Abstract
Genomic disorders are a clinically diverse group of conditions caused by gain, loss or re-orientation of a genomic region containing dosage-sensitive genes. One class of genomic disorder is caused by hemizygous deletions resulting in haploinsufficiency of a single or, more usually, several genes. For example, the heterozygous contiguous gene deletion on chromosome 22q11.2 causing DiGeorge syndrome involves at least 20-30 genes. Determining how the copy number variation (CNV) affects human variation and contributes to the aetiology and progression of various genomic disorders represents important questions for the future. Here, I will discuss the functional significance of one form of CNV, haploinsufficiency (i.e. loss of a gene copy), of DNA damage response components and its association with certain genomic disorders. There is increasing evidence that haploinsufficiency for certain genes encoding key players in the cells response to DNA damage, particularly those of the Ataxia Telangiectasia and Rad3-related (ATR)-pathway, has a functional impact. I will review this evidence and present examples of some well known clinically similar genomic disorders that have recently been shown to be defective in the ATR-dependent DNA damage response. Finally, I will discuss the potential implications of a haploinsufficiency-induced defective DNA damage response for the clinical management of certain human genomic disorders.
Key Words: DNA damage response, ATR, haploinsufficiency, genomic disorders.
1. INTRODUCTION
It has long been appreciated that changes in gene copy number are associated with phenotypes in humans. Perhaps, the most well known example of this is the trisomy 21 causative of Down syndrome. Here, increased expression of the genes on chromosome 21 results directly or indirectly in a clinically heterogeneous disorder incorporating cognitive impairment, facial dysmorphology, growth retardation, cancer predisposition, microcephaly, heart and skeletal abnormalities [1]. Interestingly, it was recently shown that CNV is in fact a common genetic trait in clinically unaffected or ‘normal’ individuals [2]. Indeed, the first complete genomic sequence from an individual, the so-called ‘Venter genome’ yielded a surprising level of CNV, highlighting the plasticity of the human genome [3]. The phenomenal recent revolution in the sensitivity and widespread usage of array-based Comparative Genomic Hybridization (array-CGH/a-CGH) techniques has led to CNV being described as the ‘Breakthrough of the Year’ by the journal Science [4]. The widespread use of a-CGH has facilitated the description of several novel genomic disorders and aided in the detailed genetic characterisation of known genomic disorders. A persistent challenge to clinical geneticists and researchers is to unravel exactly how changes in CNV of specific genes or various combinations of genes can impact on normal development. These issues have recently been extensively reviewed elsewhere and the reader is directed to these sources for an overview of a-CGH technology and its applications [5-12]. Here, I will discuss the potential role of haploinsufficiency of DNA damage response components, particularly that of the ATR-pathway, in genomic disorders. Contiguous gene deletion disorders represent a clinically diverse group of human genomic disorders caused by distinct heterozygous chromosomal deletions usually involving several genes [5, 13]. It is assumed that the various clinical manifestations of different genomic disorders arise from the combined impact of haploinsufficiency of multiple genes [14]. It is likely that there are critical genes or pathways sensitive to haploinsufficiency either alone or when combined with haploinsufficiency of other genes. There is increasing evidence from both human and murine studies suggestive of a cellular impact of haploinsufficiency of genes that control different aspects of the response to DNA damage. Since our genomes are constantly exposed to exogenously-derived (e.g. UV radiation) and endogenously-derived (e.g. metabolically generated reactive oxygen species) DNA damaging agents, an impaired ability to detect and/or respond appropriately to these threats can impact on the maintenance of genetic stability. There are many examples of human Mendelian disorders defective in the repair of or response to DNA damage [15]. The importance of these pathways is demonstrated by the increase in cancer predisposition and developmental abnormalities associated with these conditions [15]. An important DNA damage response (DDR) pathway that appears to be affected by haploinsufficiency is the ATR-dependent DDR (ATR-DDR).
2. THE ATR-DEPENDENT DNA DAMAGE RESPONSE (ATR-DDR)
The DDR can be divided into DNA repair processes and signal transduction processes that sense DNA damage and co-ordinate the appropriate response such as cell cycle checkpoint activation, DNA repair and/or apoptosis. Two phosphoinositol-3-kinase-like protein kinases (PIKK) coordinate the signal transduction response to DNA damage in mammalian cells [16-18]. Ataxia Telangiectasia Mutated (ATM) is activated following DNA double strand breaks (DSBs) whilst a related kinase, Ataxia Telangiectasia and Rad3-related (ATR), responds to single stranded regions of DNA. Single stranded DNA (ssDNA) can act as an intermediate during the activity of several excision-mediated repair pathways (nucleotide excision repair and base excision repair), during uncoupling of the transcriptional machinery from its DNA template or similarly following stalling of the DNA replication machinery at DNA lesions or single strand breaks. Following the exposure of ssDNA, it is quickly coated by RPA, a heterotrimeric protein complex (RPA1-3) that plays a role in many aspects of DNA metabolism (e.g. replication, repair, transcription). ATR is recruited to the ssDNA via its binding partner ATRIP (ATR Interacting Protein) [19, 20]. Topoisomerase binding protein 1 (TopBP1) appears to be required for optimal ATR kinase activity [21]. Phosphorylation of the histone H2A variant H2AX on Ser-139 (called γ-H2AX) is one of the earliest detectable PIKK-dependent responses to DNA damage and is required for the retention of additional damage response proteins at the damage site [22-24]. The Mre11/Rad50/NBS1 complex, the Rad17/Rfc2-5 and Rad9/Rad1/Hus1 complexes are recruited to the site of damage independently of ATR/ATRIP and are also phosphorylated by ATR [25, 26]. Retention of these complexes facilitates ATR’s ability to phosphorylate downstream substrates including Brca1, p53 and its effector kinase Chk1 [15].
There is a large amount of functional overlap between ATM and ATR. In fact, both kinase’s phosphorylate mainly the same substrates in response to DNA damage (e.g. Mre11/Rad50/NBS1 complex, p53, Brca1). DSBs can undergo exonucleolytic resection generating ssDNA overhangs, hence generating an ATR activating substrate [27]. Conversely, ssDNA generated at stalled replication forks can collapse producing overt DSBs, hence generating an ATM activating substrate [28]. Nevertheless, congenital defects in ATM and ATR are associated with clinically distinct human disorders. Mutations in ATM cause Ataxia telangiectasia, a progressive degenerative neurological disorder characterised by ataxia and lymphoid malignancy predisposition [29, 30]. Mutations in ATR result in Seckel syndrome, a disorder characterised by microcephaly and growth retardation [31, 32].
