A simple, general method to selectively introduce metal ion binding sites into polypeptides would greatly facilitate the engineering of catalytic and redox active sites, radioisotope binding sites, structural elements and spectroscopic probes into proteins.1 For example, recently it was shown that the genetic introduction of (2,2'-bipyridin-5-yl)alanine (Bpy-Ala) into the DNA binding protein catabolite activator protein generated a Cu2+ dependent oxidative DNA cleaving agent.2 Here we report that an amino acid derivative (HQ-Ala, 1) of the metal ion chelating group 8-hydroxyquinoline, which forms highly stable complexes with most transition metal ions and some lanthanides,3,4 can be genetically encoded in E. coli in response to the amber codon, TAG. Moreover, we show that addition of Zn2+ to HQ-Ala containing proteins results in the formation of both a fluorescent probe and of a heavy metal binding site for SAD phasing in protein crystallographic structure determination.
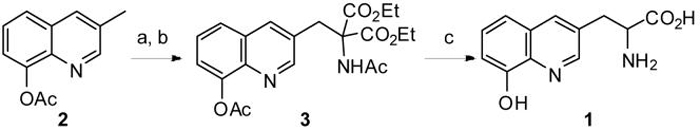
2-Amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQ-Ala, 1) was synthesized in three steps starting from 3-methylquinolin-8-yl acetate5 (2) (Scheme 1). Bromination of 2 by NBS and AIBN afforded the bromomethyl quinoline intermediate to which diethyl acetamido-malonate was added. Subsequent acidic decarboxylation and hydrolysis afforded HQ-Ala in racemic form. To genetically encode HQ-Ala in E. coli, an orthogonal Methanococcus jannaschii amber suppressor tRNA (MjtRNA) /tyrosyl-tRNA synthetase (MjTyrRS) pair was used.6 To alter the amino acid specificity of MjTyrRS, a mutant library2a was generated by randomizing (NNK) nine active site residues (Y32, L65, H70, F108, Q109, Q155, D158, I159 and L162). The library was then subjected to rounds of alternating positive and negative selections. In the positive selection, cell survival is dependent on the suppression of an amber mutation (D112) in the chloramphenicol acetyl-transferase gene in the presence of the unnatural amino acid. In the negative selection, cell survival is dependent on the suppression of amber mutations (Q2, D44 and G65) in the barnase gene in the absence of the unnatural amino acid.
Scheme 1.
Synthesis of HQ-Ala. Conditions: (a) NBS, AIBN, CCl4, reflux, 4 h; (b) NaH, diethyl acetamidomalonate, DMF, rt, 1 h, 52% in two steps.; (c) 12 N HCl, reflux, 6 h, 99%.
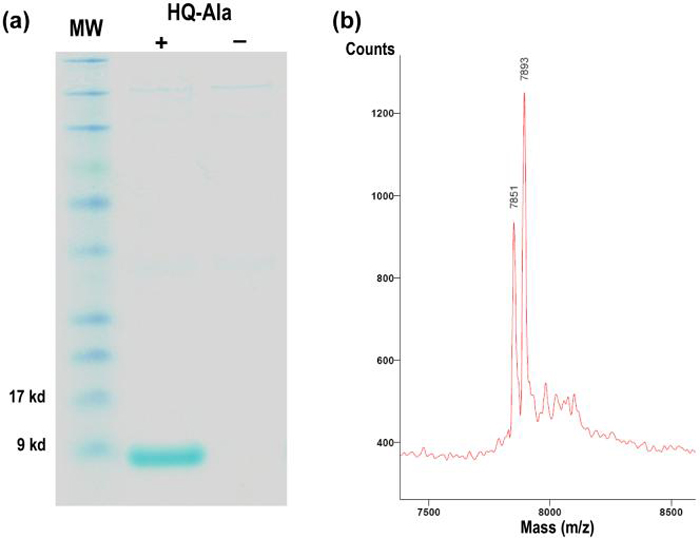
After three rounds of positive and two rounds of negative selection, 15 clones were isolated which survived on chloramphenicol only in the presence of HQ-Ala. DNA sequencing revealed eight unique mutants with HQ-3D4 corresponding to the most common sequence (Table 1). To determine the efficiency and fidelity with which the selected MjTyrRS incorporates HQ-Ala into proteins, a Z-domain protein with an amber codon at position 7 was expressed in E. coli in the presence of the MjtRNA/HQ-3D4 pair. Expression in glycerol minimal medium7 produced 1.5 mg/L full-length protein in the presence of 1 mM HQ-Ala, while no full-length protein was detected by SDS-PAGE analysis in the absence of HQ-Ala (Figure 1a). MALDI-TOF mass spectrometry (MS) analysis of the purified protein confirmed the incorporation of HQ-Ala (Figure 1b).
