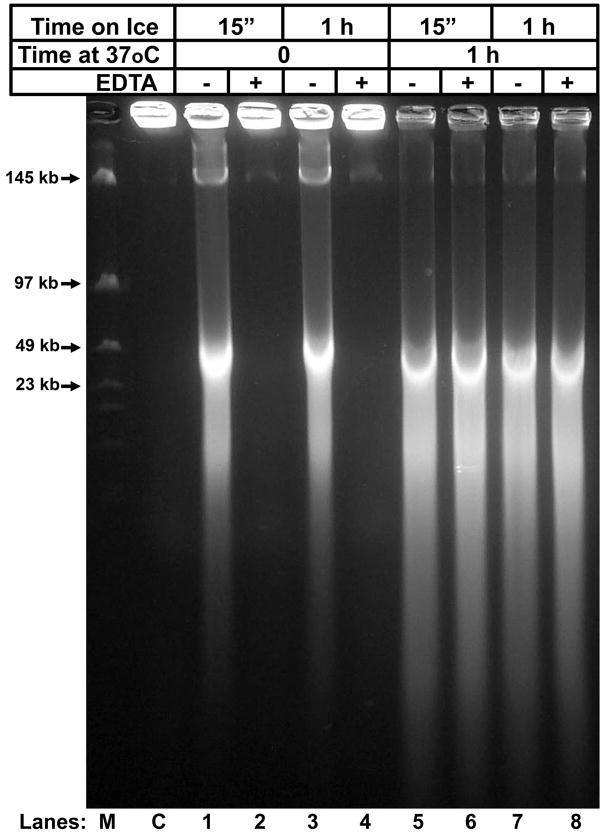
We previously demonstrated that MnCl2 with CaCl2 can activate spermatozoa to cleave its DNA to loop-sized fragments and that this process can be reversed with EDTA (Shaman et al., 2006). We suggested that this reaction proceeds by first activating TOP2B, and then activating a nuclease. Sperm DNA degradation resulting from EGTA-SDD was not reversible by EDTA, (Fig. 5B, lanes 6, 8, 12, 14, and 16). This suggested that either EGTA-SDD progressed too quickly through the activation of the TOP2B to be detected, or that EGTA-SDD does not activate TOP2B to fragment sperm DNA before DNA digestion. To discriminate between these two possibilities, we started the EGTA-SDD reaction on ice to slow it down. The spermatozoa were molded into 1% agarose plugs, as described above, and were subsequently incubated with NaCl and DTT at 37°C and then with EGTA at room temperature. They were then incubated on ice with CaCl2 for 15 min or 1h and then the reactions were moved to a 37°C water bath. At each step, some of the plugs were treated with EDTA to religate and TOP2B breaks. The results show that EGTA-activated sperm DNA digestion was reversible when the reaction proceeded at 4 °C (Fig. 7, lanes 2 and 4) and that further incubation at 37°C rendered the reaction irreversible (Fig. 7, lanes 6 and 8). These data suggest that EGTA-activated sperm DNA digestion progressed through a reversible phase before advancing to the irreversible step.
Figure 7. EGTA-SDD Progresses through a Reversible Step.
Total epididymal plasma was embedded in agarose plugs and extracted with 2 M NaCl and 2 mM DTT, and then treated with 30 mM EGTA. The plugs were then placed in 4°CTKB with 10 mM CaCl2 and incubated for either 15 min (lanes 1, 2, 5, and 6) or 1 hr (lanes 3, 4, 7 and 8) on ice. Four plugs were subsequently placed in a 37°C water bath for 1 hr (lanes 5 – 8). At the end of each reaction, the plugs were either placed directly into SDS digestion buffer to stop the reaction (lanes 1, 3, 5, and 7) or first incubated in 50 mM EDTA to test for fragmentation reversal (lanes 2, 4, 6, and 8) before being placed into SDS digestion buffer. The DNA analyzed by field inversion gel electrophoresis (FIGE).
Discussion
In this work, we demonstrated the existence of a nuclease that is activated by EGTA and then requires a divalent cation to digest DNA. To our knowledge, this is the first time a nuclease that is activated by EGTA has been described. We have also demonstrated that both the reversible sperm chromatin fragmentation (SCF) of DNA to loop-sized fragments and the more complete sperm DNA degradation (SDD) that we previously described (Shaman et al., 2006) can also be activated by EGTA. This suggests that the EGTA-activated nuclease may be involved in both processes.