3. ATR AND GENOMIC STABILITY
ATR is required to maintain genomic stability. Whilst it has a role in the stabilisation of stalled replication forks, the absence of ATR results in a very specific type of genomic instability, namely DNA Fragile Site expression. DNA Fragile Sites (DFS) are large (>100Kb) distinct genomic regions that exhibit breaks under conditions of replicative ‘stress’ [33, 34]. Such stresses can be induced in the laboratory using DNA replication inhibitors such as aphidicolin or by folate deficiency, which can indirectly impact on the availability of dNTP’s. There are about 75-80 DFS throughout the human genome but it is not really clear why DFS are so unstable. Most tend to be relatively AT-rich and contain more areas of flexibility than non-fragile site regions. Studies on the replication timing of certain fragile sites (FRA3B, FRA7H, FRAXA on chromosome 3, 7 and X respectively) indicate that they are replicated very slowly [33]. DFS are ‘hot spots’ for sister chromatid exchanges (SCE’s) and are also thought to play a role in gene amplification events via a breakage-fusion-bridge cycle [33]. Breakage at or ‘expression of’ DFS is associated with many cancers. For example, the FHIT tumour suppressor gene spans the DFS FRA3B [35]. This gene is often re-arranged or partially deleted in a wide range of tumours (lung, ovarian, breast, esophageal) [36]. Pioneering work by the Glover laboratory identified ATR as the first protein required to mediate DFS stability [37]. Subsequently, several other proteins have been implicated in maintaining DFS stability including the Brca1, SMC1, WRN helicase, Chk1 and the Fanconi anaemia pathway components FANC-A, FANC-B and FANC-D2 [33, 38-41]. Interestingly, most of these proteins are known direct ATR substrates.
4. SECKEL SYNDROME: AN ATR-PATHWAY DEFECTIVE DISORDER
Seckel syndrome, originally described as ‘Bird headed Dwarfism’ in a 1960 monograph by Dr. Helmut Seckel, is a disorder characterised by severe microcephaly, isolated skeletal abnormalities (clinodactyly, thoracic kyphosis, ivory epiphysis) and a dramatic proportionate primordial dwarfism [42]. Microcephaly is a clinical term describing a reduction in occipitofrontal (or head) circumference greater than 3 standard deviations (-3 s.d.) below the age-related mean. This reduced head circumference is a consequence of premature closure and fusion of the cranial sutures reflecting the underlying reduction in brain volume. The aetiology of microcephaly is complex. It can occur in the context of genetic (e.g. syndromal) or non-genetic (e.g. intrauterine infection) situations. Microcephaly is particularly pronounced in Seckel syndrome [31, 42, 43]. The first genetic defect associated with Seckel syndrome was described in 2003. A single synonymous hypomorphic mutation in ATR was identified in five affected individuals in two consanguineous Pakistani families [31]. The mutation (A21201G) was shown to adversely impact on splicing, resulting in dramatically reduced ATR expression in cells derived from the affected patients. Gene targeting of ATR in the murine system results in early embryonic lethality [44, 45]. Hence, these ATR-defective Seckel syndrome cells (ATR-S) proved a useful tractable model to investigate ATR-pathway function in the mammalian setting. ATR-S cells exhibited a diminished ability to phosphorylate ATR substrates following DNA damage-induced ATR-pathway activation (e.g. γH2AX formation and p53-serine-15 phosphorylation) as well as defective cell cycle checkpoint arrest and increased DFS expression [31, 46, 47].
Seckel syndrome is known to be genetically heterogeneous [43, 46, 48-50]. Interestingly, Alderton et al. showed that several unrelated non-ATR mutated Seckel syndrome cell lines all exhibited defective ATR-pathway function (e.g. failure of ATR-dependent G2-M cell cycle checkpoint arrest) [46]. Hence, in agreement with the known genetic heterogeneity of this condition, whilst not all Seckel syndrome cases are caused by mutations in ATR itself, interestingly, all Seckel syndrome cell lines (ATR-S and non-ATR-S) exhibit compromised ATR-pathway function [46]. A further interesting cellular feature of ATR-pathway defective Seckel cells is supernumerary centrosomes in 10-20% of mitotic cells [46]. A normal mitotic cell must only have two centrosomes that nucleate the microtubule spindles facilitating transfer of an equal and identical chromosome complement to the daughter cells [51, 52]. The molecular aetiology of supernumerary mitotic centrosomes in Seckel syndrome cells is currently unclear. Centrosome orientation is fundamental for determining symmetric and asymmetric stem cell division in the embryonic neuroepithelium, an essential process for normal brain development [53-55]. Hence, this cellular feature may be a relevant contributor to the severe microcephaly characteristic of Seckel syndrome. Recently, the second genetic defect associated with Seckel syndrome was described. Mutations in pericentrin (PCNT2)/kendrin which encodes a structural centrosomal protein, were identified in several Seckel syndrome patients all of which exhibited defective ATR-pathway function [56]. This exciting finding further illustrates a functional link between the DDR, the cell cycle machinery and the centrosome. Furthermore, mutations in PCNT2 were also recently found in several microcephalic osteodysplastic primordial dwarfism type II (MOPD II) patients [57, 58]. Whether PCNT2-mutated Seckel syndrome and MOPD II represent distinct disorders or allelic variants of the same condition remains an open question.
5. OTHER DISORDERS THAT EXHIBIT COMPROMISED ATR-PATHWAY FUNCTION
Compromised ATR-pathway function does not appear to be uniquely associated with Seckel syndrome. Several other known DDR disorders have been shown to be defective in aspects of ATR-pathway activity, particularly cell cycle checkpoint activation. Interestingly, whilst these disorders are all characterised by a distinct set of clinical features, there is significant clinical overlap with Seckel syndrome, particularly concerning the developmental abnormalities such as microcephaly and growth retardation (Table 1).