Table 1.
The 15 selected HQ-Ala-specific MjTyrRS mutants represent 8 unique mutants.
| MjTyrRS | Frequency | Tyr 32 | Leu 65 | His 70 | Phe 108 | Gln 109 | Gln 155 | Asp 158 | Ile 159 | Leu 162 |
|---|---|---|---|---|---|---|---|---|---|---|
| HQ-3D1 | 1 | His | Val | Met | Phe | Gln | Leu | Gly | Ser | Leu |
| HQ-3D2 | 1 | Leu | Ile | Ser | Arg | Glu | Asn | Gly | Ile | Pro |
| HQ-3D3 | 1 | Glu | Val | Thr | Arg | Glu | Gln | Ser | Ser | Ser |
| HQ-3D4 | 4 | Val | Met | Thr | Arg | Glu | Gln | Ser | Ser | Leu |
| HQ-3D5 | 3 | Met | Cys | Thr | Arg | Glu | Gln | Ser | Thr | His |
| HQ-3D6 | 2 | Leu | Ile | Thr | Arg | Glu | Gln | Ser | Ala | Arg |
| HQ-3D7 | 1 | Leu | Gln | Thr | Arg | Glu | Gln | Ser | Tyr | Leu |
| HQ-3D8 | 2 | Leu | Thr | Asn | Phe | Gln | Gln | Gly | Thr | Leu |
Figure 1.
Expression of Z-domain protein with an amber codon at position 7 in the presence of the MjtRNA/HQ-3D4 pair. (a) SDS-PAGE analysis of the mutant Z-domain protein. (b) MALDI-TOF mass analysis: expected 7848; observed 7851, 7893 (N-terminal acetylated).
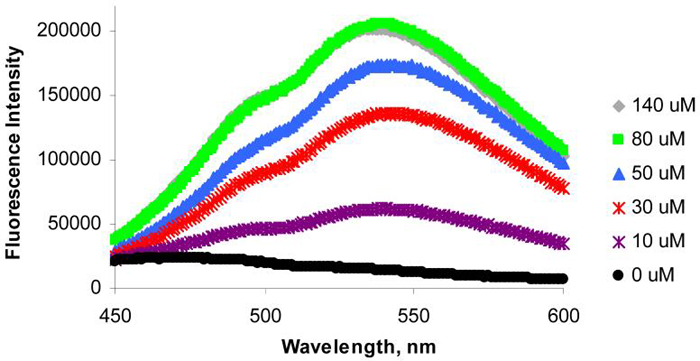
To examine whether binding of metal ions to HQ-Ala in proteins would create a site specific fluorescent reporter8, the HQ-Ala Z-domain mutant (10 μM) was titrated with Zn2+ and fluorescence from the HQ-Zn2+ complex was measured (Figure 2). The mutant protein was not fluorescent in the absence of Zn2+ when excited at 400 nm, but became fluorescent when Zn2+ was added; the fluorescence increased with increasing concentration of Zn2+. Thus the genetic incorporation of HQ-Ala into proteins should prove useful for the generation of biological metal ion sensors, and local fluorescent probes of protein structures, dynamics and ligand binding.
Figure 2.
Fluorescence spectra of the mutant Z-domain proteins (10 μM) containing HQ-Ala complexed with different concentrations of ZnSO4. Samples were excited at 400 nm.