Several different nucleases that degrade DNA have been described with widely varying dependencies on divalent cations. The list, by no means exhaustive, includes DNAse I that requires Mg+2 (Kishi et al., 2001; Suck, 1994), an endonuclease that requires Mg+2 or Mn+2 but not Ca+2 and is inhibited by Zn+2 (Kawabata et al., 1997), and a series of Ca+2/Mg+2 dependent nucleases (Durrieu et al., 2000; Stratling et al., 1984; Yakovlev et al., 1999). DNAse II does not require any divalent cations (Evans and Aguilera, 2003), and some nucleases are active in the presence of EDTA (Pohlman et al., 1993; Przykorska et al., 2004). However, to our knowledge, there has not been a nuclease that has been described with this particular activity – it requires EGTA and subsequently either Mn+2, Ca+2, or Zn+2, but not Mg+2, to digest DNA.
The sequence of this reaction suggests the possibility that a calcium binding protein is first dissociated from a regulator protein, or the nuclease, itself, and the nuclease then uses the divalent cation as a cofactor to digest the DNA. Calcium binding proteins are well known effectors in many different cell signaling pathways (Grabarek, 2006; Ikura and Ames, 2006). In most cases, however, the calcium binding protein activates an enzyme in the presence of Ca+2 by binding to it, as in the calmodulin dependent protein kinases (Chin, 2005). There is at least one precedent for a calcium binding protein inhibiting an activity which is released by the removal of Ca+2. Recoverin prevents the binding of rhodopsin to a membrane bound kinase in the presence of Ca+2, and EGTA allows the kinase to bind to rhodopsin (Ames et al., 2006). Our data are consistent with, but do not yet prove, a regulation of the nuclease in which a calcium binding protein prevents its activation. One candidate for this calcium binding protein is calmodulin, which is present in high concentrations in the male reproductive tract (Ignotz and Suarez, 2005).
These data also suggest that there are two ways to induce the degradation of sperm DNA through SDD. The first is by incubation of the spermatozoa with MnCl2 combined with CaCl2, or MnCl2 alone, as reported previously (Shaman et al., 2006), and repeated in this work (Figs. 1 and 5). This MnCl2/CaCl2 or MnCl2 activation of sperm DNA degradation is not dependent on salt extraction, and is more concentrated in vas deferens spermatozoa than in epididymal spermatozoa. CaCl2 alone cannot activate SDD with or without salt extraction. The second method is the one reported here, pretreatment with salt and DTT, then EGTA. This EGTA-SDD can be activated by CaCl2 alone, but is stronger with MnCl2.
The relationship between these two pathways for nuclease activation is not yet clear. For example, using our plasmid based nuclease assay (Fig. 6), only the luminal fluid contains a Mn+2/Ca+2 activatible nuclease. This activity is not present in the luminal pellet, nor is it extractable from spermatozoa. However, it is clear that the spermatozoa do contain a nuclease that is activated by Mn+2/Ca+2 (Fig. 5A), which is apparently not extractable. We are currently testing the hypothesis that the in vivo mechanism for activating the nuclease is a two step process that requires an “activator” which, in the presence of Mn+2, dissociates a calcium binding protein from inhibiting the nuclease, either directly or indirectly. This releases a nuclease that can employ Mn+2, Ca+2, or Zn+2, but not Mg+2, as a cofactor to digest DNA. In vitro, EGTA would dissociate the calcium binding protein without the need of the activator. Finally, in the spermatozoa, the activator must be sequestered within the spermatozoa and cannot be extracted with TX, while the nuclease can. It is also possible that the two pathways activate two different nucleases, and further work will have to be done to clarify this.
The presence of both the EGTA activated nuclease and its regulatory pathway in all three components of the vas deferens plasma bears some consideration. The epididymis contains vesicles, called epididymosomes, that provide new proteins and other components to the maturing spermatozoa emerging from the testis (Rejraji et al., 2006). These vesicles fuse with the sperm cell membrane providing new proteins that were synthesized in the epididymal epithelium. Our data, particularly the fact that the nuclease can be pelleted by high speed centrifugation are consistent with the nuclease being present within similar vesicles. It is possible that the epithelium of the vas deferens or the cauda epididymis provides the nuclease to the spermatozoa after they are fully mature.
Finally, our experiments provide the first direct link between the reversible fragmentation of sperm DNA in SCF and the more complete, irreversible degradation of DNA by a nuclease in SDD. In our previous report, we demonstrated that spermatozoa could fragment their DNA to loop-sized fragments by SCF, or completely degrade them by SDD (Shaman et al., 2006). We postulated that SCF progressed on to SDD, but could not demonstrate it directly. In this report, when salt extracted spermatozoa were treated with EGTA, then with CaCl2 on ice, the DNA was only cleaved to loop-sized fragments, and these breaks could be religated by treatment with EDTA (Fig. 7). This is identical to SCF that we had previously described. When the same samples were then moved to a 37°C incubator, the reaction progressed to the irreversible digestion of DNA, characteristic of SDD. Thus, we demonstrated that EGTA activated SDD does progress through SCF.