Table 1.
Mendelian Disorders that Exhibit Microcephaly and Growth Retardation Associated with Defective ATR-Pathway Function
| Disorder | Mutant Gene | ATR-Dependent Cellular Features |
|---|---|---|
| Seckel syndrome | ATR, PCNT2 and unknown | Defective ATR-dependent G2-M arrest, supernumerary mitotic centrosomes, DFS expression |
| Nijmegen breakage syndrome | NBS1 | Defective ATR-dependent G2-M arrest |
| Fanconi anaemia | FANC-A,B,C,D1,D2,E,F,G,H,M,J | Defective ATR-dependent G2-M arrest, DFS expression |
| MCPH1-dependent Primary Microcephaly | MCPH1 | Defective ATR-dependent G2-M arrest, supernumerary mitotic centrosomes |
5.1. Nijmegen Breakage Syndrome (NBS)
NBS is caused by hypomorphic mutations in the Nbs1 component of the Mre11/Rad50/NBS (MRN) complex [59, 60]. The MRN complex plays a central role in ATM signalling where it is thought to recruit ATM to the DSB and also facilitate its ability to phosphorylate substrates such as p53 and Chk2 [61]. In fact, all the components of this complex are ATM substrates. Historically, NBS has been described as an A-T-like disorder as cell lines from both conditions exhibit radio-sensitivity and similar cell cycle checkpoint defects in response to DSBs. But, NBS1 is also an ATR substrate and NBS patients exhibit microcephaly and growth retardation, clinical features associated with Seckel syndrome and not A-T. Recently, NBS cells were shown to be compromised for ATR-dependent checkpoint activation and other aspects of ATR-pathway function [26]. Therefore, NBS represents a human condition defective in elements of both ATM and ATR-pathway activity.
5.2. Fanconi Anaemia (FA)
FA, is a genetically heterogeneous condition characterised by a progressive aplastic anaemia, skeletal abnormalities, microcephaly and lymphoid malignancy. FA is caused by mutations in different genes whose products co-ordinately function in the cellular response to DNA cross-links [62, 63]. Several of these genes encode products that together mediate the monoubiquitylation and activation of the FANC-D2 protein in response to DNA damage, particularly during S phase [62]. ATR and NBS1 have been shown to be required for this specific modification [64-66]. In fact, ATR-S and NBS patient-derived cell lines fail to monoubiquitylate FANC-D2 following treatment with replication fork inhibitors [26]. In addition to undergoing monoubiquitylation, FANCD2 has been shown to be phosphorylated by ATM and ATR further highlighting the over lap between these DDR pathways [26, 64, 67].
5.3. MCPH1-Dependent Primary Microcephaly
Autosomal recessive Primary Microcephaly, clinically characterised by the presence of a severe microcephaly in the absence of other overt clinical features, is a genetically heterogeneous condition composed of six distinct genetic complementation groups. To date, mutations in four genes, all of which encode centrosomal proteins, have been described for this disorder (MCPH1, ASPM, CDK5RAP2, CENPJ) [55, 68-71]. Microcephalin (MCPH1), the first Primary Microcephaly gene identified encodes a BRCT-containing product that has been implicated in the response to DSBs [72, 73]. Importantly, work using MCPH1-patient derived cell lines with hypomorphic mutations in MCPH1 indicated that these cells are defective in ATR-dependent checkpoint activation and also exhibit supernumerary mitotic centrosomes [74]. Furthermore, MCPH1 was shown to interact with Chk1, a substrate and downstream effector of ATR [74].
6. THE IMPACT OF ATR HAPLOINSUFFICIENCY: MURINE STUDIES
All of the disorders described so far that collectively exhibit defective ATR-pathway function are all caused by gene mutations inherited in an autosomal recessive manner. As mentioned earlier, a knockout mouse model for ATR does not exist due to early embryonic lethality of ATR-/- blastocysts [44, 45]. Whilst ATR+/- animals are viable, re-evaluation of this murine work suggests that the ATR-DDR pathway is sensitive to gene copy number variation. Firstly, ATR+/- mice are not born at the expected Mendelian frequency suggesting a requirement for a full diploid complement of ATR protein during normal development [75]. Furthermore, decreased survival and increased tumour incidence were recorded in the ATR+/- animals compared to their ATR+/+ counterparts [75]. This is distinct to that of ATM+/- embryos and mice. Heterozygous mutations in ATR have been observed in microsatellite unstable human colorectal carcinomas further suggesting that ATR haploinsufficiency may play a role in tumourigenesis [76]. In fact, it has been suggested that ATR acts as a haploinsufficient tumour suppressor under certain circumstances. Fang and colleagues showed that ATR+/- mice when crossed into a mismatch repair defective (Mlh1-/-) background (generating ATR+/-/Mlh1-/-) were highly susceptible to embryonic lethality and premature tumour development [77].
7. THE IMPACT OF ATR HAPLOINSUFFICIENCY: HUMAN STUDIES, INCLUDING THE ASSOCIATION OF ATR+/- WITH A GENOMIC DISORDER
Fang and colleagues also showed that haploinsufficiency of ATR in human cells results in a clear DDR-defective phenotype [77]. Following gene targeting of ATR in the human HCT116 colorectal carcinoma cell system, they found a significantly increased expression of DFS as well as other gross chromosomal rearrangements and amplifications. Furthermore, these ATR+/- cells exhibited diminished ATR-mediated phosphorylation of its effector kinase Chk1.