Next we examined whether HQ-Ala could be used to introduce heavy metal ion binding sites into proteins for crystallographic structure determination. To solve the structure of macromolecules by X-ray crystallography, both the amplitude and the phase angle of the diffraction pattern have to be determined. Without a reasonable homology model or ultra high resolution data, experimental phasing methods, such as single and multi-wavelength anomalous diffraction ((S/M)AD), and single or multiple isomorphous replacement ((S/M)IR) have to be used to calculate initial phases. All of these methods require site-specific placement of heavy atom(s) into the crystal. Currently, recombinant protein expression is widely used to incorporate selenium atoms into proteins by replacement of methionine residues in the sequence allowing SeMet (S/M)AD experiments to be performed with tunable X-ray sources.9 However, introduction of too few or disordered selenium atoms are problematic in phase determination. The incorporation of selenium methionine can also adversely affect the yield and solubility of the recombinant protein. An alternative approach is to soak the protein crystal in solutions containing heavy atoms, however, the likelihood of obtaining a site-specifically bound heavy atom at high occupancy is low. It would be ideal to be able to rapidly engineer metal ion binding-sites into proteins that bind heavy atoms with high occupancy.
To this end, TM0665 (O-acetylserine sulfhydrylase), a test protein from the Thermotoga maritima Structural Genomics program10 was used to examine the utility of HQ-Ala incorporation for SAD phasing. A His-tagged TM0665 Phe22 to HQ-Ala mutant was expressed and purified by Ni2+ affinity chromatography. Crystallization of the HQ-Ala TM0665 mutant was accomplished by the hanging-drop vapor diffusion method. Upon incubation with Zn2+, the HQ-Ala TM0665 mutant was fluorescent both in solution and as crystals, which indicates successful metal ion coordination. The structure of TM0665 was determined from data collected at beamline 5.0.2 of the Advanced Light Source at 100 K using a SAD data collection strategy with an energy corresponding to the maximum value of δf″ as determined from a fluorescent scan (wavelength of 1.2815 Å). In all, 120° of data were collected in two 30° segments using an inverse beam strategy in 5° wedges to a maximum resolution of 2.1 Å (Table 2). Crystals of TM0665 belong to the space group P42212 with cell dimensions a = b = 135.31 Å, c = 74.93 Å, α = 90°, β = 90°, and γ = 90°, with two molecules in each asymmetric unit.
Table 2.
Summary of data collection statistics and model refinement.
| space Group | P42212 |
| unit cell dimensions (Å) (a,b,c) | 135.86, 135.36, 74.94 |
| X-ray radiation Wavelength (Å) | 1.2815 |
| resolution (Å) | 2.15 |
| Rmerge (highest shell) | 0.092 (0.218) |
| unique refs (observed) | 38622 (533831) |
| completeness (Highest Shell) % | 99.6 (98.0) |
| redundancy (Highest Shell) | 13.8 (13.5) |
| Rfactor (Rfree *)% | 20.7 (24.3) |
| No. protein atoms | 4376 |
| No. water atoms | 164 |
| No. Hetero Atoms | 2 |
| rmsd bonds (Å) | 0.01 |
| rmsd angles (deg) | 1.22 |
| mean B factor (Å2) | 29 |
SAD phasing was carried out with the program SOLVE in PHENIX (http://www.phenix-online.org/). The peak wavelength data was used within the resolution range 2.1 to 20 Å. The anomalous signal was high as would be expected from an almost fully occupied stable metal site; the average ratio of Bijovet pairs (<|ΔF|>/<F>) was 5.3%. This combined with the fact that were only two zinc atoms in the asymmetric unit enabled both direct methods and Patterson-based substructure determination programs such as SHELX and SOLVE to easily locate the position of the atoms, a feat that could also be performed by manual interpretation of the Patterson map. The initial mean figure of merit (FOM) is 0.34. The electron density map was further improved by the RESOLVE program (FOM = 0.66). A clearly interpretable electron density map was calculated using FFT in the CCP4 suite11. The electron density of Zn2+ became evident at the 5-σ level. The initial model was built with an automatic modeling building program, TEXAL12,13. The structure was rebuilt using XtalView and refined with the program REFMAC11,14. Atomic coordinates and structure factors of TM0665_HQ-Ala have been deposited in the Protein Data Bank (PDB code: XXX).