Reversal of double stranded DNA breaks with EDTA is a hallmark of topoisomerase II cleavage (Champoux, 2001; Wang, 2002) and our previous data have demonstrated that TOP2B is involved in mouse sperm SCF (Shaman et al., 2006). However, in these EGTA-SDD experiments we were not able to completely inhibit the reaction with topoisomerase inhibitors (data not shown) possibly because the reaction was too quick or the salt extraction interfered with the inhibition (Oestergaard et al., 2004). Whether or not TOP2B is involved, the data clearly indicate that the EGTA-SDD pathway for sperm DNA digestion progresses through a reversible loop-sized fragmentation of the chromatin before the nuclease digests the DNA. This is reminiscent of DNA degradation in apoptosis which begins with topoisomerase II cleavage of the DNA and is followed by a series of nucleases that further degrade the DNA (Gromova et al., 1995; Li and Liu, 2001).
It is important to note that the work presented here does not contradict the recent evidence that EGTA and EDTA protect spermatozoa from degrading their DNA (Kusakabe et al., 2001). EGTA, alone, does not activate the nuclease, and if no additional divalent cation is supplied, EGTA would still be expected to inhibit a host of other nucleases known to be present in the male reproductive tract.
We do not currently know the function of the EGTA-activated nuclease, but it may be part of an apoptotic mechanism that ensures that only the best sperm make it through the reproductive tract (Olson et al., 2004; Ward and Ward, 2004). The activation by EGTA suggests that the nuclease is tightly regulated by a signal transduction mechanism that involves a calcium binding protein. Such a regulation would be necessary to protect normal, healthy spermatozoa from inadvertently being destroyed or damaged by nucleases. What is clear, however, is that spermatozoa absorb a nuclease during their maturation through the male reproductive tract. It will be important to understand its role and how to inhibit its activity.
Acknowledgments
This work was supported by NIH grants HD28501 to W.S.W. and by P20 RR011091
References
- Allen MJ, Bradbury EM, Balhorn R. The natural subcellular surface structure of the bovine sperm cell. Journal of Structural Biology. 1995;114:197–208. doi: 10.1006/jsbi.1995.1019. [published erratum appears in J Struct Biol 1995 Nov–Dec;115(3):338–41] [DOI] [PubMed] [Google Scholar]
- Ames JB, Levay K, Wingard JN, Lusin JD, Slepak VZ. Structural basis for calcium-induced inhibition of rhodopsin kinase by recoverin. J Biol Chem. 2006;281:37237–45. doi: 10.1074/jbc.M606913200. [DOI] [PubMed] [Google Scholar]
- Anzar M, He L, Buhr MM, Kroetsch TG, Pauls KP. Sperm apoptosis in fresh and cryopreserved bull semen detected by flow cytometry and its relationship with fertility. Biol Reprod. 2002;66:354–60. doi: 10.1095/biolreprod66.2.354. [DOI] [PubMed] [Google Scholar]
- Baccetti B, Collodel G, Piomboni P. Apoptosis in human ejaculated sperm cells (notulae seminologicae 9) J Submicrosc Cytol Pathol. 1996;28:587–96. [PubMed] [Google Scholar]
- Blanco-Rodriguez J, Martinez-Garcia C. Apoptosis pattern elicited by several apoptogenic agents on the seminiferous epithelium of the adult rat testis. J Androl. 1998;19:487–97. [PubMed] [Google Scholar]
- Carballada R, Esponda P. Regulation of foreign DNA uptake by mouse spermatozoa. Exp Cell Res. 2001;262:104–13. doi: 10.1006/excr.2000.5079. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chin ER. Role of Ca2+/calmodulin-dependent kinases in skeletal muscle plasticity. J Appl Physiol. 2005;99:414–23. doi: 10.1152/japplphysiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- Clarke HJ. Nuclear and chromatin composition of mammalian gametes and early embryos (REVIEW) Biochemistry & Cell Biology. 1992;70:856–66. doi: 10.1139/o92-134. [DOI] [PubMed] [Google Scholar]