Work from de Ru and colleagues along with that of O’Driscoll and colleagues has provided evidence for a cellular impact of ATR haploinsufficiency associated with a human genomic disorder [78, 79]. Blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) is a disorder characterised by a reduction in the dimensions of the palpebral fissures or eye sockets (blepharophimosis), drooping eyelids (ptosis) and inverted skin folds originating from the lower eyelids (epicanthus inversus) [80]. A variant of this disorder is also associated with ovarian failure and female infertility [81]. This disorder is caused by autosomal dominant mutations in or heterozygous deletion of FOXL2, a putative forkhead transcription factor [80]. In a review of the literature, de Ru and colleagues noted that most of the BPES patients with a cytologically detectable deletion or a microdeletion on chromosome 3q, where FOXL2 resides, also exhibited microcephaly and short stature [78]. These are clinical features not typically associated with ‘classical’ BPES. Indeed, it had been previously proposed that a putative gene for microcephaly was located close to the BPES-causative gene on chromosome 3q (reviewed in [78]). De Ru and colleagues mapped the heterozygous deletion in one such BPES patient with microcephaly and short stature. They found that this patient was haploinsufficient for both FOXL2 and ATR at the genomic level. They suggested that the haploinsufficiency of ATR may be responsible for the microcephaly and short stature observed in this patient based on the occurrence of these clinical features in ATR-defective Seckel syndrome [78]. In a complementary study, O’Driscoll and colleagues subsequently showed that cells from this BPES-ATR+/- patient exhibited similar cellular defects to ATR-S cells [79]. These cells failed to show significant γH2AX formation and Chk1 phosphorylation following replication fork stalling. They also exhibited a similar ATR-dependent G2-M cell cycle checkpoint defect to ATR-S cells. Importantly, this phenotype was corrected following over-expression of ATR in the BPES-ATR+/- cells. This study reinforced the fact that haploinsufficiency of ATR has a functional impact in human cells but furthermore, that haploinsufficiency of ATR is associated with a human genomic disorder that exhibits microcephaly and short stature [79].
8. HAPLOINSUFFICIENCY OF ATR-PATHWAY COMPONENTS IN OTHER GENOMIC DISORDERS
Work from Lam and colleagues using tissue-specific conditional knockdown in mice of Chk1, an important effector kinase and substrate of ATR, showed that Chk1 haploinsufficiency enhances mammary tumourigenesis [82]. This work suggested that an ATR-pathway component and not just ATR itself is sensitive to haploinsufficiency. Following the cellular characterisation of BPES- ATR+/-, working on the presumption that the ATR-pathway as a whole is sensitive to a reduced gene copy number, O’Driscoll and colleagues examined ATR-pathway function in other human genomic disorders whose causative genomic deletions were known to result in haploinsufficiency of various ATR-pathway components [79] (Table 2).
Table 2.
Genomic Disorders that Exhibit Microcephaly and Growth Retardation Associated with Hemizygous Deletions of ATR Pathway Components
| Disorder | Chromosome Deletion | ATR-Pathway Component | ATR-Dependent Cellular Features |
|---|---|---|---|
| BPES-ATR+/- | 3q23 | ATR | Defective ATR-dependent γH2AX formation, Chk1 phosphorylation and G2-M arrest |
| Isolated Lissencephaly Sequence | 17p13.3 | RPA1 | Defective ATR-dependent γH2AX formation, Chk1 phosphorylation and G2-M arrest |
| Miller-Dieker Lissencephaly Syndrome | 17p13.3 | RPA1 | Defective ATR-dependent γH2AX formation, Chk1 phosphorylation and G2-M arrest |
| Williams-Beuren Syndrome | 7q11.23 | RFC2 | Defective ATR-dependent G2-M arrest |
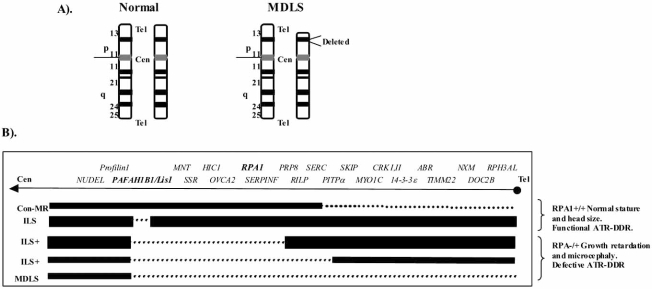
8.1. Isolated Lissencephaly Sequence (ILS) and Miller-Dieker Lissencephaly Syndrome (MDLS)
Normal human brain development involves a rapid and sustained cellular proliferation originating from the rostral end of the foetal neural tube. Cerebral cortical development is achieved via a highly regulated sequence of neuroprogenitor cell division, migration and differentiation. Platelet-activating factor acetylhydrolase, isoform B1 (PAHFAH1B1/Lis1), located on human chromosome 17p13.3, encodes a protein, Lissencephaly 1 (Lis1), which plays a central role in neuronal migration from the neuroepithelial stem cell layer during embryonic brain development [83-85]. Mutations or heterozygous deletions of PAHFAH1B1/ Lis1 alone causes Isolated Lissencephaly Sequence (ILS), a disorder characterised by reduced neuronal migration resulting in a cortical surface without significant invaginations, effectively a "smooth brain" (lissencephaly) (Fig. 1). Larger deletions identified in some ILS patients confer a more severe grade of lissencephaly associated with additional craniofacial abnormalities (ILS+) (Fig. 1). Even larger deletions extending telomerically from the PAHFAH1B1/Lis1 gene are associated with Miller-Dieker Lissencephaly Syndrome (MDLS), which is characterised by the most severe grade of lissencephaly, craniofacial abnormalities, microcephaly and growth retardation (Fig. 1) [14, 86]. Unlike ILS patients, ILS+ patients and all MDLS patients are haploinsufficient for RPA1, the largest subunit of the Replication Protein A complex (Fig. 1) [86]. As described previously, this complex coats ssDNA generated for example at a stalled replication fork allowing the recruitment and ultimate activation of ATR [19]. Similar to what was observed using BPES-ATR+/- cells, cell lines from ILS+ and MDLS patients collectively exhibited compromised ATR-pathway function [79]. Furthermore, these cellular defects could be complemented following over-expression of RPA1 in these cells. ILS patient cell lines with heterozygous deletions in PAHFAH1B1/ Lis1 only, by contrast, exhibited a functional ATR-pathway response (Fig. 1). Therefore, ILS+ and MDLS represent two further human genomic disorders with a clinical overlap with ATR-S that also exhibits compromised ATR-pathway function at the cellular level [79].
Fig. (1).