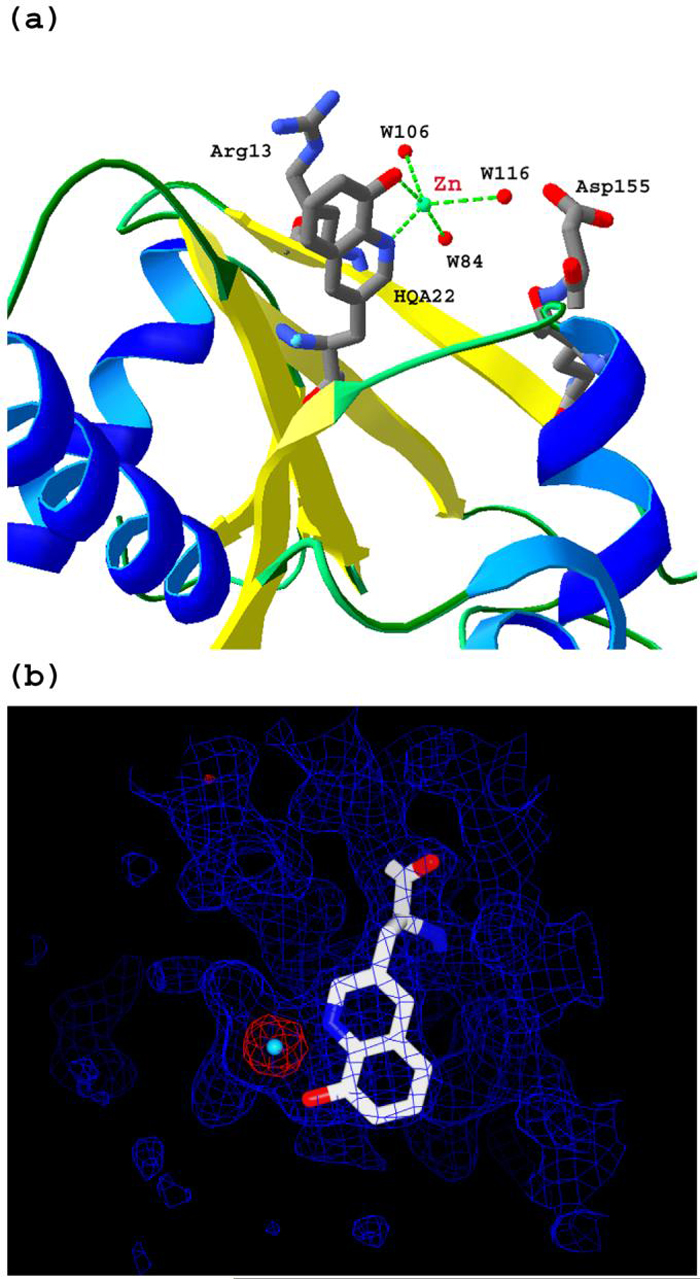
The structure of TM0665 HQ-Ala can be readily superimposed with that of the wild type protein. There is no significant difference between wild type TM0665 and the HQ-Ala mutant (root mean squared deviation (rmsd) of C alpha atoms is 0.48 Å), except for the replacement of phenylalanine by HQ-Ala at position 22. The HQ-Ala residue is located on the surface of one subunit and near the dimer interface of the two subunits in the asymmetric unit. The side chain of HQ-Ala is exposed toward the solvent and chelates Zn2+ through Zn-N (2.1 Å) and Zn-O (2.0 Å) interactions. There is no other residue directly interacting with Zn2+, indicating that the HQ-Ala group is enough to provide a well-ordered metal binding site per se.