- Durrieu F, Samejima K, Fortune JM, Kandels-Lewis S, Osheroff N, Earnshaw WC. DNA topoisomerase IIalpha interacts with CAD nuclease and is involved in chromatin condensation during apoptotic execution. Curr Biol. 2000;10:923–6. doi: 10.1016/s0960-9822(00)00620-5. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Aguilera RJ. DNase II: genes, enzymes and function. Gene. 2003;322:1–15. doi: 10.1016/j.gene.2003.08.022. [DOI] [PubMed] [Google Scholar]
- Giannakis C, Forbes IJ, Zalewski PD. Ca2+/Mg(2+)-dependent nuclease: tissue distribution, relationship to inter-nucleosomal DNA fragmentation and inhibition by Zn2+ Biochem Biophys Res Commun. 1991;181:915–20. doi: 10.1016/0006-291x(91)91278-k. [DOI] [PubMed] [Google Scholar]
- Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509–25. doi: 10.1016/j.jmb.2006.03.066. [DOI] [PubMed] [Google Scholar]
- Gromova II, Nielsen OF, Razin SV. Long-range fragmentation of the eukaryotic genome by exogenous and endogenous nucleases proceeds in a specific fashion via preferential DNA cleavage at matrix attachment sites. Journal of Biological Chemistry. 1995;270:18685–90. doi: 10.1074/jbc.270.31.18685. [DOI] [PubMed] [Google Scholar]
- Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3581–5. doi: 10.1073/pnas.92.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignotz GG, Suarez SS. Calcium/calmodulin and calmodulin kinase II stimulate hyperactivation in demembranated bovine sperm. Biol Reprod. 2005;73:519–26. doi: 10.1095/biolreprod.105.040733. [DOI] [PubMed] [Google Scholar]
- Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc Natl Acad Sci U S A. 2006;103:1159–64. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR. Replication occurs at a nucleoskeleton. Embo J. 1986;5:1403–10. doi: 10.1002/j.1460-2075.1986.tb04374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata H, Anzai N, Masutani H, Hirama T, Hishita T, Dodo M, Masuda T, Yoshida Y, Okuma M. Mg2+- or Mn2+-dependent endonuclease activities of human myeloid leukemia cells capable of producing nucleosomal-size DNA fragmentation. Biochem Biophys Res Commun. 1997;233:133–8. doi: 10.1006/bbrc.1997.6362. [DOI] [PubMed] [Google Scholar]
- Kishi K, Yasuda T, Takeshita H. DNase I: structure, function, and use in medicine and forensic science. Leg Med (Tokyo) 2001;3:69–83. doi: 10.1016/s1344-6223(01)00004-9. [DOI] [PubMed] [Google Scholar]
- Kusakabe H, Szczygiel MA, Whittingham DG, Yanagimachi R. Maintenance of genetic integrity in frozen and freeze-dried mouse spermatozoa. Proc Natl Acad Sci U S A. 2001;98:13501–6. doi: 10.1073/pnas.241517598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyprianou N, English HF, Isaacs JT. Activation of a Ca2+-Mg2+-dependent endonuclease as an early event in castration-induced prostatic cell death. Prostate. 1988;13:103–17. doi: 10.1002/pros.2990130203. [DOI] [PubMed] [Google Scholar]
- Li TK, Liu LF. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- Lin WW, Lamb DJ, Lipshultz LI, Kim ED. Demonstration of testicular apoptosis in human male infertility states using a DNA laddering technique. Int Urol Nephrol. 1999;31:361–70. doi: 10.1023/a:1007130320700. [DOI] [PubMed] [Google Scholar]
- Maione B, Pittoggi C, Achene L, Lorenzini R, Spadafora C. Activation of endogenous nucleases in mature sperm cells upon interaction with exogenous DNA. DNA Cell Biol. 1997;16:1087–97. doi: 10.1089/dna.1997.16.1087. [DOI] [PubMed] [Google Scholar]
- Oestergaard VH, Knudsen BR, Andersen AH. Dissecting the cell-killing mechanism of the topoisomerase II-targeting drug ICRF-193. J Biol Chem. 2004;279:28100–5. doi: 10.1074/jbc.M402119200. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP, NagDas SK, Melner MH. Region-specific expression and secretion of the fibrinogen-related protein, fgl2, by epithelial cells of the hamster epididymis and its role in disposal of defective spermatozoa. J Biol Chem. 2004;279:51266–74. doi: 10.1074/jbc.M410485200. [DOI] [PubMed] [Google Scholar]
- Pardoll DM, Vogelstein B, Coffey DS. A fixed site of DNA replication in eucaryotic cells. Cell. 1980;19:527–36. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Pogany GC, Corzett M, Weston S, Balhorn R. DNA and protein content of mouse sperm. Implications regarding sperm chromatin structure. Exp Cell Res. 1981;136:127–36. doi: 10.1016/0014-4827(81)90044-6. [DOI] [PubMed] [Google Scholar]