A). Chromosome 17 karyotype from a normal and MDLS individual. B). A view of a single chromatid of chromosome 17p13.3 showing the order of the genes in this region from the centromere (Cen) to the telomere (Tel). The dotted lines denote the deletions (hemizygous) of various sizes associated with respective patients listed on the left hand side. Con-MR is a patient with a telomeric deletion. This patient exhibits mental retardation but normal stature and head circumference. ILS represents an Isolated Lissencephaly Sequence patient deleted for PAFAH1B1/Lis1 alone. ILS+ denotes Isolated Lissencephaly Sequence patients with increasing deletions telomeric from PAFAH1B1/Lis1. MDLS represents Miller-Dieker Lissencephaly.
8.2. Williams-Beuren Syndrome (WBS)
WBS is caused by a hemizygous sub-microscopic deletion of 1.55-1.84Mbp on chromosome 7q11.23 encompassing around 25-30 genes. The clinical presentation of this condition is multifaceted including craniofacial, endocrinological and cardiovascular abnormalities along with microcephaly and growth retardation [87, 88]. One of the genes heterozygously deleted in WBS is ELN, which encodes elastin, a key structural component of vascular tissues. A characteristic cardiovascular abnormality of WBS, supravalvular aortic stenosis (narrowing of the ascending aorta), is thought to specifically derive from haploinsufficiency of ELN [89]. WBS patients are also haploinsufficient for RFC2 a gene that encodes a component of the Replication Factor C (RFC) complex that plays a fundamental role during DNA replication loading PCNA onto chromatin, thus facilitating DNA polymerase action [90]. RFC2 is also a component of the Rad17-RFC2-5 complex that is known to function in the DDR [91-94]. In fact, Rad17 is phosphorylated by ATM and ATR following DNA damage. Interestingly, a yeast stain (S. cerevisae) with a hypomorphic mutation in Rfc2 fails to activate cell cycle arrest following DNA damage, suggesting that the RFC complex and specifically RFC2 plays a direct role in the DDR [95]. O’Driscoll and colleagues also showed that WBS patient-derived cell lines exhibit a defective ATR-dependent DDR that could be complemented following reintroduction of RFC2 into these cells [79]. Therefore, WBS represents another genomic disorder that exhibits defective ATR-pathway activity wherein microcephaly and growth retardation are included in its clinical spectrum [79].
Of course, for all of the genomic disorders discussed above that exhibit a defective ATR-dependent DDR associated with microcephaly and short stature, the occurrence of this specific DDR defect with these particular clinical features is associative. This association, albeit strong, remains an association only. Hence, more work using complementary systems such as gene targeting or tissue-specific knockdown in the murine system will be required to definitively prove the link between ATR-pathway dysfunction and these developmental abnormalities.
9. HAPLOINSUFFICIENCY OF OTHER DDR COMPONENTS
Increasing evidence from murine gene targeting studies suggests that haploinsufficiency of various components of a diverse distinct range DDR pathways and not just ATR or ATR-pathway components has a functional impact on maintaining genomic stability. For example, haploinsufficiency of various components of the mitotic spindle checkpoint such BubR1 or MAD2 is associated with aberrant chromosome segregation and aneuploidy in mice [96, 97]. Recently, haploinsufficiency of another mitotic spindle checkpoint component, Mad1, has been shown to be associated with aneuploidy and increased constitutive tumour incidence in Mad1+/- mice compared to their wild-type (Mad1+/+) littermates [98]. Homologous recombination (HR) is an error-free DSB repair pathway used by mammalian cells when a homologous sister chromatid is available as a template for DNA repair (S- and G2-phases of the cell cycle) [99]. Haploinsufficiency of multiple genes whose products function in HR results in compromised genomic stability. Specific examples include Brca1+/-, Xrcc2+/-, Xrcc3+/-, Rad51b+/- and Rad51d+/- mice, all of which present with recombination deficiency, increased chromosomal aberrations and centrosomal fragmentation [100-105]. The Mus81-Eme1 complex functions as a structure specific endonuclease that plays a role in resolving stalling replication forks, 3’-orientated DNA flaps/overhangs and nicked HR intermediates [106]. Haploinsufficiency of both components of the structure specific endonuclease Mus81-Eme1 results in increased chromosomal aberrations and a re-replication phenotype in human and murine cells [107]. Poly(ADP-ribose) polymerase-1 (PARP-1) catalyzes the covalent attachment of long branched poly(ADP-ribose) polymers onto a diverse set of target proteins (including itself), using NAD+ as its substrate. Attachment of these negatively charged polymers changes the biological activity and properties of the target proteins. PARP1 plays an important role in sensing single stranded breaks in DNA [108-110]. The level of PARP activity is sensitive to PARP-1 gene dosage [111]. Interestingly, PARP1-/+ mouse embryonic fibroblasts were shown to exhibit increased supernumerary centrosomes relative to their PARP+/+ counterparts [111]. Furthermore, haploinsufficiency of the histone H2A sub-family member H2AX, a known ATM and ATR substrate, has been shown to compromise genomic integrity, impact on the normal response to DNA damage and enhance tumour susceptibility in the absence of p53 in mice [112].
10. IMPLICATIONS OF HAPLOINSUFFICIENCY OF DDR COMPONENTS IN HUMAN GENOMIC DISORDERS
Murine gene targeting studies have proved invaluable in identifying DDR pathways that are sensitive to haploinsufficiency (e.g. the spindle checkpoint and HR). It is likely that haploinsufficiency of these pathways will potentially contribute to the clinical features of known and/or novel genomic disorders. A major consequence of compromised DDR is increased genomic instability and cancer predisposition (reviewed in [15]). Additionally, since DNA damaging agents are the cornerstone of clinically utilised therapeutic approaches for cancer, individuals with compromised DDR are hypersensitive to such treatments. This has been observed in Ataxia telangiectasia, Nijmegen breakage syndrome, LIG4 syndrome and Fanconi anaemia patients, some of which have fatally over-responded to standard radio- and/or chemotherapy regimens in the past (reviewed in [113]). Since increased life expectancy due to improved medical supervision is now a feature of many genomic disorders, a potentially defective DDR may become more important from the perspective of tumour development and treatment. This could be particularly relevant for genomic disorders with compromised ATR-pathway function. Whilst it is not clear whether conditions such as BPES-ATR+/-, MDLS and WBS represent tumour-predisposition conditions, it is clear that they do exhibit a defective ATR-dependent DDR [79]. Provocatively, isolated reports of malignancy in these disorders, particularly in WBS exit [114-117]. Whether a compromised ATR-dependent DDR has a role here is currently unclear although worthy of deeper investigation.