In summary, a metal-chelating amino acid was site-specifically incorporated into proteins in E. coli with high efficiency and specificity in response to the amber codon. The ability to selectively add metal ions to protein crystals supplements other powerful techniques for structure determination and facilitates the introduction of well-ordered heavy metals into proteins with high occupancy for phasing techniques such as S/MIR and (S/MAS) experiments. This method becomes particularly useful for those proteins not suitable for selenomethionine phasing. Because HQ complexed with Zn2+ or Mg2+ is fluorescent, proteins with HQ-Ala can also be used as fluorescence-based sensors in vitro and in vivo.15 In addition, this method would facilitate the de novo design of metalloproteins with other novel structures and functions.
Supplementary Material
Figure 3.
(a) The overall structure of the Phe22 HQ-Ala TM0665 mutant bound with Zn2+. (b) The electron density map was calculated from the experimental phase. The model of residue HQ-Ala and Zn2+ was fit into the density.
Acknowledgment
We are grateful to the U.S. Department of Energy, Division of Materials Sciences, under Award No. DE-FG03-00ER46051, and the Skaggs Institute for Chemical Biology for support of this work. We would like to thank Heath Klock and Scott Lesley from the JCSG for providing clones for TM0665 The JCSG is supported by the NIH Protein Structure Initiative grant U54 GM074898 from the National Institute of General Medical Sciences (www.nigms.nih.gov). The work in this paper is based on experiments conducted at beamline 5.0.2 of the advanced light source (ALS). We are grateful to the staff for all of there help. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Material Sciences Division of the U.S. Department of Energy under contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
Footnotes
Supporting Information Available: Materials and methods (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).(a) Lu Y, Berry SM, Pfister TD. Chem. Rev. 2001;101:3047–3088. doi: 10.1021/cr0000574. [DOI] [PubMed] [Google Scholar]; (b) Hellinga HW. Curr. Opin. Biotech. 1996;7:437–441. doi: 10.1016/s0958-1669(96)80121-2. [DOI] [PubMed] [Google Scholar]
- (2).(a) Xie J, Liu W, Schultz PG. Angew. Chem. Int. Ed. 2007;46:9239–9242. doi: 10.1002/anie.200703397. [DOI] [PubMed] [Google Scholar]; (b) Lee HS, Schultz PG. J. Am. Chem. Soc. 2008;130:13194–13195. doi: 10.1021/ja804653f. [DOI] [PubMed] [Google Scholar]
- (3).Phillips JP. Chem. Rev. 1956;56:271–297. [Google Scholar]
- (4).Martell AE, Smith RM. Critical Stability Constants. Vol. 2. Plenum Press; New York: 1974. pp. 223–237. [Google Scholar]
- (5).Albrecht M, Blau O. Synthesis. 1997:213–216. [Google Scholar]
- (6).(a) Wang L, Brock A, Herberich B, Schultz PG. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]; (b) Xie J, Schultz PG. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- (7).Studier FW. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- (8).(a) Soroka K, Vithanage RS, Phillips DA, Walker B, Dasgupta PK. Anal. Chem. 1987;59:629–636. [Google Scholar]; (b) Pearce DA, Jotterand N, Carrico IS, Imperiali B. J. Am. Chem. Soc. 2001;123:5160–5161. doi: 10.1021/ja0039839. [DOI] [PubMed] [Google Scholar]
- (9).Henderickson WA. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- (10).Lesley SA, et al. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Collaborative Computational Project Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- (12).Ioerger TR, Sacchettini JC. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2043–2054. doi: 10.1107/s0907444902016724. [DOI] [PubMed] [Google Scholar]
- (13).Ioerger TR, Sacchettini JC. Methods Enzymol. 2003;374:244–270. doi: 10.1016/S0076-6879(03)74012-9. [DOI] [PubMed] [Google Scholar]
- (14).McRee DE. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- (15).(a) Shults MD, Pearce DA, Imperiali B. J. Am. Chem. Soc. 2003;125:10591–10597. doi: 10.1021/ja0355980. [DOI] [PubMed] [Google Scholar]; (b) Shults MD, Imperiali B. J. Am. Chem. Soc. 2003;125:14248–14249. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.