- Pohlman RF, Liu F, Wang L, More MI, Winans SC. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 1993;21:4867–72. doi: 10.1093/nar/21.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przykorska A, Solecka K, Olszak K, Keith G, Nawrot B, Kuligowska E. Wheat (Triticum vulgare) chloroplast nuclease ChSI exhibits 5′ flap structure-specific endonuclease activity. Biochemistry. 2004;43:11283–94. doi: 10.1021/bi049947u. [DOI] [PubMed] [Google Scholar]
- Rejraji H, Sion B, Prensier G, Carreras M, Motta C, Frenoux JM, Vericel E, Grizard G, Vernet P, Drevet JR. Lipid remodeling of murine epididymosomes and spermatozoa during epididymal maturation. Biol Reprod. 2006;74:1104–13. doi: 10.1095/biolreprod.105.049304. [DOI] [PubMed] [Google Scholar]
- Risley MS, Einheber S, Bumcrot DA. Changes in DNA topology during spermatogenesis. Chromosoma. 1986;94:217–27. doi: 10.1007/BF00288496. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Mariethoz E, Manicardi G, Bizzaro D, Bianchi PG, Bianchi U. Origin of DNA damage in ejaculated human spermatozoa. Rev Reprod. 1999;4:31–7. doi: 10.1530/ror.0.0040031. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Prisztoka R, Ward WS. Topoisomerase IIB and an Extracellular Nuclease Interact to Digest Sperm DNA in an Apoptotic-Like Manner. Biol Reprod. 2006;75:741–748. doi: 10.1095/biolreprod.106.055178. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The Sperm Nuclear Matrix is Required for Paternal DNA Replication. J Cell Biochem . 2007 doi: 10.1002/jcb.21321. in press. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Lino E, Ward WS. Ability of Hamster Spermatozoa to Digest Their Own DNA. Biol Reprod. 2003;69:2029–2035. doi: 10.1095/biolreprod.103.020594. [DOI] [PubMed] [Google Scholar]
- Stratling WH, Grade C, Horz W. Ca/Mg-dependent endonuclease from porcine liver. Purification, properties, and sequence specificity. J Biol Chem. 1984;259:5893–8. [PubMed] [Google Scholar]
- Suck D. DNA recognition by DNase I. J Mol Recognit. 1994;7:65–70. doi: 10.1002/jmr.300070203. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Ward MA, Ward WS. A model for the function of sperm DNA degradation. Reprod Fertil Dev. 2004;16:547–54. doi: 10.10371/RD03072. [DOI] [PubMed] [Google Scholar]
- Ward WS, Partin AW, Coffey DS. DNA loop domains in mammalian spermatozoa. Chromosoma. 1989;98:153–9. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- Widlak P, Garrard WT. Ionic and cofactor requirements for the activity of the apoptotic endonuclease DFF40/CAD. Mol Cell Biochem. 2001;218:125–30. doi: 10.1023/a:1007231822086. [DOI] [PubMed] [Google Scholar]
- Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem. 2000;275:8226–32. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- Yakovlev AG, Wang G, Stoica BA, Boulares HA, Spoonde AY, Yoshihara K, Smulson ME. A role of the Ca2+/Mg2+-dependent endonuclease in apoptosis and its inhibition by Poly(ADP-ribose) polymerase. J Biol Chem. 2000;275:21302–8. doi: 10.1074/jbc.M001087200. [DOI] [PubMed] [Google Scholar]
- Yakovlev AG, Wang G, Stoica BA, Simbulan-Rosenthal CM, Yoshihara K, Smulson ME. Role of DNAS1L3 in Ca2+- and Mg2+-dependent cleavage of DNA into oligonucleosomal and high molecular mass fragments. Nucleic Acids Res. 1999;27:1999–2005. doi: 10.1093/nar/27.9.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal Pronuclear DNA Degradation Is Functionally Linked to DNA Replication in Mouse Oocytes. Biol Reprod. 2007a doi: 10.1095/biolreprod.107.061473. In Press. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II Mediated Breaks in Spermatozoa Cause the Specific Degradation of Paternal DNA in Fertilized Oocytes. Biol Reprod. 2007b;76:666–672. doi: 10.1095/biolreprod.106.057067. [DOI] [PubMed] [Google Scholar]
- Zirkin BR, Soucek DA, Chang TS. Sperm nuclear packing and regulation during spermatogenesis and fertilization. Johns Hopkins Med J. 1982;151:101–12. [PubMed] [Google Scholar]