In conclusion, plasticity of the human genome is reflected in the high level of CNV observed in clinically normal individuals. Nevertheless, CNV is causative of many pathological conditions in humans. Gene-targeting studies in mice have shown that one form of CNV, namely haploinsufficiency, of certain DDR-pathway components is associated with compromised genomic stability. Haploinsufficiency of ATR, or some of its pathway components confers similar DDR defects to that of ATR-pathway defective Seckel syndrome cell lines. Furthermore, haploinsufficiency of ATR, RPA1 and RFC2 is associated with several human genomic disorders that exhibit microcephaly and growth retardation. Haploinsufficiency of DDR pathway components is likely to contribute to the clinical features of many genomic disorders. This will have implications for the clinical management and treatment of these conditions.
WEB RESOURCES
Online Mendelian Inheritance in Man, (http://www.ncbi.nlm.nih.gov/Omim), for reviews of the specific genes and all of the disorders described here.
DECIPHER, (https://decipher.sanger.ac.uk/), a database collating multiple genomic imbalances and their associated clinical features in human genomic disorders.
EUCARUCA, (http://agserver01.azn.nl:8080/ecaruca/whatisEc.jsp), a database of cytogenetic and clinical data of rare chromosomal aberrations from all centres that are member of the European Cytogeneticists Association (ECA).
Database of Genomic Variants, (http://projects.tcag.ca/variation/), a database listing a comprehensive summary of structural variation in the human genome.
ACKNOWLEDGEMENTS
Special thanks to Prof. Penny Jeggo for all her support over the years. Thanks also to Prof. William Dobyns and Dr. Johanna van Hagen for genomic disorder-related cell lines. M.O’D is a Cancer Research UK Senior Research Fellow whose laboratory is supported by the CRUK and UK Medical Research Council.
REFERENCES
- 1.Roper RJ, Reeves RH. Understanding the Basis for Down Syndrome Phenotypes. PLoS Genet. 2006;2:e50. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The Diploid Genome Sequence of an Individual Human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennisi E. Breakthrough of the Year: Human Genetic Variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- 5.Lupski JR. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 6.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat. Genet. 2007;39:S48–54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 7.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39(7 Suppl):S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat. Genet. 2007;39:S3–5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- 9.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 10.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 11.Conrad DF, Hurles ME. The population genetics of structural variation. Nat. Genet. 2007;39:S30–36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles ME, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007;39:S7–15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 14.Yingling J, Toyo-Oka K, Wynshaw-Boris A. Miller-Dieker syndrome: analysis of a human contiguous gene syndrome in the mouse. Am. J. Hum. Genet. 2003;73:475–488. doi: 10.1086/378096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Driscoll M, Jeggo PA. The role of double-strand break repair-insights from human genetics. Nat. Rev. Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 16.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 17.Abraham RT. PI 3-kinase related kinases: `big' players in stress-induced signaling pathways. DNA Repair. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 19.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 21.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 Activates the ATR-ATRIP Complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 23.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 25.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GCM, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 28.Stiff T, Walker S, Cerosaletti K, Goodarzi A, Petermann E, Concannon P, O'Driscoll M, Jeggo P. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NG, Taylor AM, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI 3-kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 30.Chun HH. GRA: Ataxia-telangiectasia, an evolving phenotype. DNA Repair. 2004;3:1187–1196. doi: 10.1016/j.dnarep.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 31.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 32.O'Driscoll M, Jeggo PA. Clinical Impact of ATR Checkpoint Signalling Failure in Humans. Cell Cycle. 2003;2:194–195. [PubMed] [Google Scholar]
- 33.Durkin SG, Glover TW. Chromosome Fragile Sites. Ann. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 34.Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair. 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 35.Mimori K, Druck T, Inoue H, Alder H, Berk L, Mori M, Huebner K, Croce CM. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl. Acad. Sci. USA. 1999;96:7456–7461. doi: 10.1073/pnas.96.13.7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii H, Mimori K, Inoue H, Inageta T, Ishikawa K, Semba S, Druck T, Trapasso F, Tani K, Vecchione A, Croce CM, Mori M, Huebner K. Fhit Modulates the DNA Damage Checkpoint Response. Cancer Res. 2006;66:11287–11292. doi: 10.1158/0008-5472.CAN-06-2503. [DOI] [PubMed] [Google Scholar]
- 37.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 38.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 Is Required for Common-Fragile-Site Stability via Its G2/M Checkpoint Function. Mol. Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 40.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 41.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seckel HPG. Bird-headed dwarfs: Studies in developmental anthropolgy incuding human proportions. In: Thomas CT, Basel S Karger, editors. Bird-headed dwarfs: Studies in developmental anthropology including human proportions. 1960. [Google Scholar]
- 43.Goodship J, Gill H, Carter J, Jackson A, Splitt M, Wright M. Autozygosity mapping of a seckel syndrome locus to chromosome 3q22. 1-q24. Am. J. Hum. Genet. 2000;67:498–503. doi: 10.1086/303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 45.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JHJ. Targeted disruption of the cell cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 46.Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O'Driscoll M. Seckel syndrome exhibits cellular features demonstrating defects in the ATR signalling pathway. Hum. Mol. Genet. 2004;13:3127–3138. doi: 10.1093/hmg/ddh335. [DOI] [PubMed] [Google Scholar]
- 47.Casper AM, Durkin SG, Arlt MF, Glover TW. Chromosomal instability at common fragile sites in seckel syndrome. Am. J. Hum. Genet. 2004;75:654–660. doi: 10.1086/422701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borglum AD, Balslev T, Haagerup A, Birkebaek N, Binderup H, Kruse TA, Hertz JM. A new locus for Seckel syndrome on chromosome 18p11.31 -q11.2. Eur. J. Hum. Genet. 2001;9:753–757. doi: 10.1038/sj.ejhg.5200701. [DOI] [PubMed] [Google Scholar]
- 49.Faivre L, Le Merrer M, Lyonnet S, Plauchu H, Dagoneau N, Campos-Xavier AB, Attia-Sobol J, Verloes A, Munnich A, Cormier-Daire V. Clinical and genetic heterogeneity of Seckel syndrome. Am. J. Med. Genet. 2002;112:379–383. doi: 10.1002/ajmg.10677. [DOI] [PubMed] [Google Scholar]
- 50.Kilinc MO, Ninis VN, Ugur SA, Tuysuz B, Seven M, Balci S, Goodship J, Tolun A. Is the novel SCKL3 at 14q23 the predominant Seckel locus? Eur. J. Hum. Genet. 2003;11:851–857. doi: 10.1038/sj.ejhg.5201057. [DOI] [PubMed] [Google Scholar]
- 51.Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- 52.Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat. Rev. Cancer. 2007;7:911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- 53.Golden JA. Cell migration and cerebral cortical development. Neuropathol. Appl. Neurobiol. 2001;27:22–28. doi: 10.1046/j.0305-1846.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- 54.Lu BJL, Jan YN. Control of cell divisions in the nervous system: symmetry and asymmetry. Annu. Rev. Neurosci. 2000;23:531–556. doi: 10.1146/annurev.neuro.23.1.531. [DOI] [PubMed] [Google Scholar]
- 55.Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Al Sanna N, Saggar A, Hamel B, Earnshaw WC, Jeggo PA, Jackson AP, O'Driscoll M. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauch A, Thiel CT, Schindler D, Wick U, Crow YJ, Ekici AB, van Essen AJ, Goecke TO, Al-Gazali L, Chrzanowska KH, Zweier C, Brunner HG, Becker K, Curry CJ, Dallapiccola B, Devriendt K, Dörfler A, Kinning E, Megarbane A, Meinecke P, Semple RK, Spranger S, Toutain A, Trembath RC, Voss E, Wilson L, Hennekam R, de Zegher F, Dörr HG, Reis A. Mutations in the Pericentrin (PCNT) Gene Cause Primordial Dwarfism. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 58.Delaval B, Doxsey S. Genetics: Dwarfism, Where Pericentrin Gains Stature. Science. 2008;319:732–733. doi: 10.1126/science.1154513. [DOI] [PubMed] [Google Scholar]
- 59.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 60.Digweed MSK. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair. 2004;3:1207–1217. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 61.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, D'Andrea AD. The interplay of Fanconi anemia proteins in the DNA damage response. DNA Repair. 2004;3:1063–1069. doi: 10.1016/j.dnarep.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 63.Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair. 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gregory RC, Taniguchi T, D'Andrea AD. Regulation of the Fanconi anemia pathway by monoubiquitination. Semin. Cancer Biol. 2003;13:77–82. doi: 10.1016/s1044-579x(02)00102-5. [DOI] [PubMed] [Google Scholar]
- 66.Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, Stotsky M, Mathew CG, Kastan MB, Weave DT, D'Andrea AD. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 67.Taniguchi TG-HI, Xu B, Andreassen PR, Gregory RC, Kim ST, Lane WS, Kastan MB, D'Andrea AD. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;4:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 68.Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, Costa SM, Baralle D, Raponi M, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CA, Woods CG. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 69.Woods CG. Human microcephaly. Curr. Opin. Neurobiol. 2004;14:112–117. doi: 10.1016/j.conb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Bond JRE, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, Walsh CA, Woods CG. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 71.Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, Karbani G, Jafri H, Rashid Y, Mueller RF, Markham AF, Woods CG. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin S-Y, Rai R, Li K, Xu Z-X, Elledge SJ. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc. Natl. Acad. Sci. USA. 2005;102:15105–15109. doi: 10.1073/pnas.0507722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu X, Lee J, Stern DF. Microcephalin Is a DNA Damage Response Protein Involved in Regulation of CHK1 and BRCA1. J. Biol. Chem. 2004;279:34091–34094. doi: 10.1074/jbc.C400139200. [DOI] [PubMed] [Google Scholar]
- 74.Alderton GK, Galbiati L, Griffith E, Surinya KH, Neitzel H, Jackson AP, Jeggo PA, O'Driscoll M. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat. Cell Biol. 2006;8:725–733. doi: 10.1038/ncb1431. [DOI] [PubMed] [Google Scholar]
- 75.Brown AL, Lee CH, Schwarz JK, Mitiku N, Piwnica-Worms H, Chung JH. A human Cds1-related kinase that functions downstream of ATM protein in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA. 1999;96:3745–3750. doi: 10.1073/pnas.96.7.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis KA, Mullany S, Thomas B, Chien J, Loewen R, Shridhar Viji, Cliby William A. Heterozygous ATR Mutations in Mismatch Repair-Deficient Cancer Cells Have Functional Significance. Cancer Res. 2005;65:7091–7095. doi: 10.1158/0008-5472.CAN-05-1019. [DOI] [PubMed] [Google Scholar]
- 77.Fang Y, Tsao CC, Goodman BK, Furumai R, Tirado CA, Abraham RT, Wang XF. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004;23:3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Ru M, Gille JJ, Nieuwint AWM, Bijlsma JJ, van der Blij JF, van Hagen JM. Interstitial deletion in 3q in a patient with blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) and microcephaly, mild mental retardation and growth delay: Clinical report and review of the literature. Am. J. Med. Genet. Part A. 2005;137A:81–87. doi: 10.1002/ajmg.a.30786. [DOI] [PubMed] [Google Scholar]
- 79.O' Driscoll M, Dobyns WB, van Hagen JM, Jeggo PA. Cellular and Clinical Impact of Haploinsufficiency for Genes Involved in ATR Signaling. Am. J. Hum. Genet. 2007;81:77–86. doi: 10.1086/518696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 81.De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, Gillerot Y, Mortier G, Meire F, Van Maldergem L, Courtens W, Hjalgrim H, Huang S, Liebaers I, Van Regemorter N, Touraine P, Praphanphoj V, Verloes A, Udar N, Yellore V, Chalukya M, Yelchits S, De Paepe A, Kuttenn F, Fellous M, Veitia R, Messiaen L. Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype-phenotype correlation. Hum. Mol. Genet. 2001;10:1591–1600. doi: 10.1093/hmg/10.15.1591. [DOI] [PubMed] [Google Scholar]
- 82.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 83.Cardoso C, Leventer RJ, Matsumoto N, Kuc JA, Ramocki MB, Mewborn SK, Dudlicek LL, May LF, Mills PL, Das S, Pilz DT, Dobyns WB, Ledbetter DH. The location and type of mutation predict malformation severity in isolated lissencephaly caused by abnormalities within the LIS1 gene. Hum. Mol. Genet. 2000;9:3019–3028. doi: 10.1093/hmg/9.20.3019. [DOI] [PubMed] [Google Scholar]
- 84.Leventer RJ, Cardoso C, Ledbetter DH, Dobyns WB. LIS1: from cortical malformation to essential protein of cellular dynamics. Trends Neurosci. 2001;24:489–492. doi: 10.1016/s0166-2236(00)01887-7. [DOI] [PubMed] [Google Scholar]
- 85.Cardoso C, Leventer RJ, Dowling JJ, Ward HL, Chung J, Petras KS, Roseberry JA, Weiss AM, Das S, Martin CL, Pilz DT, Dobyns WB, Ledbetter DH. Clinical and molecular basis of classical lissencephaly: Mutations in the LIS1 gene (PAFAH1B1) Hum. Mutat. 2002;19:4–15. doi: 10.1002/humu.10028. [DOI] [PubMed] [Google Scholar]
- 86.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, Gross A, Martin CL, Allanson J, Pilz DT, Olney AH, Mutchinick OM, Hirotsune S, Wynshaw-Boris A, Dobyns WB, Ledbetter DH. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 2003;72:918–930. doi: 10.1086/374320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tassabehji M. Williams-Beuren syndrome: a challenge for genotype-phenotype correlations. Hum. Mol. Genet. 2003;12:229–237. doi: 10.1093/hmg/ddg299. [DOI] [PubMed] [Google Scholar]
- 88.Wu YQ, Sutton VR, Nickerson E, Lupski JR, Potocki L, Korenberg JR, Greenberg F, Tassabehji M, Shaffer LG. Delineation of the common critical region in Williams syndrome and clinical correlation of growth, heart defects, ethnicity, and parental origin. Am. J. Med. Genet. 1998;78:82–89. doi: 10.1002/(sici)1096-8628(19980616)78:1<82::aid-ajmg17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 89.Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette MJ, Grant J, Dennis N, Reardon W, Splitt M, Read AP, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am. J. Hum. Genet. 1999;64:118–125. doi: 10.1086/302214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ellison V, Stillman B. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J. Biol. Chem. 1998;273:5979–5987. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- 91.Griffith J, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 2002;277:15233–15236. doi: 10.1074/jbc.C200129200. [DOI] [PubMed] [Google Scholar]
- 92.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang XZL, Zheng H, Wei Q, Elledge SJ, Li L. Genomic instability and endoreduplication triggered by RAD17 deletion. Genes Dev. 2003;17:965–970. doi: 10.1101/gad.1065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noskov VN, Araki H, Sugino A. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol. Cell Biol. 1998;18:4914–4923. doi: 10.1128/mcb.18.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 97.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 98.Iwanaga Y, Chi Y-H, Miyazato A, Sheleg S, Haller K, Peloponese J-M Jr, Li Y, Ward JM, Benezra R, Jeang K-T. Heterozygous Deletion of Mitotic Arrest-Deficient Protein 1 (MAD1) Increases the Incidence of Tumors in Mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 99.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 100.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 101.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 103.Deans B, Griffin CS, O'Regan P, Jasin M, Thacker J. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003;63:8181–8187. [PubMed] [Google Scholar]
- 104.Date O, Katsura M, Ishida M, Yoshihara T, Kinomura A, Sueda T, Miyagawa K. Haploinsufficiency of RAD51B Causes Centrosome Fragmentation and Aneuploidy in Human Cells. Cancer Res. 2006;66:6018–6024. doi: 10.1158/0008-5472.CAN-05-2803. [DOI] [PubMed] [Google Scholar]
- 105.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive Chromosomal Instability in Rad51d-Deficient Mouse Cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 106.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1017. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 107.Hiyama T, Katsura M, Yoshihara T, Ishida M, Kinomura A, Tonda T, Asahara T, Miyagawa K. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Res. 2006;34:880–892. doi: 10.1093/nar/gkj495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeggo PA. DNA repair: PARP - another guardian angel? Curr. Biol. 1998;8:49–51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 109.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schreiber V, Dantzer FO, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 111.Kanai M, Tong W-M, Wang Z-Q, Miwa M. Haploinsufficiency of poly(ADP-ribose) polymerase-1-mediated poly(ADP-ribosyl)ation for centrosome duplication. Biochem. Biophys. Res. Commun. 2007;359:426–430. doi: 10.1016/j.bbrc.2007.05.108. [DOI] [PubMed] [Google Scholar]
- 112.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O'Driscoll M, Jeggo PA. CsA can induce DNA double-strand breaks: implications for BMT regimens particularly for individuals with defective DNA repair. Bone Marrow Transplant. 2008 doi: 10.1038/bmt.2008.18. in press [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 114.Shizuyo U, Masaru K, Shigekazu K, Michihiko W. Gallbladder cancer in a patient with Miller-Dieker syndrome. Acta Paediatrica. 2006;95:113–114. doi: 10.1080/08035250510046731. [DOI] [PubMed] [Google Scholar]
- 115.Amenta S, Moschovi M, Sofocleous C, Kostaridou S, Mavrou A, Fryssira H. Non-Hodgkin lymphoma in a child with Williams syndrome. Cancer Genet. Cytogenet. 2004;154:86–88. doi: 10.1016/j.cancergencyto.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 116.Thornburg C, Roulston D, Castle V. Burkitt lymphoma and Williams syndrome: a model for children with a multisystem disorder and malignancy. J. Pediatr. Hematol. Oncol. 2005;27:109–111. doi: 10.1097/01.mph.0000153444.43816.ea. [DOI] [PubMed] [Google Scholar]
- 117.Semmekrot BA, Rotteveel JJ, Bakker-Niezen SHFL. Occurrence of an astrocytoma in a patient with Williams syndrome. Pediatr. Neurosci. 1985-1986;12:188–191. doi: 10.1159/000120245. [DOI] [PubMed] [Google Scholar]